ABSTRACT
Asymptomatic infant carriers of toxigenic Clostridium difficile are suggested to play a role in the transmission of C. difficile infection (CDI) in adults. However, the mode of C. difficile carriage in infants remains to be fully elucidated. We investigated longitudinal changes in carriage rates, counts, and strain types of toxigenic C. difficile in infants. Stools collected from 111 healthy infants in Belgium periodically from birth until the age of 6 months were examined by quantitative PCR targeting 16S rRNA and toxin genes. Toxigenic C. difficile was detected in 18 of 111 infants (16%) in the period up to the age of 6 months. The carriage rate of toxigenic C. difficile remained below 5% until the age of 3 months. The carriage rate increased to 13% 1 week after weaning (average age, 143 days) and reached 16% at the age of 6 months. Counts of toxigenic C. difficile bacteria ranged from 104 to 108 cells/g of stool. Notably, two infants retained >108 cells/g of stool for at least several weeks. Average counts in the 18 infants hovered around 107 cells/g of stool from the age of 3 days until the age of 6 months, showing no age-related trend. Genotyping of toxigenic C. difficile isolates from the 18 infants revealed that 11 infants each retained a particular monophyletic strain for at least a month. The genotype most frequently identified was the same as that frequently identified in symptomatic adult CDI patients. Thus, toxigenic C. difficile strains—potential causes of CDI in adults—colonized the infants' intestines.
IMPORTANCE Our study provides longitudinal data on counts and strain types of toxigenic C. difficile in infants. We found that considerable numbers of toxigenic C. difficile bacteria colonized the infants' intestines. The results of strain typing suggest that toxigenic C. difficile carried by healthy infants could be potentially pathogenic to adults. These results and findings are informative not only for ecological studies but also for efforts to prevent or control the spread of CDI in adults.
INTRODUCTION
Toxigenic Clostridium difficile is the main cause of health care-associated gastrointestinal infections. In general, C. difficile is not a normal inhabitant of the adult gastrointestinal tract, because its colonization is considered to be prevented by the presence of commensal microbiota (1–3). Colonization of the intestines by toxigenic C. difficile is the first crucial step in the development of C. difficile infection (CDI). Accumulating evidence suggests that disruption of the microbiota and frequent exposure to toxigenic C. difficile could increase the risk of CDI (4–6). Whereas asymptomatic carriage of toxigenic C. difficile is rare in adults, occurring in <5% of adults (7, 8), it is very common in infants (9, 10), probably due to the immaturity of the microbiota composition. Regardless of the high prevalence, a great majority of infants rarely show CDI-related abdominal symptoms. The presumed reason for this is that infants are not susceptible to C. difficile toxins owing to the poor development of the toxin receptors or cellular signaling pathways in the intestines (11). However, asymptomatic infantile carriers of toxigenic C. difficile are suggested to be potential reservoirs of pathogenic strains and to play a role in the transmission of CDI in adults (12–14). Investigations of the carriage rates of toxigenic C. difficile in infants can be informative for preventive control of CDI in adults.
According to a review by Jangi and Lamont (15), the carriage rates of C. difficile are 37% in healthy infants less than 1 month of age and 30% in those between 1 and 6 months of age. This rate drops considerably to 14% at between 6 and 12 months of age and decreases to 10% after 1 year of age. Because the carriage rate is likely to peak by 6 months of age, infants of these ages are thought to have a greater chance of spreading C. difficile strains. Therefore, it is of great importance to better understand the colonization patterns of C. difficile during the first 6 months of life.
Our aim was to investigate time-dependent changes in carriage rates, counts, and strain types of toxigenic C. difficile in infants. For this purpose, we examined stool specimens collected from 111 infants at 0 days (meconium), 3 days, 7 days, 30 days, 90 days, and 180 days of age and after weaning by using TaqMan-based quantitative PCR (qPCR) that was previously developed for the selective quantification of both total C. difficile strains (i.e., all types) and toxigenic strains (16). We isolated toxigenic C. difficile from stool specimens and genotyped the isolates by using capillary gel electrophoresis-based PCR ribotyping (CGE-PCR ribotyping).
