ABSTRACT
Riemerella anatipestifer is a major bacterial pathogen that causes septicemic and exudative diseases in domestic ducks. In our previous study, we found that deletion of the AS87_01735 gene significantly decreased the bacterial virulence of R. anatipestifer strain Yb2 (mutant RA625). The AS87_01735 gene was predicted to encode a nicotinamidase (PncA), a key enzyme that catalyzes the conversion of nicotinamide to nicotinic acid, which is an important reaction in the NAD+ salvage pathway. In this study, the AS87_01735 gene was expressed and identified as the PncA-encoding gene, using an enzymatic assay. Western blot analysis demonstrated that R. anatipestifer PncA was localized to the cytoplasm. The mutant strain RA625 (named Yb2ΔpncA in this study) showed a similar growth rate but decreased NAD+ quantities in both the exponential and stationary phases in tryptic soy broth culture, compared with the wild-type strain Yb2. In addition, Yb2ΔpncA-infected ducks showed much lower bacterial loads in their blood, and no visible histological changes were observed in the heart, liver, and spleen. Furthermore, Yb2ΔpncA immunization of ducks conferred effective protection against challenge with the virulent wild-type strain Yb2. Our results suggest that the R. anatipestifer AS87_01735 gene encodes PncA, which is an important virulence factor, and that the Yb2ΔpncA mutant can be used as a novel live vaccine candidate.
IMPORTANCE Riemerella anatipestifer is reported worldwide as a cause of septicemic and exudative diseases of domestic ducks. The pncA gene encodes a nicotinamidase (PncA), a key enzyme that catalyzes the conversion of nicotinamide to nicotinic acid, which is an important reaction in the NAD+ salvage pathway. In this study, we identified and characterized the pncA-homologous gene AS87_01735 in R. anatipestifer strain Yb2. R. anatipestifer PncA is a cytoplasmic protein that possesses similar PncA activity, compared with other organisms. Generation of the pncA mutant Yb2ΔpncA led to a decrease in the NAD+ content, which was associated with decreased capacity for invasion and attenuated virulence in ducks. Furthermore, Yb2ΔpncA immunization of ducks conferred effective protection against challenge with the virulent wild-type strain Yb2. Altogether, these results suggest that PncA contributes to the virulence of R. anatipestifer and that the Yb2ΔpncA mutant can be used as a novel live vaccine candidate.
INTRODUCTION
Riemerella anatipestifer is a causative agent of an epizootic disease in poultry, especially in ducks (1). R. anatipestifer infection has a worldwide distribution, and it occurs as acute or chronic septicemia characterized by fibrinous pericarditis, airsacculitis, perihepatitis, caseous salpingitis, and vegetative disorder. To date, 21 serotypes have been identified by agglutination tests (2, 3), and serotypes 1, 2, and 10 have been responsible for most of the major outbreaks in China (4). There are large variations in virulence among different serotypes of R. anatipestifer, and even within a given serotype (5). Because R. anatipestifer is of major economic significance to the poultry industry, there has been much research regarding the molecular pathogenic mechanisms of this species. To date, several virulence factors have been identified, including VapD (6), cohemolysin of Actinobacillus pleuropneumoniae (7), OmpA (8), and putative genes associated with lipopolysaccharide synthesis (9–11).
We previously reported the identification of 49 virulence-associated genes via random transposon mutagenesis (12). Transposon Tn4351 disrupted the AS87_01735 gene, which encodes a predicted nicotinamidase (PncA), a key enzyme that catalyzes the conversion of nicotinamide (NAM) to nicotinic acid (NA) (an important reaction in the NAD+ salvage pathway). The disruption of AS87_01735 resulted in a 488,000-fold attenuation of virulence, compared with the wild-type strain. PncA, also known as nicotinamidase, nicotinamide deaminase, or nicotinamide amidase, hydrolyzes NAM to NA for the production of NAD via the Preiss-Handler pathway. NAD is a coenzyme that is found in all living cells, and it has several essential roles in metabolism. It can be synthesized either via a de novo pathway from amino acids or via salvage pathways that use NAM, NA, or nicotinamide riboside (NR) as precursors (13). In lower organisms, including bacteria and yeast, NAM is converted to NA by PncA. Recent studies have indicated the importance of PncA activity in the interactions between pathogens and their hosts, as it affects intracellular replication and pathogen dissemination (14–16). In addition, Leishmania infantum PncA is essential for NAD+ production and parasite proliferation (17).
To our knowledge, no studies have yet elucidated the importance of PncA activity in R. anatipestifer. This study describes the expression and identification of the predicted R. anatipestifer PncA-encoding gene, AS87_01735, as well as the virulence and immunization protection of an AS87_01735 gene deletion mutant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The plasmids and primers used in this study are presented in Table 1. R. anatipestifer Yb2 (GenBank accession no. CP007204) is the wild-type virulent strain; the mutant strain RA625 (named Yb2ΔpncA in this study), which was derived from that strain, has a Tn4351 transposon inserted into the AS87_01735 gene (12). All R. anatipestifer strains were maintained as frozen glycerol stocks, and they were cultured in tryptic soy agar (TSA) (BD Difco, Franklin Lakes, NJ, USA) at 37°C in 5% CO2 for 24 h or in tryptic soy broth (TSB) (BD Difco) at 37°C for 8 to 12 h, with shaking at 200 rpm. The Escherichia coli-Flavobacterium johnsoniae shuttle plasmid pCP29 and E. coli strain S17-1 were kindly provided by Mark J. McBride (University of Wisconsin-Milwaukee, Milwaukee, WI, USA). E. coli strains were cultured on Luria-Bertani (LB) plates at 37°C for 12 to 16 h or in LB broth at 37°C for 6 to 8 h, with shaking at 200 rpm. When necessary, antibiotics were added to the medium at the following concentrations: ampicillin at 100 μg/ml for E. coli strain S17-1, kanamycin at 50 μg/ml for E. coli strain BL21(DE3), kanamycin at 50 μg/ml and erythromycin at 0.5 μg/ml for the mutant strain Yb2ΔpncA, and kanamycin at 50 μg/ml and cefoxitin at 5 μg/ml for the complemented strain cYb2ΔpncA.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Characteristicsa | Source or reference |
|---|---|---|
| Strains and plasmids | ||
| Yb2 | Riemerella anatipestifer serotype 2 strain | 4 |
| Escherichia coli S17-1 | λpir hsdR pro thi; chromosomally integrated RP4-2, Tc::Mu Km::Tn7 | 18 |
| RA625 | Tn4351 insertion mutant of R. anatipestifer Yb2, pncA::Tn | 12 |
| cYb2ΔpncA | Mutant RA625 carrying plasmid pCP29-pncA | This study |
| pCP29 | ColE1 ori (pCP1 ori), Apr (Emr); E. coli-F. johnsoniae shuttle plasmid | 18 |
| pCP29-pncA | pCP29 containing ompA promoter and pncA ORF, cfxA (Apr) | This study |
| Primers | ||
| ompA promoter P1 | 5′-CAGGTACCATAGCTAAAATTTTGGCAGTAAC-3′ (KpnI site underlined) | 18 |
| ompA promoter P2 | 5′-CGACTCGAGCATTCCAATTCTCTTATTATC-3′ (XhoI site underlined) | 18 |
| pncAorf-F | 5′-CCGCTCGAGATGAAAAAAGCACTTATCGTGG-3′ (XhoI site underlined) | This study |
| pncAorf-R | 5′-CATGCATGCTTAACCTAAAATGGTAACACCA-3′ (SphI site underlined) | This study |
| PncA-F | 5′-CGGGATCCATGAAAAAAGCACTTATCGTGGT-3′ (BamHI site underlined) | This study |
| PncA-R | 5′-CCCAAGCTTACCTAAAATGGTAACACCATTT-3′ (HindIII site underlined) | This study |
ORF, open reading frame; cfxA, cefoxitin resistance gene.
