ABSTRACT
Thanks to their wide host range and virulence, staphylococcal bacteriophages (phages) belonging to the genus Twortlikevirus (staphylococcal Twort-like phages) are regarded as ideal candidates for clinical application for Staphylococcus aureus infections due to the emergence of antibiotic-resistant bacteria of this species. To increase the usability of these phages, it is necessary to understand the mechanism underlying host recognition, especially the receptor-binding proteins (RBPs) that determine host range. In this study, we found that the staphylococcal Twort-like phage ΦSA012 possesses at least two RBPs. Genomic analysis of five mutant phages of ΦSA012 revealed point mutations in orf103, in a region unique to staphylococcal Twort-like phages. Phages harboring mutated ORF103 could not infect S. aureus strains in which wall teichoic acids (WTAs) are glycosylated with α-N-acetylglucosamine (α-GlcNAc). A polyclonal antibody against ORF103 also inhibited infection by ΦSA012 in the presence of α-GlcNAc, suggesting that ORF103 binds to α-GlcNAc. In contrast, a polyclonal antibody against ORF105, a short tail fiber component previously shown to be an RBP, inhibited phage infection irrespective of the presence of α-GlcNAc. Immunoelectron microscopy indicated that ORF103 is a tail fiber component localized at the bottom of the baseplate. From these results, we conclude that ORF103 binds α-GlcNAc in WTAs, whereas ORF105, the primary RBP, is likely to bind the WTA backbone. These findings provide insight into the infection mechanism of staphylococcal Twort-like phages.
IMPORTANCE Staphylococcus phages belonging to the genus Twortlikevirus (called staphylococcal Twort-like phages) are considered promising agents for control of Staphylococcus aureus due to their wide host range and highly lytic capabilities. Although staphylococcal Twort-like phages have been studied widely for therapeutic purposes, the host recognition process of staphylococcal Twort-like phages remains unclear. This work provides new findings about the mechanisms of host recognition of the staphylococcal Twort-like phage ΦSA012. The details of the host recognition mechanism of ΦSA012 will allow us to analyze the mechanisms of infection and expand the utility of staphylococcal Twort-like phages for the control of S. aureus.
INTRODUCTION
Staphylococcus aureus, a Gram-positive coccus, is a commensal and pathogenic bacterium that causes opportunistic infections in humans and animals. Currently, antibiotic resistance in this species poses a threat to public health. Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of hospital-acquired infections around the world (1). The emergence of such antibiotic-resistant bacteria requires development of alternatives to antibiotic-based therapies. One promising alternative is phage therapy, in which bacteriophages (phages) are used to treat bacterial infections (2, 3). In preclinical trials performed in mice, S. aureus infections (including MRSA infections) were successfully treated by use of phages (4). Therefore, phage therapy has attracted great interest as an alternative to antibiotics.
Previously, we isolated the virulent staphylococcal phage ΦSA012, which exerts lytic effects on a wide range of S. aureus isolates from bovine mastitis cases (5). Genomic analysis of ΦSA012 revealed that it belongs to the genus Twortlikevirus, which contains the Staphylococcus phages Twort, K, and G, whereas the genus SPO1likevirus contains the Bacillus phage SPO1 as well as Listeria phages P100 and A511. Lactobacillus phage LP65 and Enterococcus phage ΦEF24C were classified as orphans within the subfamily (6). Representatives of the two genera share the following features: they (i) have large genomes (127 to 140 kb), (ii) are strictly virulent, (iii) infect Gram-positive, low-GC-content bacteria, (iv) have a wide host range among strains of the susceptible bacterial genus, and (v) share considerable similarities at the protein sequence level (7). Thanks to their wide host range and highly lytic capabilities, phages in these genera, including ΦSA012, are considered to be promising candidates for therapeutic use and as detection agents (8, 9).
The infection process of phages can be divided into the following steps: phage adsorption to the host, DNA injection into the host cell, DNA replication, assembly of phage particles, and lysis of the host cell. Because adsorption of a phage to the host, caused by the interaction between a phage receptor on the bacterial surface and receptor-binding proteins (RBPs) in the tips of the tail fibers or tail spikes, is extremely specific, RBPs determine the target bacteria for phage infection (10). Therefore, application of phages as therapy necessitates an understanding of the interaction between RBPs and phage receptors on the bacterial surface.
The interaction of RBPs with phage receptors on the bacterial surface has been studied widely with Escherichia coli phages T4 and lambda (λ). In T4, gp37, located in the tips of long tail fibers, reversibly binds to lipopolysaccharides (LPS) or OmpC in E. coli K-12 (11). This first adsorption activates a second adsorption, in which gp12 (the short tail fiber) irreversibly binds to LPS on the cell surface (12). Although many studies of RBPs in phages of Gram-negative bacteria have been carried out, knowledge of RBPs in the phages of Gram-positive bacteria remains limited. To date, RBPs have been identified for Bacillus subtilis phages SPP1 and φ29, Streptococcus thermophilus phages DT1 and MD4, Lactococcus lactis phages bIL67 and CHL92 of the c2 species, sk1, bIL170, and p2 of the 936 species, and TP901-1 and Tuc2009 of the P335 species, and Listeria phage A118 (13–22). Among S. aureus phages, RBPs have also been identified in Siphoviridae phage φSLT and Podoviridae AHJD-like phages S24-1 and S13′ (23, 24). Remarkably, Habann et al. identified that the RBPs of Listeria phage A511 and staphylococcal Twort-like phages ISP and Twort (Gp108, gp40, and gp17, respectively) are located in short tail fibers (25). Among Gram-positive bacteria, cell wall-associated carbohydrates, such as teichoic acids, are regarded as phage receptors of B. subtilis, S. aureus, and Listeria monocytogenes (15, 25–29). In S. aureus, siphoviruses need N-acetylglucosamine (GlcNAc) in wall teichoic acids (WTAs) for adsorption, whereas myoviruses, such as staphylococcal Twort-like phages, adsorb to the WTA backbone (27). A recent study suggested that staphylococcal podoviruses require α-GlcNAc, but not β-GlcNAc, for adsorption (30). Nevertheless, the mechanism of host recognition by staphylococcal Twort-like phages remains unclear. In this study, we focused on the mechanism of host recognition by staphylococcal Twort-like phage ΦSA012 at the molecular level.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions.
Bacterial strains, phages, and plasmids are listed in Table 1. S. aureus strain RN4220 was kindly supplied by Motoyuki Sugai (Hiroshima University Graduate School of Biomedical & Health Sciences, Hiroshima, Japan), with the permission of Richard P. Novick (Skirball Institute of Biomolecular Medicine, New York, NY), and was used for gene manipulations (31). S. aureus strain SA003, which was isolated from raw milk samples from a mastitic cow, was used for propagation and enumeration of staphylococcal phages. The S. aureus virulent phage ΦSA012 was isolated from sewage in a previous study (5). ΦSA012 and SA003 were deposited in the culture collection of the NITE Biological Resource Center, Kisarazu, Japan (accession no. NBRC110649 and NBRC110650, respectively). Shuttle vectors pNL9164 and pLI50 were purchased from Sigma-Aldrich (St. Louis, MO) and Addgene (Cambridge, MA), respectively. The shuttle vector pKOR1 was kindly supplied by Taeok Bae (Indiana University School of Medicine-Northwest, Indianapolis, IN). All S. aureus and E. coli strains were grown at 37°C in Luria-Bertani (LB) medium overnight, unless otherwise stated.
TABLE 1.