MATERIALS AND METHODS
Stool specimens.
Stool specimens collected from subjects in an observational study conducted in Belgium (ISRCTN66704989) (17) were used in this study. The study subjects were 111 infants (including 2 pairs of twins), 82 of whom were delivered vaginally and 29 of whom were delivered by cesarean section. Stool samples were taken at 0 days (meconium), 3 days, 7 days, 30 days, 90 days, and 180 days of age and after weaning (1 week after the introduction of solids, at 143 days of age on average) (Table 1).
TABLE 1.
Stool specimens used in this study
| Stool specimen | No. of specimens | Mean day of stool sampling ± SDa |
|---|---|---|
| Meconium | 99 | 0.7 ± 1.0 |
| Day 3 | 83 | 2.8 ± 1.0 |
| Day 7 | 100 | 8.4 ± 2.1 |
| Day 30 | 108 | 31 ± 3.4 |
| Day 90 | 107 | 91 ± 5.9 |
| Weaning | 75 | 143 ± 20 |
| Day 180 | 105 | 182 ± 7.9 |
The day of delivery of each infant was defined as day 0.
This study was approved by the ethics committee of the hospital network of Antwerp (Ziekenhuisnetwerk Antwerpen) and conducted in compliance with ethical principles from the Declaration of Helsinki, guidelines of good clinical practice, and applicable regulations. Written informed consent was obtained from the infants' mothers.
Standard strain.
Clostridium difficile DSM 1296T was selected as the standard strain for the generation of analytical curves for qPCR and was cultured anaerobically at 37°C in modified Gifu anaerobic medium broth (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan) supplemented with 1% glucose. The number of bacterial cells in fresh cultures was determined by using the 4′,6-diamidino-2-phenylindole (DAPI) staining method. The suspension was stored at −80°C until use for DNA extraction. DNA was extracted from bacterial suspensions as described previously (18).
Quantification of C. difficile by qPCR.
Clostridium difficile in stool specimens was quantified by using TaqMan-based qPCR with three primer-probe sets, namely, 16SrRNA-F/R/P, targeting toxin A-positive and toxin B-positive (A+ B+), toxin A-negative and toxin B-positive (A− B+), and toxin A-negative and toxin B-negative (A− B−) types; tcdB-F/R/P, targeting A+ B+ and A− B+ types; and tcdA-F/R/P, targeting the A+ B+ type (Table 2), as previously described (16), with some modifications. Stool DNA extracted previously from stool samples of infants (17) was used for the analysis. In brief, for the preparation of a standard DNA solution, bacterial DNA was extracted from 2 × 108 cells of a pure culture of the DSM 1296T strain (A+ B+ type) and dissolved in 1 ml of Tris-EDTA (TE) buffer. Five microliters of 10-fold serial dilutions of standard DNA was applied for PCR to obtain a standard analytical curve ranging from 100 to 105 cells/5 μl of template. Five microliters of the extracted stool DNA solution (10 mg stool DNA/ml) and 2- and 4-fold dilutions of this solution were applied for PCR as a template containing the corresponding amounts of DNA from 50, 25, and 12.5 μg of stool. The ABI Prism 7900HT sequence detection system (Thermo Fisher Scientific Inc., Waltham, MA) with 384-well optical plates was used as the qPCR platform. qPCR counts (cells per gram of stool) were calculated by applying the obtained threshold cycle (CT) values for the stool specimens to the standard analytical curve. The lower detection limit was 2 × 104 cells (4.3 log10 cells)/g of stool.
TABLE 2.