Complementation was accomplished using the shuttle plasmid pCP29, which contains an expression cassette with the R. anatipestifer ompA promoter (18). The pncA open reading frame was amplified from the wild-type strain Yb2 by using the primers pncAorf-F and pncAorf-R. It was then digested with XhoI and SphI and cloned into pCP29 to generate plasmid pCP29-pncA. Expression of the pncA gene was under the control of the ompA promoter, as described previously (18). The recombinant plasmid pCP29-pncA was then conjugated into the mutant strain Yb2ΔpncA. Transconjugants were screened on TSA supplemented with cefoxitin and kanamycin and were verified by PCR amplification. The complemented strain was designated cYb2ΔpncA.
Ethics.
One-day-old Cherry Valley ducklings were purchased from Zhuanghang Duck Farm (Shanghai, China) and housed at our animal facilities. Animal handling was conducted according to the standards specified by the Institutional Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (CAAS). The protocol was approved by the Committee on the Ethics of Animal Experiments of the Shanghai Veterinary Research Institute, CAAS (permit no. Shvri-po-0145). When necessary, the ducklings were euthanized humanely with sodium pentobarbital to minimize suffering.
Expression and purification of recombinant predicted PncA protein.
The open reading frame of the predicted pncA-encoding gene was amplified from R. anatipestifer Yb2 genomic DNA and cloned into the pET-28a(+) vector, using the BamHI and HindIII cloning sites, to construct the recombinant plasmid pET-pncA. The insert of pET-pncA was confirmed by DNA sequencing (12). Expression of histidine-tagged recombinant predicted PncA protein (rPncA) was induced in E. coli strain BL21(DE3) cells by treatment with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 6 h at 37°C, with shaking. Cells were harvested by centrifugation at 10,000 × g for 5 min at 4°C, resuspended in lysis buffer (20 mM Na3PO4, 0.5 M NaCl [pH 7.4]), lysed by sonication, and purified using HisTrap affinity columns (GE Healthcare, Uppsala, Sweden), according to the manufacturer's protocol. The purified fractions were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentrations were measured with a bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China), with bovine serum albumin as a standard.
Enzymatic activity assay.
PncA activity was measured as described previously (19), with some modifications. Purified rPncA (1.0 μg/reaction) was incubated with 32 mM NAM for 45 min at 37°C, in a total volume of 400 μl containing final concentrations of 10 mM Tris (pH 7.5), 150 mM NaCl, and 1 mM MgCl2. The reaction system without the addition of NAM was used as a control. The ammonia produced from the catalytic hydrolysis of NAM by rPncA was determined with an ammonia diagnostic kit (Megazyme, Wicklow, Ireland). A correction for baseline production of ammonia was made by subtracting the ammonia produced by rPncA incubated without the NAM substrate. One unit of activity was defined as the amount of enzyme that converted 1 μmol substrate/min. The specific activity was expressed as units per milligram of protein.
Immunoblot analysis.
Two New Zealand rabbits were injected subcutaneously with 1 mg of purified rPncA or the TonB-dependent receptor from R. anatipestifer, in Montanide ISA 50 V (SEPPIC, Paris, France) adjuvant, to develop rabbit antiserum against rPncA or the TonB-dependent receptor. The injections were administered three times, at 3-week intervals. Blood samples were collected before injection and 2 weeks after the third injection, and antibody titers against rPncA and the TonB-dependent receptor in the sera were determined with an enzyme-linked immunosorbent assay. Qualified antiserum was used for Western blot analysis.
Subcellular fractions (cytosolic and membrane fractions) of the wild-type strain Yb2 were obtained using a bacterial membrane protein extraction kit (BestBio, Shanghai, China), according to the manufacturer's protocol. Proteins were quantified with a BCA protein assay kit (Beyotime). For Western blot analysis, the proteins were separated by SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked for 2 h at 37°C in phosphate-buffered saline (PBS) containing 5% nonfat milk, washed with PBS containing 0.05% Tween 20, and then incubated for 2 h with the rabbit anti-rPncA polyclonal antibody, followed by incubation for 1 h with an IRDye 800CW-conjugated donkey anti-rabbit IgG polyclonal antibody (LI-COR Biosciences, Lincoln, NE, USA). The blots were visualized with an Odyssey two-color infrared imaging system (LI-COR Biosciences).
To determine PncA expression in the complemented strain cYb2ΔpncA, whole-protein extracts of the wild-type strain Yb2 (positive control), the mutant strain Yb2ΔpncA (negative control), and the complemented strain cYb2ΔpncA were separated by SDS-PAGE and electrotransferred to a PVDF membrane. Immunoblots were probed with the rabbit anti-rPncA polyclonal antibody and a donkey anti-rabbit IgG polyclonal antibody conjugated to IRDye 800CW (LI-COR Biosciences), as described above. The rabbit anti-TonB-dependent receptor antibody was used to control for protein loading.
Quantification of intracellular NAD+ concentrations.