Bacterial strains, phages, and plasmids
| Bacterial strain, phage, or plasmid | Relevant feature(s) | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. aureus strains | ||
| SA003 | Host strain, isolated from bovine mastitis, lacks tarM | 5 |
| SA003R11 | ΦSA012-resistant derivative | 60 |
| SA003R20 | ΦSA012-resistant derivative | 60 |
| RN4220 | Restriction-deficient, transformable strain | 61 |
| RN4220 ΔtarM | In-frame deletion mutant of tarM in RN4220 | This study |
| RN4220 ΔtarM::pLIP3_tarM | RN4220 ΔtarM complemented with tarM via the plasmid pLIP3_tarM | This study |
| E. coli strains | ||
| JM109 | Used for plasmid construction | TaKaRa Bio |
| Rosetta-gami 2(DE3) | Used for expression of recombinant proteins | Novagen |
| Phages | ||
| ΦSA012 | S. aureus lytic phage | 5 |
| ΦSA012M1 | Spontaneous mutant phage | This study |
| ΦSA012M2 | Spontaneous mutant phage | This study |
| ΦSA012M11 | Spontaneous mutant phage | This study |
| ΦSA012M20 | Spontaneous mutant phage | This study |
| ΦSA012M38 | Spontaneous mutant phage | This study |
| ΦSA012TM103 | Phage harboring three mutations in orf103 | This study |
| Plasmids | ||
| pKOR1 | E. coli-S. aureus shuttle vector, temperature sensitive | 33 |
| pLI50 | E. coli-S. aureus shuttle vector | Addgene |
| pLIP3_tarM | tarM expression plasmid driven by pLI50 | This study |
| pNL9164 | E. coli-S. aureus shuttle vector | Sigma |
| pET-29a | Expression vector for production of recombinant proteins | Novagen |
Phage preparation.
All phages were propagated by the plate lysate method (32). Briefly, 100 μl of phage lysate (>105 PFU/ml) was mixed with 100 μl of overnight bacterial culture in 3 ml of 0.5% top agar, plated on LB agar, and incubated at 37°C overnight. After 5 ml of salt magnesium (SM) buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5], 0.01% gelatin) was added to the plate and the overlayer was scraped off to extract phage, the supernatant was collected by centrifugation (5,000 × g, 15 min, 4°C). The obtained phage lysate was purified by polyethylene glycol (PEG) sedimentation and CsCl density gradient centrifugation (32). Each phage culture was titrated and stored at 4°C until use.
Molecular cloning in S. aureus.
The primers used in this study are shown in Table 2. Deletion of tarM in S. aureus RN4220 was performed using the shuttle vector pKOR1 as described before (33). A DNA fragment for allelic exchange was prepared by splicing by overlap extension (SOE) PCR, digested with EagI and EcoRV, and inserted into pKOR1 (New England BioLabs, Ipswich, MA). The resultant plasmid was constructed in E. coli JM109 and electroporated into S. aureus RN4220 (34). An S. aureus strain in which the plasmid had integrated into the genome was selected at 43°C in the presence of chloramphenicol (10 μg/ml). Subsequently, the plasmid was excised and cured by culturing the strain in brain heart infusion (BHI) medium at 30°C or 43°C. To screen for plasmid-free strains, colonies were replica plated onto tryptic soy broth (TSB) agar, with or without chloramphenicol (10 μg/ml). Anhydrotetracycline (1 μg/ml) was used for counterselection. Gene deletion was confirmed by PCR with the appropriate primer set. For complementation of tarM, the plasmid pLIP3_tarM for ectopic expression of tarM was constructed by integrating the P3 promoter, which is constitutive in S. aureus, and the tarM gene into the shuttle vector pLI50 (35, 36). DNA fragments corresponding to the P3 promoter and tarM were amplified from genomic DNA of RN4220 and inserted into pLI50. The plasmid pLIP3_tarM was constructed in E. coli JM109 and then electroporated into S. aureus RN4220 ΔtarM (34).
TABLE 2.
Primers used in this study
| Primer | Direction | Sequence (5′ → 3′)a | Purpose |
|---|---|---|---|
| PKOR1 insert_c_F | Forward | CACAGGAAACAGCTATGACATAG | Insert check in pKOR1 |
| PKOR1 insert_c_R | Reverse | CAGGTACATCATTCTGTTTGTG | Insert check in pKOR1 |
| tarM_A_Eag1 | Forward | AAACGGCCGTGAAATTGAAGAGAGTAAAGGTATTTC | SOE PCR for deletion of tarM |
| tarM_B | Reverse | TTTTGGAAAACTCCCTGGTCC | SOE PCR for deletion of tarM |
| tarM_C | Forward | GGACCAGGGAGTTTTCCAAAAGGTCAAGGGTTAAGTATGATAGAAG | SOE PCR for deletion of tarM |
| tarM_D_EcoRV | Reverse | AAAGATATCAGTAGTTACAGCTGGAAGAAA | SOE PCR for deletion of tarM |
| tarM_Fw | Forward | AATGGATCGAAGAACGAAAATGT | Check for deletion of tarM |
| tarM_Rv | Reverse | ACGCCTTATGTTAATGTTTTTTATATTTG | Check for deletion of tarM |
| pLI50-insertcheck-fw | Forward | TAATACCGCGCCACATAGCAGAAC | Insert check in pLI50 or pLIP3 |
| pLI50-insertcheck-rv | Reverse | TAGCTCACGCTATGCCGACATTC | Insert check in pLI50 or pLIP3 |
| P1075_EcoR1 | Forward | AAAGAATTCGAGTGGTATAAGTGGTTTTTCG | Amplify DNA fragment of P3 promoter |
| P697_Kpn1 | Reverse | AAAGGTACCTTCACCTCTGTTCTTACGACCTC | Amplify DNA fragment of P3 promoter |
| TarM_e_p3_Fw | Forward | AAACCCGGGGAGGTAAAGGAATAATTATAATGAAAAAAA | Complementation of tarM |
| TarM_e_Rv | Reverse | AAAGTCGACTTAGCTATTGAAAAGATTTAACCATTTTTC | Complementation of tarM |
| ORF103M-F | Forward | CCCAAGCTTGGGGGGTTGATTGACCCCTCTTT | Introduction of mutations into a recombinant phage |
| ORF103M-R | Reverse | CCCAAGCTTGGGCCCTAGCTCCTTGTCATACCC | Introduction of mutations into a recombinant phage |
| ORF103b-F | Forward | TGAATCCACAACTCAATATGCAAC | Check of mutations in a recombinant phage |
| ORF103c-F | Forward | TCATCTAGTAAAGGTAATGGTGC | Check of mutations in a recombinant phage |
| ORF103r-F | Forward | GGGAATTCCATATGGCATTTAACTACACGCCTC | Expression of ORF103 |
| ORF103r-R | Reverse | CCGCTCGAGTCCTCTATTAATTCCCATAATATTGTATACC | Expression of ORF103 |
| ORF105r-F | Forward | GGGAATTCCATATGGCATTTAACTACACGCCTC | Expression of ORF103 |
| ORF105r-R | Reverse | CCGCTCGAGTCCTCTATTAATTCCCATAATATTGTATACC | Expression of ORF103 |
| pET29a_Insert-c_F | Forward | CATGAGCCCGAAGTGGCGAGCCCGATCTTC | Insert check in pET29a |
| pET29a_Insert-c_R | Reverse | CGCTGCGCGTAACCACCACACCCGCCGCGC | Insert check in pET29a |
Bold underlined residues indicate restriction sites.
Isolation of mutant phages from coculture experiments.
In order to isolate ΦSA012-resistant derivatives and mutant phages, SA003 and ΦSA012 were cocultured. SA003 was inoculated into 4.5 ml of LB medium and cultured until early exponential phase (optical density at 660 nm [OD660] = 0.1) in a TVS062CA compact rocking incubator (Advantec, Tokyo, Japan). Approximately 4.5 × 108 PFU of ΦSA012 was added (multiplicity of infection [MOI] of 1) and cultured at 37°C with shaking at 40 rpm. After 2 to 10 days, bacterium-phage mixed cultures were collected. Forty-five microliters of bacterium-phage mixed culture was transferred to 4.5 ml of fresh LB medium (1:100 dilution) and cultured under the same conditions. After 2 to 10 days, the bacterium-phage mixed culture was collected, and a 1:100 dilution was performed again.