Oligonucleotides used in this study
| Target | Gene | Oligonucleotide | Sequence (5′–3′)a |
|---|---|---|---|
| Total Clostridium difficile (A+ B+, A− B+, and A− B+ types) | 16S rRNA gene | CD16SrRNA-F | GCAAGTTGAGCGATTTACTTCGGT |
| CD16SrRNA-P | FAM-TGCCTCTCAAATATATTATCCCGTATTAG-TAMRA | ||
| CD16SrRNA-R | GTACTGGCTCACCTTTGATATTYAAGAG | ||
| TcdB-producing Clostridium difficile (A+ B+ and A− B+ types) | tcdB | tcdB-F | TACAAACAGGTGTATTTAGTACAGAAGATGGA |
| tcdB-P | FAM-TTTKCCAGTAAAATCAATTGCTTC-TAMRA | ||
| tcdB-R | CACCTATTTGATTTAGMCCTTTAAAAGC | ||
| TcdA-producing Clostridium difficile (A+ B+ type) | tcdA | tcdA-F | CAGTCGGATTGCAAGTAATTGACAAT |
| tcdA-P | FAM-TTGAGATGATAGCAGTGTCAGGATTG-TAMRA | ||
| tcdA-F | AGTAGTATCTACTACCATTAACAGTCTGC |
All oligonucleotides were described previously (16). FAM, 6-carboxyfluorescein; TAMRA, 6-carboxymethyltetramethylrhodamine.
Identification of toxin types of predominant C. difficile strains in individual stool specimens.
The toxin type of the predominant C. difficile strains in each stool specimen was determined by comparing the three qPCR counts targeting the 16S rRNA gene for total C. difficile strains (A+ B+, A− B+, and A− B− types), tcdB for TcdB-producing strains (A+ B+ and A− B+ types), and tcdA for TcdA-producing strains (A+ B+ type), as previously described (16).
Isolation of toxigenic C. difficile.
Clostridium difficile was isolated by stool culture with cefoxitin-cycloserine-egg yolk (CCEY) agar as previously described (16). In brief, plates of CCEY agar (BioConnections Ltd., Knypersley, United Kingdom) supplemented with 40 ml of egg yolk emulsion (BioConnections), two vials of cefoxitin-cycloserine (BioConnections), and 10 ml of lysed horse blood per liter of medium were prepared in-house and stored at 4°C for a maximum of 1 week before use. Frozen stool specimens were thawed and mixed with an equal volume of absolute ethanol. After incubation of the mixture at room temperature for 30 to 60 min, 100 μl of alcohol-treated stool or its 10-fold dilution was inoculated onto CCEY plates and cultured under anaerobic conditions at 37°C for 48 to 72 h in an anaerobic glove box (Coy Laboratory Products Inc., Grass Lake, MI). At least three suspected colonies were subjected to rapid identification of C. difficile by real-time PCR using the CD16SrRNA-F/R/P primer-probe set.
PCR identification of toxin types of isolates.
The toxin type of each C. difficile isolate was identified by PCR for the tcdA and tcdB genes, as described previously by Lemee et al. (19). tcdA-specific primers (forward primer 5′-AGATTCCTATATTTACATGACAATAT-3′ and reverse primer 5′-GTATCAGGCATAAAGTAATATACTTT-3′) and tcdB-specific primers (forward primer 5′-GGAAAAGAGAATGGTTTTATTAA-3′ and reverse primer 5′-ATCTTTAGTTATAACTTTGACATCTTT-3′) were used for amplification. If the sizes of the respective amplicons of tcdA and tcdB were 370 bp and 160 bp, the isolate was determined to be of the A+ B+ type. If the respective amplicons were 110 bp and 160 bp, the isolate was determined to be of the A− B+ type.
Total bacterial counts in stool.
The numbers of total stool bacteria were determined by the DAPI staining method as previously described (20).
TcdB production by C. difficile isolates.
The TcdB production capacities of C. difficile isolates were determined by a cell cytotoxicity neutralization assay (CCNA) with a C. difficile Toxin/Antitoxin kit (Techlab, Blacksburg, VA), as previously described (16). In brief, identified C. difficile colonies were subcultured anaerobically at 37°C in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD) for 5 days. The suspension was then centrifuged at 16,000 × g for 5 min, and the supernatant was filtered with a 0.45-μm membrane filter. The filtrate was added to a Vero-B4 cell culture with or without C. difficile antitoxin. After incubation of the culture at 37°C for 24 or 48 h, the presence or absence of TcdB was determined by judging the cytopathic effects.