Cellular NAD+ was extracted for quantification from the wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA, as described in the manufacturer's protocol (BioAssay Systems, Hayward, CA, USA). Briefly, bacteria were grown in TSB at 37°C for 6, 9, and 12 h, with shaking. Each culture sample (1 ml) was centrifuged, washed with cold PBS, and treated with 100 μl of NAD+ extraction buffer. After incubation at 60°C for 5 min, the samples were cooled on ice and then 20 μl of assay buffer and 100 μl of NADH extraction buffer were added to neutralize the extracts. Following centrifugation at 16,000 × g for 5 min, the supernatants were collected to quantify the NAD+ concentrations using an EnzyChrom NAD+/NADH assay kit (BioAssay Systems).
Bacterial adherence and invasion assays.
Vero cells (ATCC CCL-81) were used to evaluate the effects of the R. anatipestifer pncA gene on bacterial adherence and invasion (9). The cells were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (HyClone; GE Healthcare, Little Chalfont, United Kingdom) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) and were seeded (105 cells per well) into 24-well tissue culture plates for 20 h prior to infection. Cells were infected with bacteria, at a multiplicity of infection of 50, by centrifugation at 100 × g for 5 min at room temperature. The infected cells were incubated for 1.5 h at 37°C in 5% CO2, washed three times with sterile PBS, and lysed with 0.1% trypsin (100 μl/well). Serial dilutions of the cell suspensions were plated onto TSA plates to determine the number of viable bacterial cells. For the invasion assay, the cell culture, bacterial infection, and plating procedures were performed as described above for the bacterial adherence assay, except that extracellular bacteria were killed with 100 μg/ml gentamicin, which was added to the medium for an additional 1 h after the bacterial infection, followed by three washes with PBS.
Bacterial growth curves and virulence determination.
The wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA were grown in TSB at 37°C for 8 h, with shaking. Equal amounts of each bacterial culture were then transferred into fresh TSB at a ratio of 1:100 (vol/vol) and incubated at 37°C, with shaking at 200 rpm. Bacterial growth was measured as described previously (33), by counting the number of bacterial CFU at 3-h intervals for 15 h.
The 50% lethal dose (LD50) for the mutant strain Yb2ΔpncA was measured as described in our previous study (12). To further assess whether R. anatipestifer pncA is involved in systemic invasion and dissemination, experimental infection of Cherry Valley ducks was performed. Eighteen 21-day-old ducks were divided randomly into three groups (six ducks per group) and infected with 107 CFU (in 0.5 ml of PBS) of the wild-type strain Yb2, the mutant strain Yb2ΔpncA, or the complemented strain cYb2ΔpncA. Blood samples were collected at 6, 12, 24, and 48 h postinfection (hpi), serially diluted with PBS, and plated on TSA plates for counting of viable bacterial numbers.
For histological examination, ducks from each group were sacrificed at 48 hpi and heart, liver, and spleen samples were collected. Tissues were fixed in formalin, sectioned, and stained with hematoxylin and eosin, as described previously (20). Samples from ducks that were not infected were used as pathogen-negative controls.
Vaccination and challenge studies.
Seven-day-old Cherry Valley ducks were divided randomly into four groups (10 ducks per group). Each duck in group 1 or group 3 received one injection of the mutant strain Yb2ΔpncA (group 1) or two injections of the mutant strain Yb2ΔpncA (group 3), at a dose of 2.5 × 108 CFU. The ducks in group 2 received one injection of saline, and those in group 4 received two injections of saline; they were used as the controls. The two injections were given 7 days apart. On day 14 postimmunization, the ducks in each group were challenged with 106 CFU (10 times the LD50) of the wild-type virulent strain Yb2, to evaluate the level of protection conferred by the Yb2ΔpncA mutant. The protection rate was calculated using the following formula: [1 − (no. of dead ducklings per group/total no. of ducklings per group)] ×100.
Statistical analyses.
Statistical analyses were performed by one-way analysis of variance using Prism software (version 6; GraphPad Software, Inc., San Diego, CA, USA), and P values of <0.05 were considered to be significant.
RESULTS
R. anatipestifer AS87_01735 encodes a protein with nicotinamidase activity.
Sequencing and Basic Local Alignment Search Tool (BLAST) analysis revealed that the Tn4351 insertion in strain Yb2ΔpncA was located at bp 184 of the AS87_01735 gene, which was identified in our previous study (12). The AS87_01735 gene is 606 nucleotides in length, and it encodes a predicted 201-amino acid nicotinamidase, PncA. A similarity search of nucleotide databases at the National Center for Biotechnology Information using the BLAST program showed that the AS87_01735 gene is highly conserved in R. anatipestifer, as it shares 99% identity with R. anatipestifer strain 153 (GenBank accession no. CP007504.1), R. anatipestifer strain 17 (GenBank accession no. CP007503.1), RA-CH-2 (GenBank accession no. CP004020.1), ATCC 11845 (GenBank accession no. CP003388.1), RA-GD (GenBank accession no. CP002562.1), CH3 (GenBank accession no. CP006649.1), and RA-CH-1 (GenBank accession no. CP003787.1). The predicted protein encoded by AS87_01735 shares sequence identity with putative nicotinamidases from other bacteria, including Chryseobacterium sp. FH2 (GenBank accession no. WP_048510700.1) (80% identity), Flavobacteriaceae bacterium 3519-10 (GenBank accession no. WP_015806149.1) (78% identity), Elizabethkingia (GenBank accession no. WP_021346903.1) (74% identity), Epilithonimonas lactis (GenBank accession no. WP_034978491.1) (75% identity), and Bergeyella zoohelcum (GenBank accession no. WP_034986151.1) (68% identity). A sequence alignment indicated that PncA shared 22.6% to 45.5% sequence identity with crystallized nicotinamidases from other organisms, including Mycobacterium tuberculosis (PDB accession no. 3PL1 [36.3% identity]), Streptococcus pneumoniae (PDB accession no. 3O94 [22.6% identity]), Acinetobacter baumannii (PDB accession no. 2WTA [45.5% identity] and PDB accession no. 2WT9 [45.5%]), Pyrococcus horikoshii (PDB accession no. 1ILW [41.6%]), Saccharomyces cerevisiae (PDB accession no. 2H0R [36.4%]), and Leishmania infantum (PDB accession no. 3R2J [35.5%]) (see Fig. S1A in the supplemental material). In addition, the sequence alignment demonstrated that the R. anatipestifer PncA protein harbored the catalytic residues (D9, K108, and C153), metal binding sites (D49, H51, and H83), and conserved cis-peptide bond (involving L148 and A149) that are required for nicotinamidase activity (see Fig. S1B).