For isolation of ΦSA012-resistant derivatives and mutant phages, 1.5 ml of mixed culture was separated by centrifugation (9,730 × g, 5 min, 4°C) at each passage step. After washing four times with phosphate-buffered saline (PBS) to remove free phage, the pellet was resuspended in PBS and spread onto LB plates; one colony was picked as a ΦSA012-resistant derivative after overnight incubation. Supernatant from the coculture was used for a plaque assay with SA003. After overnight incubation, one plaque was also picked as a mutant phage.
This procedure was repeated continuously until the 38th passage. ΦSA012-resistant derivatives and mutant phages were defined as SA003R1 to SA003R38 and ΦSA012M1 to ΦSA012M38, respectively. Each number in the names of the ΦSA012-resistant derivatives and mutant phages represents the number of passages in coculture (e.g., SA003R11 refers to the phage-resistant derivative isolated from the coculture at the 11th passage, and ΦSA012M20 refers to the mutant phage isolated from the coculture at the 20th passage).
Extraction and analysis of phage genomic DNA.
Genomic DNAs of ΦSA012 and five mutant phages (ΦSA012M1, ΦSA012M2, ΦSA012M11, ΦSA012M20, and ΦSA012M38) were extracted from purified phages by use of a phage DNA isolation kit (Norgen Biotek Corp., Thorold, ON, Canada). Whole-genome sequencing was performed on a Genome Sequencer FLX+ system (Roche, Basel, Switzerland). Sequencing results were assembled and aligned using GS De Novo Assembler v2.8 (Roche, Basel, Switzerland) and Tablet (The James Hutton Plant Bioinformatics Group, Invergowrie, Scotland), respectively. Open reading frames (ORFs) were predicted with myRAST (http://blog.theseed.org/servers/presentations/t1/running-a-job-with-the-desktop-rast.html). Nucleotide and amino acid sequences were scanned for homologs by BLAST searches (37). Phage-carried tRNA genes were identified using Aragorn and tRNA Scan SE ver. 1.21 software (38, 39).
Generation and isolation of a recombinant phage harboring three mutations in orf103.
In order to construct a plasmid harboring three mutations in orf103, a DNA fragment was amplified from the genomic DNA of ΦSA012M20 by a PCR using primers ORF103M-F and ORF103M-R. This PCR fragment contained the nucleotide sequence between 200 bp upstream and 110 bp downstream of orf103. The recombinant fragment was digested with HindIII (TaKaRa Bio) and inserted into the shuttle vector pNL9164 (40). The plasmid was constructed and cloned in E. coli JM109. The constructed plasmid (named pNL9164::TMorf103) was then electroporated into S. aureus strain RN4220 (34).
A recombinant ΦSA012 phage harboring three mutations in orf103 (ΦSA012TM103) was generated by the following procedure. Transformed RN4220 harboring pNL9164::TMorf103 was grown to early exponential phase (OD660 = 0.1; 108 CFU/ml) at 32°C with shaking at 120 rpm in 10 ml of LB medium. ΦSA012 (106 PFU/ml) was then added at an MOI of 0.01. During phage infection, homologous recombination between phage DNA and the plasmid in transformed RN4220 harboring ORF103 of ΦSA012M20 might happen at a low frequency. After overnight incubation at 32°C with shaking, the supernatant was collected by centrifugation (6,230 × g, 10 min, 4°C) and plated for a plaque assay for isolation of ΦSA012TM103. Briefly, 500 μl of phage lysate and 250 μl of overnight culture of phage-resistant derivative SA003R11 (susceptible to ΦSA012M20 but not to ΦSA012) were added to 6 ml of 0.5% top agar and poured onto LB agar plates. After incubation overnight at 37°C, single plaques were picked and resuspended in 100 μl of SM buffer. These suspensions were purified by two rounds of plaque assay with SA003R11. Mutations in orf103 of ΦSA012TM103 were identified by Sanger sequencing using primers ORF103b-F and ORF103c-F.
Protein expression and purification.
C-terminally His-tagged recombinant ORF103 and ORF105 were expressed in E. coli Rosetta-gami 2(DE) from vector pET-29a and purified by immobilized-metal affinity chromatography (IMAC). To construct plasmids for expression of ORF103 and ORF105, DNA fragments were amplified by PCR with the appropriate primer sets (Table 2). Recombinant fragments for protein expression were digested with NdeI and XhoI (New England BioLabs) and inserted into pET-29a. The plasmids were constructed and cloned into E. coli JM109, and the clones were then electroporated into E. coli Rosetta-gami 2(DE).
To produce the ORF103 and ORF105 proteins, expression was induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an OD600 of 0.5. After incubation at 28°C with shaking at 160 rpm overnight, the cells were collected by centrifugation (6,230 × g, 10 min, 4°C), resuspended in phosphate buffer (20 mM phosphate [pH 7.4], 0.5 M NaCl), and disrupted by sonication for 40 min in a VP60-S sonicator (Taitec, Koshigaya, Japan). Target proteins were purified using a HisTrap HP column (GE Healthcare Life Sciences, Buckinghamshire, United Kingdom). After His-tagged proteins were eluted from the Ni column with elution buffer (40 to 500 mM imidazole in phosphate buffer), the imidazole was removed by dialysis against phosphate buffer.
Preparation of antibodies.
Polyclonal rabbit antibodies against the recombinant ORF103 and ORF105 proteins were generated by Japan Bio Serum (Hiroshima, Japan). Briefly, rabbits were immunized with 0.3 mg of protein once every 2 weeks for 8 weeks; in total, 1.5 mg of protein was used. After blood was collected in week 10, anti-ORF103 serum was purified on a protein A column and stored at −20°C until use. The binding ability of the anti-ORF103 and anti-ORF105 polyclonal antibodies was confirmed by Western blotting.
Immunoelectron microscopy.
Immunoelectron microscopy was conducted as described previously (23). A freshly purified phage sample (1010 PFU/ml) was mixed with purified anti-ORF103 antibody diluted in SM buffer (1:100) and incubated at room temperature for 30 min. The samples were loaded onto ester-carbon-coated copper grids (EMJapan, Tokyo, Japan). The copper grids were washed twice with SM buffer and incubated with 12-nm colloidal gold-labeled AffiniPure goat anti-rabbit IgG(H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA) in PBS (1:50) at 37°C for 30 min. After washing with SM buffer and Milli-Q water twice each, the grids were stained with 2% uranyl acetate and observed on a JEM-1400 Plus microscope (JEOL, Akishima, Japan).
Spot test and assay of the efficiency of plating (EOP) with antibodies.
The infectivity of phages was evaluated as previously described (5). Briefly, 2-μl aliquots of serially diluted phage lysate (107 to 1010 PFU/ml) were dropped onto LB plates overlaid with S. aureus strains mixed with 0.5% top agar and then incubated overnight to assess plaque formation.
The EOP of phages on S. aureus strains with antibodies was measured by plaque assay, with the phage lysate adjusted with SA003. For assay with an antibody to evaluate the role of ORF103 or ORF105 in infection, 10 μl of anti-ORF103 or anti-ORF105 serum was added to 100 μl of phage lysate. After incubation for 1 h at room temperature, the number of infectious phage was enumerated by a plaque assay with SA003. Serum collected from a rabbit before immunization (preimmune serum) was used as a control.
Adsorption assay.
The adsorption efficiency of phages on S. aureus strains was measured by titrating free phage present in the supernatant after defined periods of cell-phage contact. S. aureus cells were prepared by 10% inoculation of overnight culture into 4.5 ml of LB medium; the culture was then incubated at 37°C with shaking at 120 rpm to an OD660 of 1.0 (∼109 CFU/ml). Phage lysate (107 PFU/ml) was then added to the bacterial culture. After infection at 37°C with shaking at 120 rpm, free phage was collected by centrifugation (9,730 × g, 1 min) at defined times and titrated using SA003. For longer incubation, 50 μg/ml of chloramphenicol or erythromycin was added, and cells were equilibrated for 10 min at 37°C before infection to inhibit cell growth and phage development during incubation with phages (41). Adsorption efficiency was calculated by dividing the number of adsorbed phage by the initial number of phage.
Statistical analysis.
Two-tailed Student's t test was used to determine statistical significance.