Capillary gel electrophoresis-based PCR ribotyping.
Strain types of C. difficile isolates were characterized by CGE-PCR ribotyping as described previously by Indra et al. (21), with some modifications. In brief, DNA was extracted from pure cultures of each isolate. The 16S rRNA-to-23S rRNA intergenic spacer region was amplified in a 25-μl PCR mixture composed of 0.5 U of Ex Taq DNA polymerase (TaKaRa Bio, Shiga, Japan), 1× Ex Taq buffer, 200 μM deoxyribonucleoside triphosphates, 0.5 μM primers, and 1 μl of template DNA. PCR amplification consisted of 1 cycle of 95°C for 30 s; 25 cycles of 95°C for 1 min, 57°C for 1 min, and 72°C for 1 min; and 1 cycle of 72°C for 30 min. One microliter of the amplicon or its 10-fold dilution was mixed with 1 μl of GeneScan 600 LIZ (Thermo Fisher Scientific) and 9 μl of Hi-Di formamide (Thermo Fisher Scientific). After heating at 95°C for 3 min and subsequent incubation on ice for 5 min, the amplicon was analyzed by capillary gel electrophoresis using ABI Prism 3130xl genetic analyzers (Thermo Fisher Scientific). The fragment files (.fsa) obtained were imported into BioNumerics software version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium). Genetic relatedness based on fragment patterns was determined for fragments ranging in length from 200 to 600 nucleotides. A dendrogram was created by using the multiscale setting for comparisons and the Dice coefficient with the unweighted pair group method with arithmetic means (UPGMA) for clustering. The PCR ribotypes (PRs) were determined by querying the fragment files (.fsa) against the Web-based database WEBRIBO (http://webribo.ages.at/). If no PCR ribotype was matched, a new PCR ribotype (NPR plus one digit) specific to this study was allocated to the strain.
Multilocus sequence typing.
Strain types of C. difficile isolates were characterized by multilocus sequence typing (MLST) with primers for seven housekeeping genes, aroE, dutA, gmk, groEL, recA, sodA, and tpi, as described previously by Rousseau et al. (13). In brief, DNA was extracted from pure cultures of each isolate in BHI broth. Each target gene was amplified in a 25-μl PCR mixture composed of 2 U Fast Taq polymerase (Roche, Basel, Switzerland), 1× PCR buffer, 200 μM deoxyribonucleoside triphosphates, 2 mM MgCl2, 0.4 μM each primer, 10 μl of GC-RICH solution, and 1 μl of template DNA. PCR amplification consisted of 1 cycle of 95°C for 5 min; 30 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min; and 1 cycle of 72°C for 10 min. The amplicons were purified with ExoSAP-IT for PCR product cleanup (USB, Cleveland, OH) and sequenced with primers for each gene by using ABI Prism 3130xl genetic analyzers. The sequences obtained for the seven genes were analyzed by using BioNumerics. BioNumerics queried a database (C. difficile MLST database at http://bigsdb.pasteur.fr/) for the sequences of the seven genes from each strain and allocated a specific sequencing type (ST) to each strain automatically. A dendrogram was created by using the multiscale setting for comparisons and the UPGMA for clustering.
Statistical analysis.
Characteristics of C. difficile-positive infants and C. difficile-negative infants were compared by using Fisher's exact test or a χ2 test.
Accession number(s).
Nucleotide sequences determined by MLST were deposited in the GenBank/EMBL/DDBJ database under accession numbers LC151619 to LC151870.
RESULTS
Carriage rates and bacterial counts of C. difficile in stool samples, as determined by qPCR.