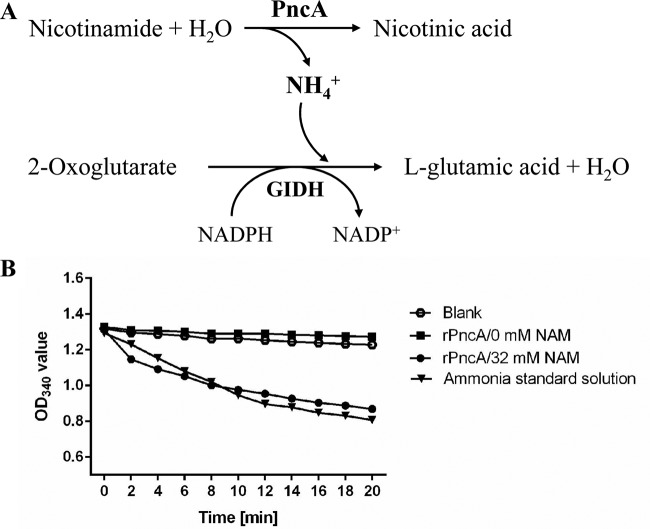
Histidine-tagged rPncA was efficiently expressed in E. coli BL21(DE3), as shown by the presence of a 25-kDa band in a Coomassie blue-stained SDS-PAGE gel (Fig. 1A, lane 2). After purification with HisTrap affinity columns, rPncA was identified as a single band in the gel (Fig. 1A, lane 3). Western blotting showed a 25-kDa band when rPncA was expressed in E. coli BL21(DE3) (Fig. 1B, lane 2), further indicating that rPncA was expressed successfully in E. coli BL21(DE3). The complemented strain cYb2ΔpncA was identified by Western blot analysis using an antibody against the TonB-dependent receptor of R. anatipestifer, as a control for protein loading (Fig. 1C, lane 3). The enzymatic activity of purified rPncA was assayed by quantifying the release of ammonia from the conversion of NAM to NA (Fig. 2A). The ammonia released from the rPncA-catalyzed reaction was determined by monitoring the decrease in absorbance at 340 nm associated with the oxidation of NADPH to NADP+ by glutamate dehydrogenase, which produces l-glutamic acid from 2-oxoglutarate (Fig. 2A). As shown in Fig. 2B, the rPncA-catalyzed reaction released a level of ammonia similar to that of the ammonia standard solution, leading to the conversion of NADPH to NADP+, whereas the reaction buffer without NAM (negative control) or a blank control exhibited no change in ammonia release. The enzymatic activity (mean ± standard deviation) of purified rPncA was 54.15 ± 7.93 U/mg protein, based on the absorbance at 340 nm. These results indicate that rPncA has high catalytic activity in vitro.
FIG 1.
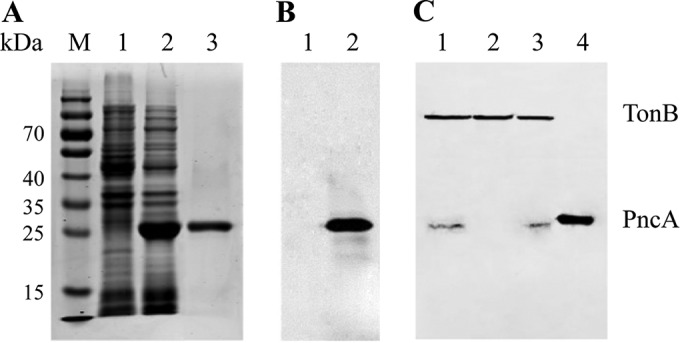
Expression and identification of R. anatipestifer PncA. (A) SDS-PAGE analysis of rPncA. Lane M, molecular weight markers; lane 1, negative control, i.e., E. coli BL21(DE3) transformed with pET-28a(+), with IPTG induction; lane 2, supernatant from E. coli BL21(DE3) transformed with pET-pncA, with IPTG induction; lane 3, purified rPncA. (B) Western blot profile. Lane 1, negative control, i.e., E. coli BL21(DE3) transformed with pET-28a(+), with IPTG induction; lane 2, E. coli BL21(DE3) transformed with pET-pncA, with IPTG induction. (C) Identification of the complemented strain cYb2ΔpncA by Western blot analysis. A second antibody against the TonB-dependent receptor of R. anatipestifer was used to control for protein loading. Lane 1, whole-cell proteins from the wild-type strain Yb2; lane 2, whole-cell proteins from the mutant strain Yb2ΔpncA; lane 3, whole-cell proteins from the complemented strain cYb2ΔpncA; lane 4, positive control, i.e., supernatant from E. coli BL21(DE3) transformed with pET-pncA, with IPTG induction.
FIG 2.
Enzymatic activity of R. anatipestifer rPncA. (A) Schematic of the coupled assay for measurement of the enzyme kinetics of rPncA. GIDH, glutamate dehydrogenase. (B) Assay results. One microgram of purified rPncA was added to reaction buffer to measure the enzyme kinetics. The optical density at 340 nm (OD340) decreased similarly with time for the rPncA reaction product and the ammonia standard solution, whereas no change was observed for the reaction buffer without NAM. Distilled water was used as the blank control.
Plasmid pCP29-pncA, which carries the AS87_01735 gene under the control of the R. anatipestifer ompA promoter, was transformed into the Yb2ΔpncA mutant strain for complementation, and PncA expression was determined. Whole-cell proteins of the wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA were transferred to a PVDF membrane and incubated with anti-PncA serum for Western blot analysis. The cYb2ΔpncA and Yb2 strains showed a positive band at 23 kDa, indicating that PncA is encoded by the AS87_01735 gene and that it was expressed in the cYb2ΔpncA strain (Fig. 1C). Based on the results described above, the AS87_01735 gene was designated pncA.
Localization of PncA in R. anatipestifer.
Anti-PncA polyclonal antibodies generated against rPncA specifically recognized a 23-kDa protein in whole-protein extracts of the wild-type strain Yb2, which is consistent with the estimated size of the protein (Fig. 3, lane 1). To determine the subcellular location of PncA in R. anatipestifer, the cytoplasmic and membrane fractions were extracted and analyzed by Western blotting. A single 23-kDa band corresponding to PncA was detected in the purified cytoplasm (Fig. 3, lane 3), indicating that PncA was expressed in the cytoplasm of R. anatipestifer.