Accession number(s).
The complete genome of ΦSA012 has been deposited in the GenBank database under accession number AB903967.
RESULTS
Genomic analysis of ΦSA012 and its mutant phages.
Whole-genome sequencing of ΦSA012 revealed that its genome is 142,094 bp long and contains 207 ORFs (see Table S1 in the supplemental material). Terminally redundant regions, a common feature of Twortlikevirus and Spounalikevirus, were found at both ends of the genome (7, 62–65). Three tRNA genes (Met-tRNA gene, bp 8,180 to 8,109; Asp-tRNA gene, bp 30,496 to 30,423; and Phe-tRNA gene, bp 30,416 to 30,344) are carried in the ΦSA012 genome.
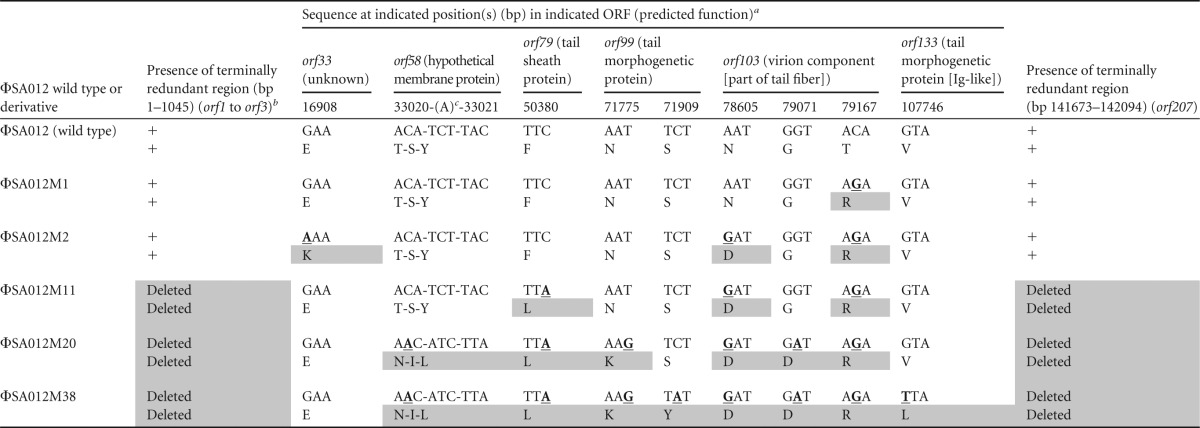
Mutant phages isolated from cocultures with SA003 showed infectivities different from that of wild-type ΦSA012 by the spot test with ΦSA012-resistant derivatives (data not shown). In order to find the candidate genes responsible for host recognition, all mutations in five mutant phages isolated from cocultures were identified by whole-genome sequencing (Table 3). Mutations were found in six ORFs (orf33, orf58, orf79, orf99, orf103, and orf133) and the terminally redundant regions located at both ends of the genome.
TABLE 3.
Distribution of mutation sites in mutant phages
All positions are relative to the genome of ΦSA012. For each phage variant, upper sequences are nucleotide sequences, and lower sequences are amino acid sequences. Bold underlined residues indicate nucleotide mutations, and gray boxes represent mutations in amino acids.
+, the region is present.
An insertion of adenine (A) was discovered between bp 33020 (adenine) and bp 33021 (cytosine), resulting in a premature stop codon and partial deletion of the product (aa 85 to 108).
Comparisons of structural proteins among Twort-like viruses and relatives.
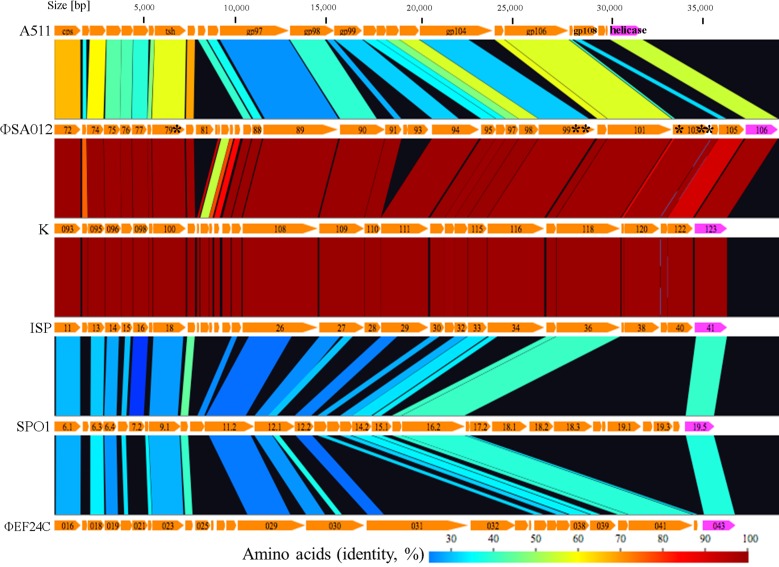
Comparisons of structural proteins (from capsid to helicase) among ΦSA012, K, and ISP (Twortlikevirus), SPO1 (Spounalikevirus), and ΦEF24C revealed that the region from ORF103 to ORF105 is unique to staphylococcal Twort-like phages (ΦSA012, K, and ISP) at the protein level (Fig. 1). The basic structural components, such as the capsid (ORF72 in ΦSA012), tail sheath (ORF79 in ΦSA012), and tape major protein (ORF89 in ΦSA012), as well as a helicase involved in DNA replication (ORF106 in ΦSA012), share higher degrees of similarity, probably due to functional similarities between the respective proteins. ORF101 in ΦSA012, which was predicted to be an adsorption-associated tail protein facilitating infection of Gram-positive bacteria by digesting sialic acid residues in their slime or capsules due to the presence of a putative conserved neuraminidase/sialidase domain, was also conserved among related phages (7, 45).
FIG 1.
Comparisons of main structural proteins among Twortlikevirus and related phages. Host bacteria for each phage are as follows: A511, Listeria monocytogenes; ΦSA012, K, and ISP, Staphylococcus aureus; SPO1, Bacillus subtilis; and ΦEF24C, Enterococcus faecalis. Asterisks (*) represent the locations of mutations. Gp108 in A511 is an RBP (25). The figure was generated using GenomeMatcher (59).
Infectivity of a recombinant phage harboring three mutations in orf103.
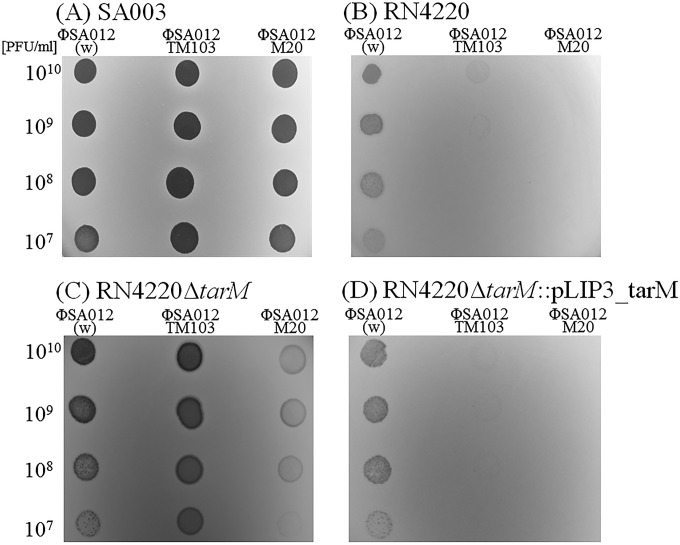
The results of the spot test revealed that the infectivities of the recombinant phage ΦSA012TM103 and the mutant phage ΦSA012M20 differed from that of the wild-type phage ΦSA012 (Fig. 2). All three phages (ΦSA012, ΦSA012TM103, and ΦSA012M20) could produce plaques in S. aureus SA003. However, ΦSA012TM103 produced very turbid plaques, whereas ΦSA012M20 could not produce plaques in RN4220, indicating that the three mutations in orf103 inhibited infection of RN4220. Because ORF103 is likely to bind the WTA polymer due to the existence of a carbohydrate-binding domain in ORF103, we compared the genes related to WTA synthesis between SA003 and RN4220. We found that SA003 lacks the tarM gene, whose product is responsible for glycosylation of α-GlcNAc of WTAs, whereas RN4220 has this gene (46). To investigate the importance of the encoded protein, we knocked out tarM in RN4220. Deletion of tarM made RN4220 susceptible to ΦSA012TM103 and ΦSA012M20, harboring mutated ORF103. Thus, TarM-mediated α-GlcNAc modification of WTAs inhibited infection by ΦSA012TM103 and ΦSA012M20 but not that by ΦSA012. Complementation of tarM in the deletion mutant restored the infectivity of phages, indicating that the role of ORF103 is related to α-GlcNAc on WTAs.