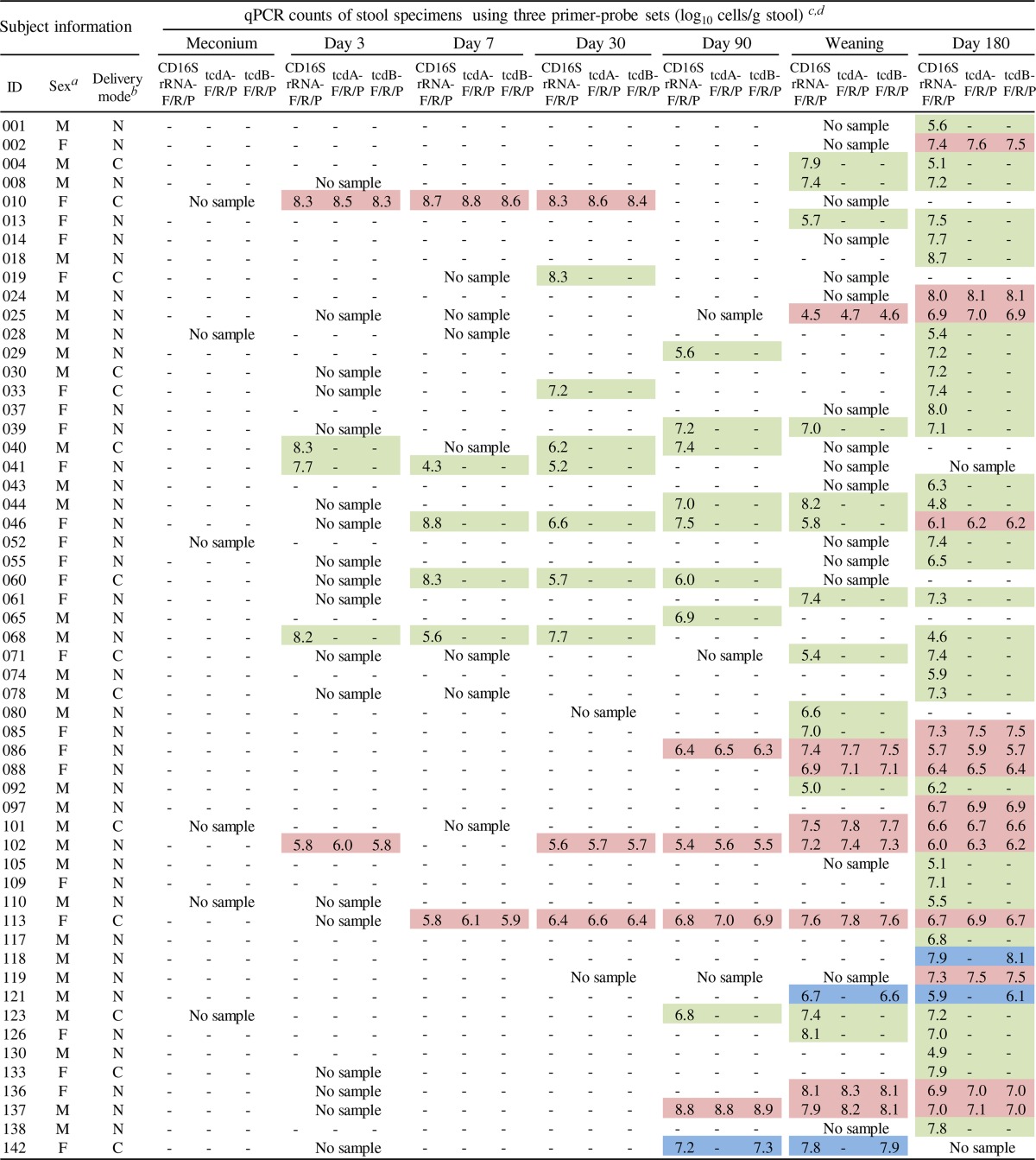
Infant stool samples collected 7 times from birth until 180 days of age were examined by qPCR with the CD16SrRNA-F/R/P, tcdB-F/R/P, and tcdA-F/R/P primer-probe sets to quantify total C. difficile strains (A+ B+, A− B+, and A− B− types), TcdB-producing strains (A+ B+ and A− B+ types), and TcdA-producing strains (A+ B+ type), respectively (see Table S1 in the supplemental material). In the entire period up to the age of 6 months, C. difficile was detected in 55 of 111 infants (50%), and toxigenic C. difficile was found in 18 infants (16%). Details of the counts of C. difficile bacteria in the 55 C. difficile-positive infants are shown in Table 3. The number of toxigenic C. difficile bacteria ranged from 104 to 108 cells/g of stool. Certain infants (e.g., subjects 010 and 137) retained large quantities of toxigenic strains (108 cells/g of stool) for several weeks. The number of total stool bacteria in these infants ranged from ∼109 to 1011 cells/g of stool (see Table S2 in the supplemental material). The proportion of C. difficile strains among the total bacteria reached as high as 10% in subject 010 at 3 days of age.