FIG 3.
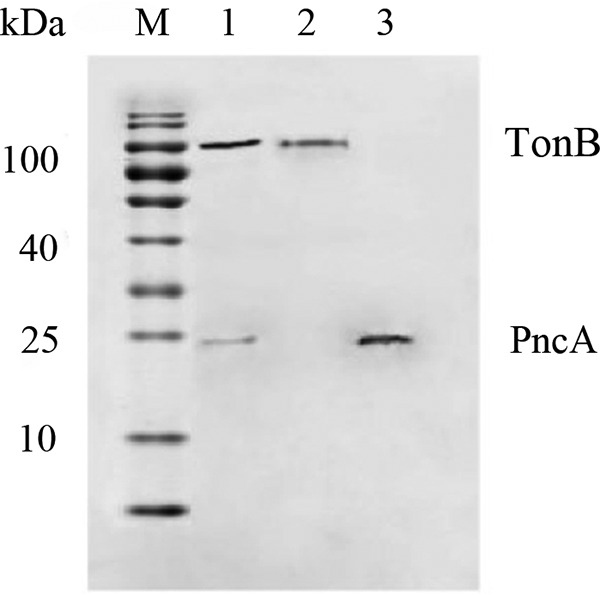
Immunoblot analysis of the subcellular location of PncA in R. anatipestifer. An antibody against the TonB-dependent receptor of R. anatipestifer was used to control for protein loading and membrane localization. Lane M, molecular size markers; lane 1, whole-cell proteins from R. anatipestifer strain Yb2; lane 2, membrane fraction from R. anatipestifer strain Yb2; lane 3, cytoplasmic fraction from R. anatipestifer strain Yb2.
The intracellular NAD+ concentration is lower in the R. anatipestifer mutant strain Yb2ΔpncA.
Given the central position of PncA in the bacterial NAD+ salvage pathway, the intracellular NAD+ concentrations in the wild-type and mutant strains were measured. In both the exponential and stationary growth phases, the intracellular NAD+ concentrations in the mutant strain Yb2ΔpncA were 2-fold lower than those in the wild-type strain Yb2 (Table 2). For exponential-phase cultures grown in TSB, the intracellular NAD+ concentrations in the mutant strain Yb2ΔpncA and the complemented strain cYb2ΔpncA were comparable; in the stationary phase, however, the Yb2ΔpncA strain contained 2-fold less NAD+ than the cYb2ΔpncA strain. Thus, in the complemented strain, plasmid pCP29-pncA restored NAD+ concentrations to wild-type levels in both growth phases.
TABLE 2.
Determination of intracellular NAD+ concentrations
| Growth phase | NAD+ concentration (μM) |
||
|---|---|---|---|
| Yb2 | Yb2ΔpncA | cYb2ΔpncA | |
| Exponential | 4.29 ± 0.19 | 2.69 ± 0.14a | 3.42 ± 0.25 |
| Stationary | 13.04 ± 1.30 | 6.46 ± 0.13a,b | 11.3 ± 1.41 |
P < 0.05, compared with the wild-type strain Yb2.
P < 0.05, compared with the complemented strain cYb2ΔpncA.
Deletion of the pncA gene decreases bacterial invasion.
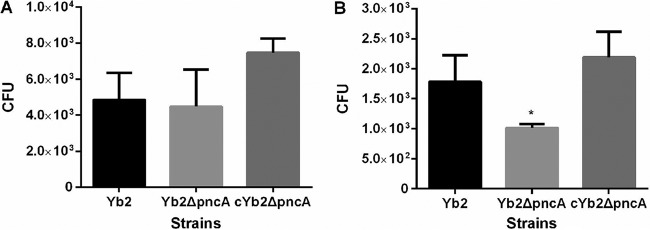
To investigate whether PncA plays a role in bacterial adhesion to biotic surfaces, the adherence and invasion capacities of the wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA were assessed in Vero cells. After incubation with Vero cells, the host cell-associated bacterial CFU value for strain Yb2ΔpncA was comparable to that of the wild-type strain Yb2 (Fig. 4A); however, the invasion capacity of strain Yb2ΔpncA was significantly reduced, compared with that of strain Yb2 (P < 0.05). Strain cYb2ΔpncA exhibited a wild-type level of invasion (Fig. 4B).
FIG 4.
Bacterial adherence (A) and invasion (B) assays. The assays were performed with Vero cells. Data represent the number of cell-associated bacteria (A) or bacteria invaded into Vero cells (B) in each well of 24-well plates. Error bars, standard deviations from three independent experiments that were performed in triplicate. *, P < 0.05.
Deletion of the pncA gene significantly attenuates bacterial virulence.