FIG 2.
Spot tests of phages on S. aureus strains. Two microliters of concentrated phage lysate (107 to 1010 PFU/ml) was dropped onto an LB plate overlaid with S. aureus cells. (A) SA003 (ΔtarM); (B) RN4220; (C) RN4220 ΔtarM; (D) RN4220 ΔtarM::pLIP3_tarM. w, wild type.
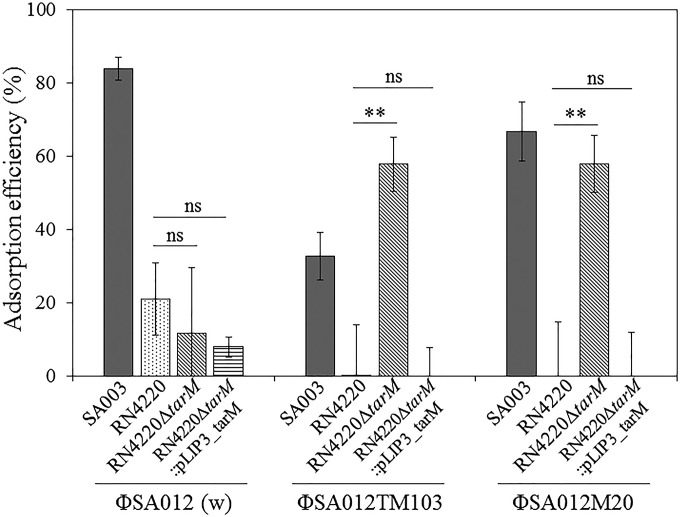
The behaviors of S. aureus strains in adsorption assays corresponded with the results of spot tests (Fig. 2 and 3). All phages (ΦSA012, ΦSA012TM103, and ΦSA012M20) had the ability to adsorb to SA003, although the adsorption efficiency of ΦSA012TM103 was lower than those of ΦSA012 and ΦSA012M20. For RN4220, on the other hand, ΦSA012TM103 and ΦSA012M20 had lower adsorption efficiencies than that of ΦSA012. Thus, mutations in orf103 changed the adsorption efficiency, indicating that ORF103 is involved in adsorption. Deletion of tarM in RN4220 enhanced the adsorption efficiencies of the ORF103 mutant phages ΦSA012TM103 and ΦSA012M20 but did not affect adsorption of ΦSA012. Complementation of tarM confirmed that α-GlcNAc of WTAs inhibited adsorption of ΦSA012TM103 and ΦSA012M20 but not that of ΦSA012.
FIG 3.
Adsorption efficiencies with 108 CFU/ml of S. aureus strains in 60 min. Error bars indicate standard deviations (SD). Three biological replicates were conducted. **, P < 0.01; ns, not significant.
From these results, we concluded that ORF103 is the RBP that binds to α-GlcNAc on WTAs and that mutations in ORF103 altered the protein's function, resulting in deficient adsorption of ORF103 mutant phages to S. aureus strains whose WTAs contain α-GlcNAc due to the activity of TarM. It is plausible that the absence of α-GlcNAc in WTAs did not contribute to phage resistance due to the presence of ORF105, previously shown to be an RBP.
Effects of anti-ORF103 and anti-ORF105 antibodies on phage infection.
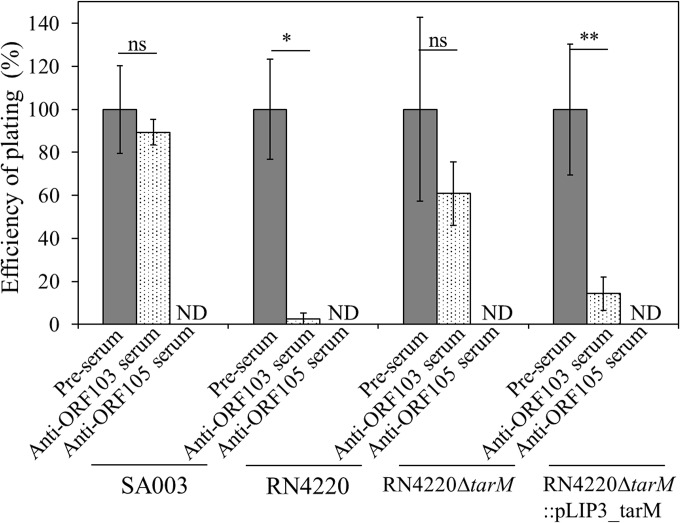
To test our hypothesis, we evaluated the effects of polyclonal antibodies raised against ORF103 and ORF105 on phage infection (Fig. 4). Infection of RN4220 by ΦSA012 was drastically inhibited by anti-ORF103 serum, but infection of SA003 was not. Inhibition by anti-ORF103 serum was not observed for infection of RN4220 ΔtarM by ΦSA012, whereas anti-ORF103 serum inhibited infection of a tarM-complemented strain. This indicated that the anti-ORF103 serum inhibited the interaction between ORF103 and α-GlcNAc, supporting the idea that ORF103 binds to α-GlcNAc on WTAs and plays an important role in infection only in S. aureus strains whose WTAs contain α-GlcNAc.
FIG 4.
EOP of ΦSA012 with anti-ORF103 and anti-ORF105 antibodies. Preimmune serum (pre-serum) was used in controls. The number of plaques observed in the presence of preimmune serum was set as 100%. Plaques were not detected (ND) in the presence of anti-ORF105 serum. Three biological replicates were conducted. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ns, not significant.
In contrast, inhibition by anti-ORF105 serum was observed for all S. aureus strains (SA003, RN4220, RN4220 ΔtarM, and RN4220 ΔtarM::pLIP3_tarM) irrespective of the presence of α-GlcNAc, indicating that ORF105 is the primary RBP for infection by ΦSA012.
Location of ORF103 in ΦSA012.
Immunoelectron microscopy using anti-ORF103 antibody and a gold-conjugated anti-rabbit antibody revealed that ORF103 in ΦSA012 is localized on the tail fiber at the bottom of the baseplate (Fig. 5). This observation supported the idea that ORF103 interacts with components on the cell surface during infection.
FIG 5.