TABLE 3.
Bacterial counts of Clostridium difficile in 55 C. difficile-positive infants by qPCR
M, male; F, female.
N, natural delivery; C, cesarean section.
−, not detected.
Toxin types of C. difficile predominating in each specimen were identified on the basis of qPCR counts for the three genes according to the criteria described in Materials and Methods. On the basis of toxin type, each specimen is highlighted as follows: red, A+ B+ type; blue, A− B+ type; green, A− B− type.
Clostridium difficile was not detected in meconium samples, but 6% of infants had acquired C. difficile by the age of 3 days. Thereafter, the carriage rate of C. difficile rose to 12% at 90 days of age, then rose to 31% at weaning, and finally rose to 45% at 180 days of age (Fig. 1). The rate of carriage of toxigenic C. difficile (A+ B+ type and A− B+ type) similarly increased from 2% at 3 days of age to 15% at 180 days of age (Fig. 1). Regardless of the age of infants, approximately two-thirds of all C. difficile carriers were carriers of a nontoxigenic strain (A− B− type), and one-third were carriers of a toxigenic strain (mostly of the A+ B+ type).
FIG 1.
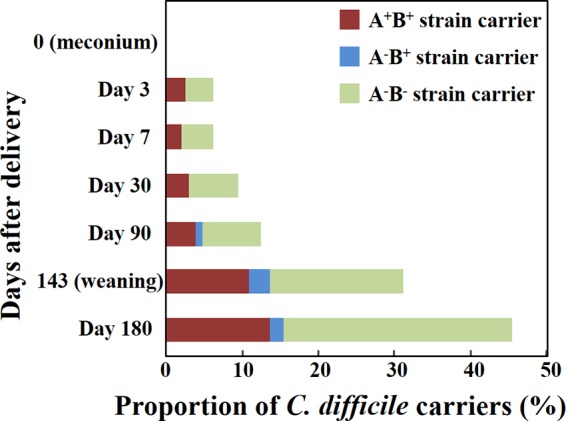
Carriage rates of each toxin type of Clostridium difficile in infants. On the basis of the qPCR counts for three genes (16S rRNA, tcdA, and tcdB), the toxin types (A+ B+, A− B+, or A− B−) of predominant C. difficile strains in individual specimens were identified. The rates of carriage of each toxin type of C. difficile were calculated with respect to each stool specimen group.
Isolation of toxigenic C. difficile from stool samples and characterization of the isolates.
For further characterization of toxigenic C. difficile in infantile intestines, we isolated C. difficile strains from stool specimens that were positive for toxigenic C. difficile upon qPCR analysis. Clostridium difficile was isolated from 36 of the 38 positive stool specimens (Table 4). PCR amplification of tcdA and tcdB revealed that the isolates from 31 of the 36 specimens were of the A+ B+ type, and those from the other 5 specimens were of the A− B+ type, in agreement with the toxin types of the predominant C. difficile strains identified by qPCR with stool DNA (Table 3). There were no specimens from which strains of two or more different toxin types were isolated. All the isolates from the 36 specimens were confirmed by CCNAs to be capable of producing the TcdB toxin.
TABLE 4.