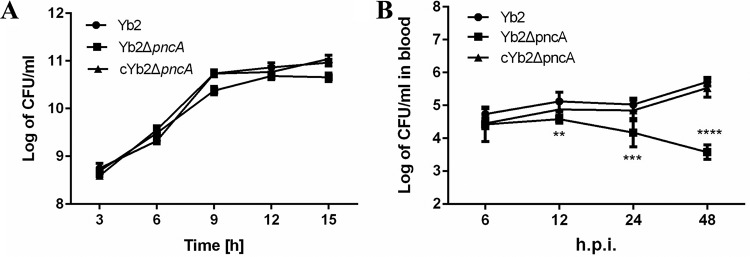
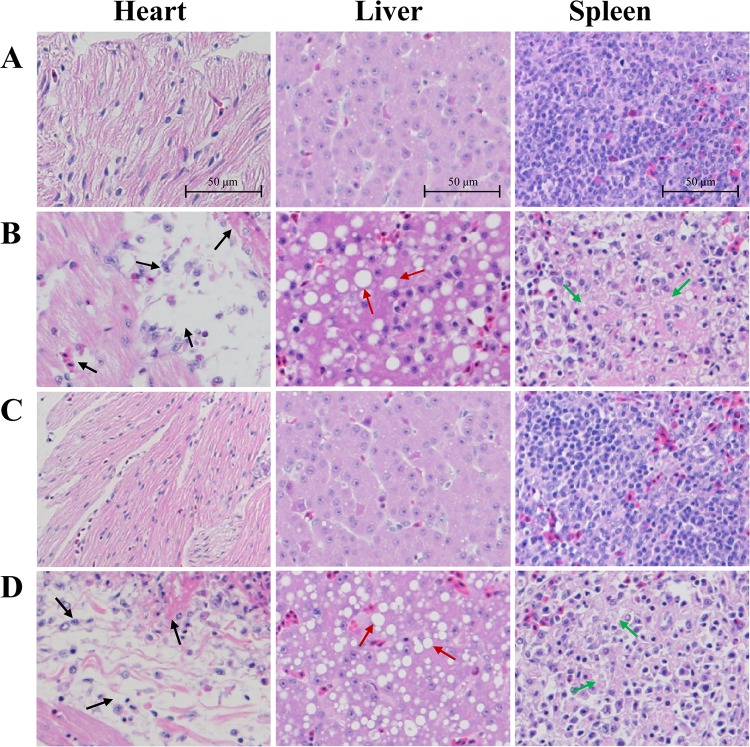
When bacterial growth in vitro was examined, no significant differences were found among the wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA (Fig. 5A). Our previous study revealed that the virulence of the Yb2ΔpncA strain was attenuated approximately 488,000-fold, compared with that of strain Yb2. To assess the role of PncA in systemic invasion and dissemination, bacterial loads in the blood of ducks infected with the wild-type strain Yb2, the mutant strain Yb2ΔpncA, or the complemented strain cYb2ΔpncA were determined. As shown in Fig. 5B, significantly fewer bacteria were recovered from the blood of ducks infected with strain Yb2ΔpncA, compared with ducks infected with the wild-type strain, at 12, 24, and 48 hpi. Moreover, the bacterial load in the blood of ducks infected with strain Yb2ΔpncA gradually decreased as the infection time increased. The hearts, livers, and spleens of ducks infected with strain Yb2ΔpncA were examined histologically at 48 hpi and showed no pathological changes; they were almost the same as those obtained from normal ducks that had not been challenged (Fig. 6C). The samples from ducks infected with the wild-type strain Yb2 were collected at 48 hpi, and they showed severe gross lesions of fibrinous exudates in the pericardial cavity, as well as histopathological changes such as cellular edema, fibrin deposition, inflammatory cell infiltration, and vesicular degeneration (Fig. 6B). Infection with the complemented strain cYb2ΔpncA resulted in similar histopathological changes in the heart, liver, and spleen (Fig. 6D).
FIG 5.
Determination of bacterial growth in vitro and in vivo. (A) Bacterial growth curves in TSB. Bacterial CFU were counted at 3-h intervals, and the data are presented as the means of three replicates. No significant difference was observed among the R. anatipestifer wild-type strain Yb2, the mutant strain Yb2ΔpncA, and the complemented strain cYb2ΔpncA. (B) Bacterial loads in the blood of infected ducks. Six ducks each were infected with strain Yb2, strain Yb2ΔpncA, or strain cYb2ΔpncA at a dose of 1 × 107 CFU. The bacterial loads were counted at 6, 12, 24, and 48 hpi. Data are means ± standard deviations. At 12, 24, and 48 hpi, the bacterial loads in the blood samples from ducks infected with strain Yb2ΔpncA were significantly smaller than those in samples from ducks infected with strain Yb2. The data were analyzed using one-way analysis of variance. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FIG 6.
Histopathological examinations. (A) Samples from a normal duck. (B) Samples from a duck infected with the wild-type strain Yb2, at 48 hpi. (C) Samples from a duck infected with the mutant strain Yb2ΔpncA, at 48 hpi. (D) Samples from a duck infected with the complemented strain cYb2ΔpncA, at 48 hpi. No pathological changes were observed in the tissues of Yb2ΔpncA-infected ducks. In Yb2- or cYb2ΔpncA-infected ducks, pathological changes, including inflammatory cell infiltration, hemorrhage, cellular edema, and fibrinous exudate, were observed in the heart (black arrows), while vacuolar degeneration in hepatocytes was observed in the liver (red arrows) and diffuse vacuolation and decreased cellularity were found in the spleen (green arrows). The tissues were stained with hematoxylin and eosin. Magnification, ×200.
Vaccination with the mutant strain Yb2ΔpncA protects against virulent Yb2 challenge.
To determine whether the mutant strain Yb2ΔpncA can be used as a live vaccine candidate, a protection experiment was conducted with ducks. At day 14 postvaccination, ducks were challenged with 106 CFU (10 times the LD50) of the virulent wild-type strain Yb2. The ducks in group 1 (which received one immunization) and group 3 (which received two immunizations) were 80% and 90% protected from challenge, respectively (Table 3). The ducks in groups 2 and 4 (control groups) showed clinical symptoms and were killed within 7 days postchallenge. These results indicate that the attenuated mutant strain Yb2ΔpncA can be used as a live vaccine candidate.
TABLE 3.
Animal challenge experiment
| Groupa | No. of injections | Inoculation typeb | Challenge strainc | No. of duck deaths | Protection rate (%)d |
|---|---|---|---|---|---|
| 1 | 1 | Yb2ΔpncA | Yb2 | 2 | 80 |
| 2 | 1 | Saline | Yb2 | 10 | 0 |
| 3 | 2 | Yb2ΔpncA | Yb2 | 1 | 90 |
| 4 | 2 | Saline | Yb2 | 10 | 0 |
Each group contained 10 ducks.
Seven-day-old ducks were immunized once or twice. Ducks that received two immunizations were immunized 7 days apart.
The challenge dose was 10 times the LD50.
The protection rate was calculated as follows: [1 − (no. of dead ducklings per group/total no. of ducklings per group)] × 100.
DISCUSSION
This study was initially designed to discover the pncA gene in R. anatipestifer strain Yb2 and to explore the potential of the mutant for live vaccine development. The mutant strain Yb2ΔpncA was screened from a library of random transposon mutants in animals. A BLAST search indicated that the transposon-disrupted gene AS87_01735 showed high levels of identity among R. anatipestifer strains of different serotypes, suggesting that the AS87_01735 gene is highly conserved in R. anatipestifer. The predicted protein encoded by AS87_01735 exhibits sequence identity to characterized nicotinamidases from other organisms ranging from 22.6% to 45.5%. To confirm that the product of the AS87_01735 gene in R. anatipestifer is an active nicotinamidase, the AS87_01735 gene was cloned into the expression vector pET28a(+), and rPncA was expressed in E. coli BL21(DE3), purified, and characterized in an enzymatic assay. As shown in Fig. 3, rPncA exhibited nicotinamidase activity that hydrolyzed NAM to NA and ammonia. Additionally, Western blot analysis revealed that the Yb2ΔpncA mutant did not react with anti-PncA serum, while the wild-type strain Yb2 and the complemented strain cYb2ΔpncA showed a positive band at 23 kDa, suggesting that PncA is lacking in strain Yb2ΔpncA. These results indicate that the R. anatipestifer AS87_01735 gene is the PncA-encoding gene and that PncA is responsible for the aforementioned characteristics of the Yb2ΔpncA strain.