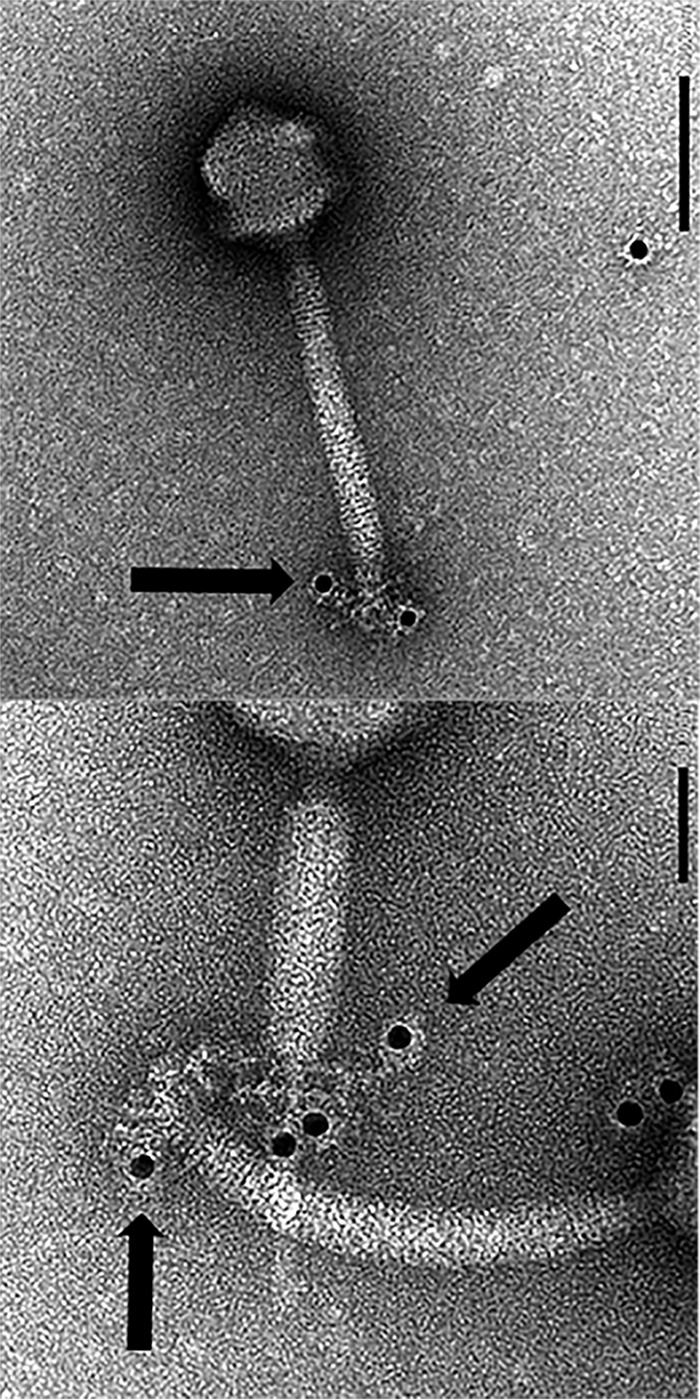
Localization of ORF103 in ΦSA012 stained with a gold-conjugated secondary antibody. Bars, 100 nm (top) and 50 nm (bottom). The size of the gold particles is 12 nm. Black spots in the images represent gold particles conjugated to the secondary antibody.
Role of the three mutations in orf103 in mutant phages during coevolution.
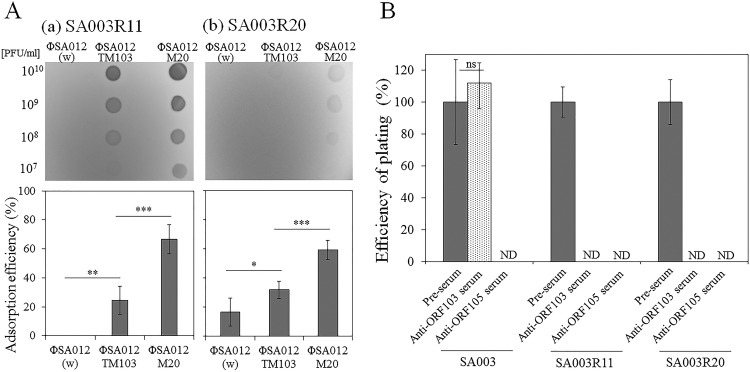
Apart from the function of ORF103, i.e., binding to α-GlcNAc, mutations in ORF103 seemed to be important during coevolution between ΦSA012 and SA003. The results of spot tests and adsorption assays confirmed that ΦSA012TM103 and ΦSA012M20 could infect the ΦSA012-resistant derivatives SA003R11 and SA003R20 due to the increase in adsorption efficiency caused by the three mutations in orf103 (Fig. 6A). The inconsistency in adsorption efficiency between ΦSA012TM103 and ΦSA012M20 suggested that mutations in other genes besides orf103 also affected adsorption.
FIG 6.
Effects of mutations in ORF103. (A) Spot tests and adsorption assays with ΦSA012-resistant strains. Upper and lower panels show the results of adsorption assays (109 CFU/ml of cells in 270 min) and spot tests, respectively, with SA003R11 (a) and SA003R20 (b). Three or five biological replicates were conducted. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) EOP of ΦSA012M20 with anti-ORF013 and anti-ORF105. Preimmune serum was used in controls. The number of plaques formed in the presence of preimmune serum was defined as 100%. ND, not detected. Three biological replicates were conducted. Error bars indicate SD. ns, not significant.
The inhibitory effect of anti-ORF103 serum on infection by ΦSA012M20 also suggested that mutated ORF103 is essential for infection of SA003R11 and SA003R20. In contrast, anti-ORF103 serum did not inhibit infection of SA003 (Fig. 6B). Because WTAs in SA003R11 and SA003R20 do not contain α-GlcNAc due to the absence of tarM in these strains, this finding implied that mutations in ORF103 changed the original function of ORF103 to enhance adsorption on ΦSA012-resistant derivatives. Thus, mutations in ORF103 were necessary to adapt to phage resistance arising during coevolution.
DISCUSSION
In silico analysis of ΦSA012 and its mutant phages suggests that ORF103 is responsible for host recognition.
Deletions of bp 1 to 1,045 (containing orf1 to orf3) and bp 141,673 to 142,094 (containing orf207) were detected in ΦSA012M11 to ΦSA012M38 (Table 3). In SPO1, these terminally redundant regions contain the host takeover module (47). For staphylococcal Twort-like phages, it has also been suggested that some or all genes located in these regions play a role in host takeover, by analogy to the corresponding regions of SPO1 (7). Insertion of one nucleotide in orf58 caused a frameshift and generated a premature stop codon, leading to partial deletion of the gene product (amino acids [aa] 85 to 108). orf58 encodes a protein homologous to the membrane protein MbpC of staphylococcal Twort-like phage A5W; it is predicted to play a role in attachment of the complex of replicating phage DNA to a cell membrane, analogously to membrane protein p16.7 of Bacillus phage φ29 (7, 48). Although substitutions of amino acids in ORF33 were observed only in ΦSA012M2, they did not accumulate among later mutant phages. Other ORFs in which mutations were observed (orf79, orf99, orf103, and orf133) encode putative tail proteins. orf79 is predicted to encode the tail sheath protein and has homologs in a number of phages. The functions of orf99 and orf103, predicted to encode tail morphogenetic proteins, remain unknown. Due to the presence of an immunoglobulin (Ig)-like domain conserved among hundreds of phages, orf133 is predicted to encode a tail protein that plays an accessory role by interacting weakly with carbohydrates on the bacterial surface (7, 49, 50).
Given that the largest number of mutations accumulated in orf103, we hypothesized that orf103 plays a crucial role in determining infectivity. Due to the selection pressures imposed by phage-bacterium coevolution, phage RBP genes are among the most diverse genes (51). It is worth mentioning that a carbohydrate-binding domain (1,4-β-glucanase CenC from Cellulomonas fimi) was identified in the central position of ORF103 (aa 221 to 337) by InterPro 56.0, and one of the three mutations was located within this domain (52, 53). This observation implied that ORF103 encodes a possible RBP that binds to sugar components in WTAs on the cell surface. Since ORF105, a homolog of gp40 in ISP that acts as an RBP (25), is located in the unique region from ORF103 to ORF105 revealed by comparisons of structural proteins among ΦSA012 and its related phages, this unique region from ORF103 to ORF105 determines the host range for staphylococcal Twort-like phages (Fig. 1). Thus, we focused mainly on orf103 as a host determinant gene in this study.
Presence of two RBPs in staphylococcal Twort-like phages.
Our findings showed that at least two adsorption apparatuses (ORF103 and ORF105) are present in ΦSA012. Due to the carbohydrate-binding domain in ORF103, ORF103 recognizes sites in WTAs different from those recognized by ORF105. The N-terminal 37 residues of ORF103 and ORF105 are 41% similar. In most cases, the N termini of RBPs are conserved due to the connection to the virus particle, whereas the C termini, responsible for recognition of the host receptor, are diverse. The conserved N termini of ORF103 and ORF105 may suggest that ORF103 and ORF105 are conjugated to the same virion component.
Several phages are known to possess multiple adsorption apparatuses (51). The E. coli lytic myovirus phi92 has at least five different tail spikes and tail fiber proteins identified by cryoelectron microscopy, allowing it to infect a wide range of E. coli and Salmonella strains (54). Two different RBPs, LtfA and LtfB of the T5-like siphoviruses DT57C and DT571/2, recognize different O antigens, i.e., the O22 or O87 type and the O81 type, respectively (55). Two different carbohydrate-binding modules have been identified in Lactococcus lactis phage Tuc2009; the first is in a classical bona fide RBP (BppL), and the other is in an accessory protein, BppA (56). BppA enhances adsorption to cells, although its true contribution is not fully understood (57).
The distribution of tarM in S. aureus strains has been addressed in previous studies. Many human-associated S. aureus lineages lost tarM over the course of evolution (30). However, the presence of multiple RBPs may explain the wide host range of staphylococcal Twort-like phages.
Role of ORF103 in infection.
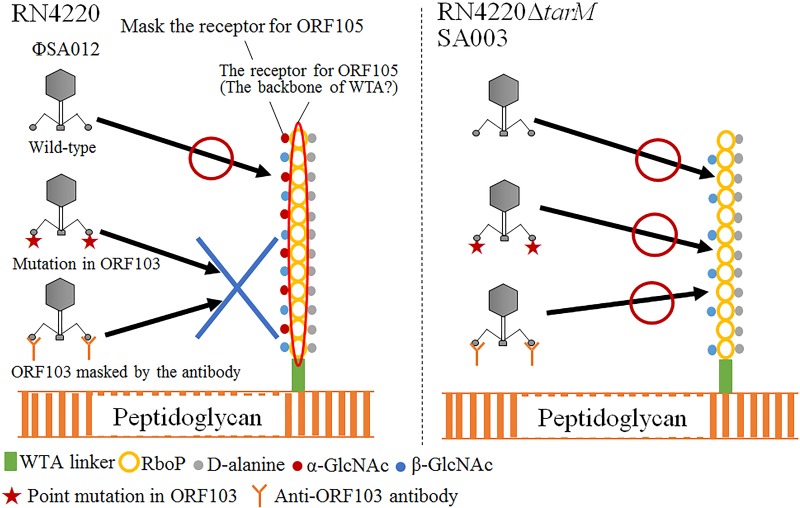
From the results of spot tests, EOP tests with antibodies, and adsorption assays, it was clear that ORF103 is involved in adsorption and binds to α-GlcNAc in WTAs (Fig. 2 to 4). SA003 and RN4220 ΔtarM were still susceptible to ΦSA012 upon inhibition of ORF103 by anti-ORF103 antibody (Fig. 4); thus, in the absence of α-GlcNAc, the function of ORF103 is not essential for infection of host cells by ΦSA012. Deletion of tarM or tarS, encoding an enzyme that glycosylates α-GlcNAc or β-GlcNAc on WTAs, respectively, did not affect infection of S. aureus strains by phage K, whereas the lack of WTAs by deletion of tagO, a gene related to the initial step of WTA synthesis, rendered S. aureus strains resistant to phages (30). Therefore, staphylococcal myovirus is thought to recognize the backbone of WTAs (27). In an infection by S. aureus podovirus, the presence of α-GlcNAc inhibited the adsorption of phages to β-GlcNAc, the podovirus phage receptor, and deletion of TarM enhanced phage adsorption (30). Likewise, it is possible that α-GlcNAc masks the backbone of WTAs, which is the binding site for the primary RBP ORF105, and that ORF103 helps the phage to bind the WTA backbone by binding α-GlcNAc (Fig. 7). Hence, phages that lacked wild-type ORF103 function due to point mutations or antibodies were not able to infect S. aureus strains in the presence of α-GlcNAc, whereas a loss of ORF103 function did not affect phage infection in the absence of α-GlcNAc. The influence of β-GlcNAc on adsorption may be minor, as no inhibition of infection by the anti-ORF103 antibody was observed in SA003 irrespective of the presence of tarS, whose product is responsible for glycosylation of β-GlcNAc.
FIG 7.
Scheme of putative adsorption mechanism of ΦSA012. In most S. aureus strains, the WTA polymer consists of 11 to 40 ribitol phosphate (RboP) repeats substituted with d-alanine and α- and β-GlcNAc, covalently attached to a peptidoglycan with a WTA linkage unit. ΦSA012 recognizes the WTA backbone. α-GlcNAc masks the WTA backbone, which is the binding site of the primary RBP ORF105, and ORF103 helps the phage to bind the WTA backbone by binding α-GlcNAc. Hence, a phage in which ORF103 is disabled by point mutations or antibodies cannot infect S. aureus strains in the presence of α-GlcNAc, whereas the loss of ORF103 function does not affect phage infection in the absence of α-GlcNAc.
Changes in ORF103 function by point mutations arising during coevolution.
Three mutations in ORF103 contributed to adsorption to the ΦSA012-resistant derivatives SA003R11 and SA003R20, whereas adsorption to RN4220 was inhibited by these mutations. The effect of these three mutations on adsorption enhanced the infectivity of ΦSA012TM103, as shown in the spot test and the adsorption assay. It is intriguing that mutant phages derived from ΦSA012 acquired mutations in orf103 to adapt to ΦSA012-resistant derivatives of SA003, despite the absence of α-GlcNAc. Mutations in RBPs enable phages to recognize a new phage receptor and to circumvent inhibition of adsorption during coevolution (51). A recent study on phage λ and its host, E. coli B, demonstrated that this phage can evolve to target a new receptor, OmpF, when expression of the cognate receptor, LamB, is reduced by mutations (58). In order to survive during coevolution, it is reasonable for the phage to expand its host range by acquiring point mutations in ORF103 without altering ORF105, allowing mutant phages to infect both wild-type and resistant derivatives. The results of adsorption assays confirmed the strong inhibition of adsorption of ΦSA012 to SA003R11 and SA003R20, suggesting that the structure of the cell surface was altered during coevolution (Fig. 6). Thus, it is plausible that the three mutations altered the function of ORF103 to allow the phage to survive during coevolution. From the complete genomes of SA003R11 and SA003R20, we also identified point mutations in genes responsible for WTA and peptidoglycan synthesis (A. H. Azam, I. Takeuchi, K. Miyanaga, and Y. Tanji, unpublished data). This implies that alterations in the structure of the cell surface induced mutations in orf103, resulting in acquisition of a new function by ORF103: specifically, in SA003R11 and SA003R20, the protein acquired the ability to bind mutated WTA structures at the expense of the ability to bind α-GlcNAc. However, further study of the effects of point mutations in ORF103 is still necessary.
Supplementary Material
ACKNOWLEDGMENT
This research was financially supported by grant 24246133 from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01385-16.