NAD+, acting as both a coenzyme for hydride-transfer enzymes and a substrate for NAD+-consuming enzymes, is widely distributed throughout biological systems, and it is involved in several biological functions, such as signaling, control of gene expression, and aging (21, 22). Canonically, the two basic types of NAD+ synthesis consist of a de novo biosynthetic pathway and a salvage biosynthetic pathway. In de novo biosynthesis, nicotinic acid mononucleotide is synthesized from aspartic acid in three enzymatic steps in prokaryotes or from l-tryptophan in five steps in eukaryotes, followed by two enzymatic steps to complete the synthesis of NAD+. R. anatipestifer lacks two homologues of the nadABC genes that are involved in the de novo pathway. In contrast, R. anatipestifer possesses all of the putative members of a functional salvage pathway. A genomically based reconstitution of NAD+ metabolism suggested that R. anatipestifer relies largely on the salvage pathway to synthesize NAD+. PncA is a key enzyme in the cyclical salvage pathway for the production of NAD+ from NAM (23, 24). In this study, the putative nicotinamidase PncA, which is encoded by the AS87_01735 gene, is assumed to have an important role in R. anatipestifer biology by salvaging excess NAM while maintaining elevated NAD+ levels. Disruption of the pncA gene led to a 2-fold decrease in the intracellular NAD+ content, which was restored in the complemented strain cYb2ΔpncA (which expressed PncA). Although it lacks two homologues of the nadABC genes that are involved in the de novo synthesis pathway, R. anatipestifer possesses a putative quinolinate synthase (AS87_04445) that catalyzes the conversion of iminoaspartate to quinolinate (25), the second step of the de novo NAD+ biosynthetic pathway. In addition, R. anatipestifer harbors putative enzymes that are involved in the last two steps of de novo NAD+ biosynthesis, i.e., a nicotinate-nucleotide adenylyltransferase (AS87_07940), a phosphodiesterase (AS87_08680), and an NAD+ synthetase (AS87_08685). It is interesting that the aforementioned three enzymes are also involved in salvage NAD+ biosynthesis. Thus, defective NAD+ recycling caused by PncA inactivation has a large effect on intracellular NAD+ levels. In some organisms, supplementation of pncA mutants with increasing concentrations of NAM or NR can reverse the growth defect phenotype and increase the intracellular NAD+ contents. For example, Mycobacterium bovis can support the deletion of the de novo pathway because it has a salvage pathway that is still functional when NAM is supplied at high concentrations (26). In R. anatipestifer, such a perturbation of the intracellular NAD+ contents has no effect on bacterial growth in vitro. One possible reason for this is that TSB contains precursors (NAM, NA, or NR) for NAD+, and another is that R. anatipestifer may rely on the de novo NAD+ synthesis pathway for growth in vitro.
In addition to its role in regulating NAD+ biosynthesis, PncA is involved in bacterial virulence (27, 28). Brucella abortus PncA contributes to the utilization of nutrients that are required for intracellular replication and infectivity in mice (14). Plasmid-encoded PncA of Borrelia burgdorferi is essential for its infectivity in a mammalian host (16). In our previously study, the virulence of the Yb2ΔpncA mutant strain was >488,000-fold attenuated, compared with that of the wild-type strain Yb2, based on LD50 determinations. The present study showed that the bacterial loads in the blood of ducks infected with the Yb2ΔpncA strain were significantly lower than those of ducks that were infected with the wild-type strain Yb2. Furthermore, ducks that were infected with the Yb2ΔpncA strain did not show any histological changes in the heart, liver, or spleen, whereas ducks infected with the wild-type strain Yb2 showed severe gross lesions of fibrinous exudates in the pericardial cavity and on the surface of the liver, as well as histopathological changes such as fibrinous exudation, vesicular degeneration, diffuse vacuolation, and decreased cellularity. Therefore, we demonstrated that the pncA gene is a virulence factor of R. anatipestifer.
Various antibiotics are currently used to prevent and to control R. anatipestifer infections in ducks, but the increasing prevalence of drug-resistant strains is posing a serious challenge for antibiotic treatment (29, 30). Therefore, immunization should be the ideal way to prevent ducks from being infected with R. anatipestifer. One advantage of live attenuated vaccines is that they can stimulate protracted cell-mediated immunity and confer good protection (31, 32). Our study showed that ducks that were immunized with the mutant strain Yb2ΔpncA were protected from challenge with wild-type virulent R. anatipestifer, suggesting that the Yb2ΔpncA strain is an effective vaccine candidate for protecting ducks against serotype 2 R. anatipestifer infections in China.
In conclusion, we identified the R. anatipestifer AS87_01735 gene as a PncA-encoding gene that contributes to bacterial NAD+ biosynthesis and virulence. The mutant strain Yb2ΔpncA could be used as a novel vaccine candidate.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Shanghai Laboratory Animal Research Center (Shanghai, China) for preparation of tissue sections and histopathological diagnosis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01829-16.