REFERENCES
- 1.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 3.Ryan EM, Gorman SP, Donnelly RF, Gilmore BF. 2011. Recent advances in bacteriophage therapy: how delivery routes, formulation, concentration and timing influence the success of phage therapy. J Pharm Pharmacol 63:1253–1264. doi: 10.1111/j.2042-7158.2011.01324.x. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 187:613–624. doi: 10.1086/374001. [DOI] [PubMed] [Google Scholar]
- 5.Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. 2009. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 75:4483–4490. doi: 10.1128/AEM.02641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobocka M, Hejnowicz MS, Dabrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dabrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Glowacka A. 2012. Genomics of staphylococcal Twort-like phages—potential therapeutics of the post-antibiotic era. Adv Virus Res 83:143–216. doi: 10.1016/B978-0-12-394438-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 8.Loessner MJ, Rees CE, Stewart GS, Scherer S. 1996. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol 62:1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins ATA. 2014. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol 80:6694–6703. doi: 10.1128/AEM.01789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. doi: 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 11.Henning U, Hashemolhosseini S. 1994. Receptor recognition by T-even-type coliphages, p 291–298. In Karam JD. (ed), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC. [Google Scholar]
- 12.Riede I. 1987. Receptor specificity of the short tail fibres (gp12) of T-even type Escherichia coli phages. Mol Gen Genet 206:110–115. doi: 10.1007/BF00326544. [DOI] [PubMed] [Google Scholar]
- 13.Duplessis M, Levesque CM, Moineau S. 2006. Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol 72:3036–3041. doi: 10.1128/AEM.72.4.3036-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol 41:325–336. doi: 10.1046/j.1365-2958.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 15.Bielmann R, Habann M, Eugster MR, Lurz R, Calendar R, Klumpp J, Loessner MJ. 2015. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 477:110–118. doi: 10.1016/j.virol.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Vegge CS, Vogensen FK, McGrath S, Neve H, van Sinderen D, Brondsted L. 2006. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J Bacteriol 188:55–63. doi: 10.1128/JB.188.1.55-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuer-Lauridsen B, Janzen T, Schnabl J, Johansen E. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10–17. doi: 10.1016/S0042-6822(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 18.Guo S, Shu D, Simon MN, Guo P. 2003. Gene cloning, purification, and stoichiometry quantification of phi29 anti-receptor gp12 with potential use as special ligand for gene delivery. Gene 315:145–152. doi: 10.1016/S0378-1119(03)00729-7. [DOI] [PubMed] [Google Scholar]
- 19.Dupont K, Vogensen FK, Neve H, Bresciani J, Josephsen J. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl Environ Microbiol 70:5818–5824. doi: 10.1128/AEM.70.10.5818-5824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sciara G, Blangy S, Siponen M, McGrath S, van Sinderen D, Tegoni M, Cambillau C, Campanacci V. 2008. A topological model of the baseplate of lactococcal phage Tuc2009. J Biol Chem 283:2716–2723. doi: 10.1074/jbc.M707533200. [DOI] [PubMed] [Google Scholar]
- 21.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichiere J, van Heel M, Campanacci V, Moineau S, Cambillau C. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci U S A 107:6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Haard HJW, Bezemer S, Ledeboer AM, Muller WH, Boender PJ, Moineau S, Coppelmans M-C, Verkleij AJ, Frenken LGJ, Verrips CT. 2005. Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J Bacteriol 187:4531–4541. doi: 10.1128/JB.187.13.4531-4541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchiyama J, Takemura-Uchiyama I, Kato S, Sato M, Ujihara T, Matsui H, Hanaki H, Daibata M, Matsuzaki S. 2014. In silico analysis of AHJD-like viruses, Staphylococcus aureus phages S24-1 and S13′, and study of phage S24-1 adsorption. Microbiologyopen 3:257–270. doi: 10.1002/mbo3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko J, Narita-Yamada S, Wakabayashi Y, Kamio Y. 2009. Identification of ORF636 in phage φSLT carrying Panton-Valentine leukocidin genes, acting as an adhesion protein for a poly(glycerophosphate) chain of lipoteichoic acid on the cell surface of Staphylococcus aureus. J Bacteriol 191:4674–4680. doi: 10.1128/JB.01793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habann M, Leiman PG, Vandersteegen K, Van den Bossche A, Lavigne R, Shneider MM, Bielmann R, Eugster MR, Loessner MJ, Klumpp J. 2014. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol Microbiol 92:84–99. doi: 10.1111/mmi.12539. [DOI] [PubMed] [Google Scholar]
- 26.Brown S, Santa Maria JP, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 193:4006–4009. doi: 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutter E, Sulakvelidze A. 2004. Bacteriophages: biology and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 29.Xiang Y, Leiman PG, Li L, Grimes S, Anderson DL, Rossmann MG. 2009. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell 34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Gerlach D, Du X, Larsen J, Stegger M, Kühner P, Peschel A, Xia G, Winstel V. 2015. An accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci Rep 5:17219. doi: 10.1038/srep17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:705–712. [DOI] [PubMed] [Google Scholar]
- 32.Carlson K. 2005. Working with bacteriophages: common techniques and methodological approaches, p 437–494. In Kutter E, Sulakvelidze A (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 33.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. [DOI] [PubMed] [Google Scholar]
- 35.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 36.Jeong D-W, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. 2011. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol 193:4672–4684. doi: 10.1128/JB.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baptista C, Santos MA, Sao-Jose C. 2008. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J Bacteriol 190:4989–4996. doi: 10.1128/JB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Reference deleted.
- 44.Reference deleted.
- 45.Eyer L, Pantucek R, Zdrahal Z, Konecna H, Kasparek P, Ruzickova V, Hernychova L, Preisler J, Doskar J. 2007. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 7:64–72. [DOI] [PubMed] [Google Scholar]
- 46.Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A. 2010. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem 285:13405–13415. doi: 10.1074/jbc.M109.096172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart CR, Gaslightwala I, Hinata K, Krolikowski KA, Needleman DS, Peng AS, Peterman MA, Tobias A, Wei P. 1998. Genes and regulatory sites of the “host-takeover module” in the terminal redundancy of Bacillus subtilis bacteriophage SPO1. Virology 246:329–340. doi: 10.1006/viro.1998.9197. [DOI] [PubMed] [Google Scholar]
- 48.Alcorlo M, Gonzalez-Huici V, Hermoso JM, Meijer WJ, Salas M. 2007. The phage phi29 membrane protein p16.7, involved in DNA replication, is required for efficient ejection of the viral genome. J Bacteriol 189:5542–5549. doi: 10.1128/JB.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser JS, Yu Z, Maxwell KL, Davidson AR. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol 359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 50.Fraser JS, Maxwell KL, Davidson AR. 2007. Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol 10:382–387. doi: 10.1016/j.mib.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 51.Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 52.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brun E, Johnson PE, Creagh AL, Tomme P, Webster P, Haynes CA, McIntosh LP. 2000. Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C. Biochemistry 39:2445–2458. doi: 10.1021/bi992079u. [DOI] [PubMed] [Google Scholar]
- 54.Schwarzer D, Buettner FFR, Browning C, Nazarov S, Rabsch W, Bethe A, Oberbeck A, Bowman VD, Stummeyer K, Muhlenhoff M, Leiman PG, Gerardy-Schahn R. 2012. A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis. J Virol 86:10384–10398. doi: 10.1128/JVI.00801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golomidova A, Kulikov E, Prokhorov N, Guerrero-Ferreira R, Knirel Y, Kostryukova E, Tarasyan K, Letarov A. 2016. Branched lateral tail fiber organization in T5-like bacteriophages DT57C and DT571/2 is revealed by genetic and functional analysis. Viruses 8:26. doi: 10.3390/v8010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. 2016. The atomic structure of the phage Tuc2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. mBio 7:e01781-15. doi: 10.1128/mBio.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J Virol 87:8429–8440. doi: 10.1128/JVI.00907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. 2012. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osada K. 2014. Master's thesis. Tokyo Institute of Technology, Yokohama, Japan. [Google Scholar]
- 61.Fox E, O'Mahony T, Clancy M, Dempsey R, O'Brien M, Jordan K. 2009. Listeria monocytogenes in the Irish dairy farm environment. J Food Prot 72:1450–1456. [DOI] [PubMed] [Google Scholar]
- 62.O'Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol 186:2862–2871. doi: 10.1128/JB.186.9.2862-2871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui Z, Song Z, Wang Y, Zeng L, Shen W, Wang Z, Li Q, He P, Qin J, Guo X. 2012. Complete genome sequence of wide-host-range Staphylococcus aureus phage JD007. J Virol 86:13880–13881. doi: 10.1128/JVI.02728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu J, Liu X, Lu R, Li Y, Song J, Lei L, Sun C, Feng X, Du C, Yu H, Yang Y, Han W. 2012. Complete genome sequence of Staphylococcus aureus bacteriophage GH15. J Virol 86:8914–8915. doi: 10.1128/JVI.01313-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vandersteegen K, Kropinski AM, Nash JH, Noben JP, Hermans K, Lavigne R. 2013. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J Virol 87:3237–3247. doi: 10.1128/JVI.02763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.