REFERENCES
- 1.Leavitt S, Ayroud M. 1997. Riemerella anatipestifer infection of domestic ducklings. Can Vet J 38:113. [PMC free article] [PubMed] [Google Scholar]
- 2.Sandhu TS, Leister ML. 1991. Serotypes of ‘Pasteurella’ anatipestifer isolates from poultry in different countries. Avian Pathol 20:233–239. doi: 10.1080/03079459108418760. [DOI] [PubMed] [Google Scholar]
- 3.Pathanasophon P, Phuektes P, Tanticharoenyos T, Narongsak W, Sawada T. 2002. A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 31:267–270. doi: 10.1080/03079450220136576. [DOI] [PubMed] [Google Scholar]
- 4.Hu Q, Zhang Z, Miao J, Liu Y, Liu X, Ding S. 2001. The epidemiology study of Riemerella anatipestifer infection in Jiangsu and Anhui provinces. Chinese J Vet Sci Technol 31:12–13. [Google Scholar]
- 5.Subramaniam S, Huang B, Loh H, Kwang J, Tan HM, Chua KL, Frey J. 2000. Characterization of a predominant immunogenic outer membrane protein of Riemerella anatipestifer. Clin Diagn Lab Immunol 7:168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C, Hung P, Chang Y. 1998. Molecular characterization of a plasmid isolated from Riemerella anatipestifer. Avian Pathol 27:339–345. doi: 10.1080/03079459808419349. [DOI] [PubMed] [Google Scholar]
- 7.Crasta KC, Chua K-L, Subramaniam S, Frey J, Loh H, Tan H-M. 2002. Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J Bacteriol 184:1932–1939. doi: 10.1128/JB.184.7.1932-1939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Q, Han X, Zhou X, Ding C, Zhu Y, Yu S. 2011. OmpA is a virulence factor of Riemerella anatipestifer. Vet Microbiol 150:278–283. doi: 10.1016/j.vetmic.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Ding C, Wang S, Han X, Hou W, Yue J, Zou J, Yu S. 2014. The AS87_04050 gene is involved in bacterial lipopolysaccharide biosynthesis and pathogenicity of Riemerella anatipestifer. PLoS One 9:e109962. doi: 10.1371/journal.pone.0109962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou J, Wang X, Tian M, Cao S, Hou W, Wang S, Han X, Ding C, Yu S. 2015. The M949_1556 gene plays a role on the bacterial antigenicity and pathogenicity of Riemerella anatipestifer. Vet Microbiol 177:193–200. doi: 10.1016/j.vetmic.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, Wang X, Ding C, Tian M, Han X, Wang S, Yu S. 2015. Characterization and cross-protection evaluation of M949_1603 gene deletion Riemerella anatipestifer mutant RA-M1. Appl Microbiol Biotechnol 99:10107–10116. doi: 10.1007/s00253-015-6848-y. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Ding C, Wang S, Han X, Yu S. 2015. Whole-genome sequence analysis and genome-wide virulence gene identification of Riemerella anatipestifer strain Yb2. Appl Environ Microbiol 81:5093–5102. doi: 10.1128/AEM.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belenky P, Bogan KL, Brenner C. 2007. NAD+ metabolism in health and disease. Trends Biochem Sci 32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Kurokawa D, Watanabe K, Makino S, Shirahata T, Watarai M. 2004. Brucella abortus nicotinamidase (PncA) contributes to its intracellular replication and infectivity in mice. FEMS Microbiol Lett 234:289–295. doi: 10.1111/j.1574-6968.2004.tb09546.x. [DOI] [PubMed] [Google Scholar]
- 15.Ma B, Pan SJ, Zupancic ML, Cormack BP. 2007. Assimilation of NAD+ precursors in Candida glabrata. Mol Microbiol 66:14–25. doi: 10.1111/j.1365-2958.2007.05886.x. [DOI] [PubMed] [Google Scholar]
- 16.Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 17.Gazanion E, Garcia D, Silvestre R, Gerard C, Guichou JF, Labesse G, Seveno M, Cordeiro-Da-Silva A, Ouaissi A, Sereno D, Vergnes B. 2011. The Leishmania nicotinamidase is essential for NAD+ production and parasite proliferation. Mol Microbiol 82:21–38. doi: 10.1111/j.1365-2958.2011.07799.x. [DOI] [PubMed] [Google Scholar]
- 18.Hu Q, Zhu Y, Tu J, Yin Y, Wang X, Han X, Ding C, Zhang B, Yu S. 2012. Identification of the genes involved in Riemerella anatipestifer biofilm formation by random transposon mutagenesis. PLoS One 7:e39805. doi: 10.1371/journal.pone.0039805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghislain M, Talla E, Francois JM. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 20.He S, Chen H, Jiao F. 2011. Improved preparation method for animal histopathological section. Prog Vet Med 32:130–132. [Google Scholar]
- 21.Lin SJ, Guarente L. 2003. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol 15:241–246. doi: 10.1016/S0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler M, Niere M. 2004. NAD+ surfaces again. Biochem J 382:e5–e6. doi: 10.1042/BJ20041217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tritz GJ, Matney TS, Gholson RK. 1970. Mapping of the nadB locus adjacent to a previously undescribed purine locus in Escherichia coli K-12. J Bacteriol 102:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frothingham R, Meeker-O'Connell WA, Talbot EA, George JW, Kreuzer KN. 1996. Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother 40:1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ceciliani F, Caramori T, Ronchi S, Tedeschi G, Mortarino M, Galizzi A. 2000. Cloning, overexpression, and purification of Escherichia coli quinolinate synthetase. Protein Expr Purif 18:64–70. doi: 10.1006/prep.1999.1153. [DOI] [PubMed] [Google Scholar]
- 26.Vilcheze C, Weinrick B, Wong KW, Chen B, Jacobs WR Jr. 2010. NAD+ auxotrophy is bacteriocidal for the tubercle bacilli. Mol Microbiol 76:365–377. doi: 10.1111/j.1365-2958.2010.07099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raynaud C, Etienne G, Peyron P, Laneelle MA, Daffe M. 1998. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144:577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 28.Jewett MW, Jain S, Linowski AK, Sarkar A, Rosa PA. 2011. Molecular characterization of the Borrelia burgdorferi in vivo-essential protein PncA. Microbiology 157:2831–2840. doi: 10.1099/mic.0.051706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YP, Tsao MY, Lee SH, Chou CH, Tsai HJ. 2010. Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol 39:333–338. doi: 10.1080/03079457.2010.507761. [DOI] [PubMed] [Google Scholar]
- 30.Chen YP, Lee SH, Chou CH, Tsai HJ. 2012. Detection of florfenicol resistance genes in Riemerella anatipestifer isolated from ducks and geese. Vet Microbiol 154:325–331. doi: 10.1016/j.vetmic.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Girard MP, Steele D, Chaignat CL, Kieny MP. 2006. A review of vaccine research and development: human enteric infections. Vaccine 24:2732–2750. doi: 10.1016/j.vaccine.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Venkatesan MM, Ranallo RT. 2006. Live-attenuated Shigella vaccines. Expert Rev Vaccines 5:669–686. doi: 10.1586/14760584.5.5.669. [DOI] [PubMed] [Google Scholar]
- 33.Peñuelas-Urquides K, Villarreal-Treviño L, Silva-Ramírez B, Rivadeneyra-Espinoza L, Said-Fernández S, Bermúdez de León M. 2013. Measuring of Mycobacterium tuberculosis growth. A correlation of the optical measurements with colony forming units. Braz J Microbiol 44:287–290. doi: 10.1590/S1517-83822013000100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.