Isolation and characterization of toxigenic Clostridium difficile
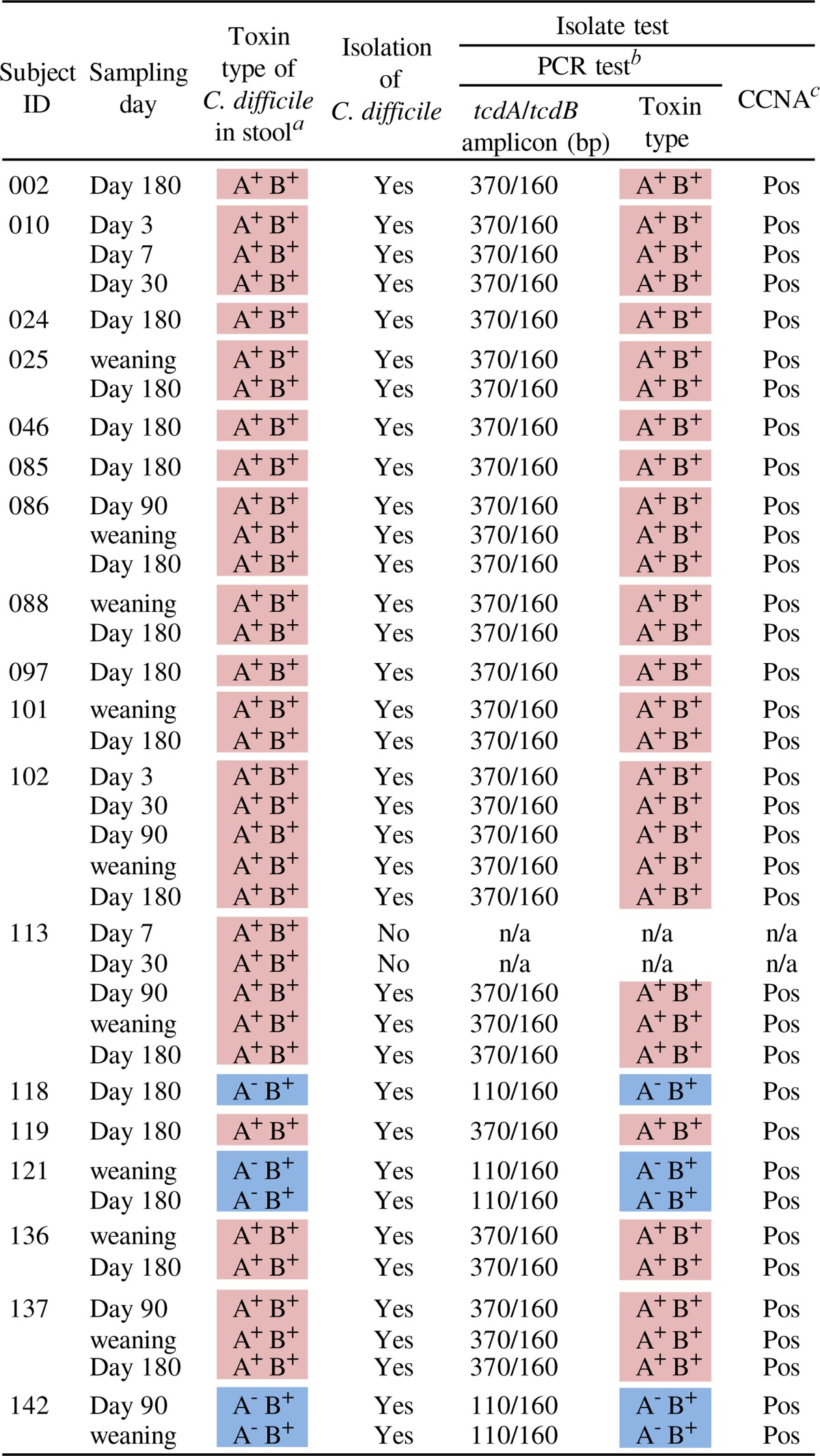
Toxin types of C. difficile predominating in each specimen were identified on the basis of qPCR counts for the three genes.
n/a, not applicable.
CCNA, cell cytotoxicity neutralization assay.
Comparison of genotypes of toxigenic C. difficile isolates.
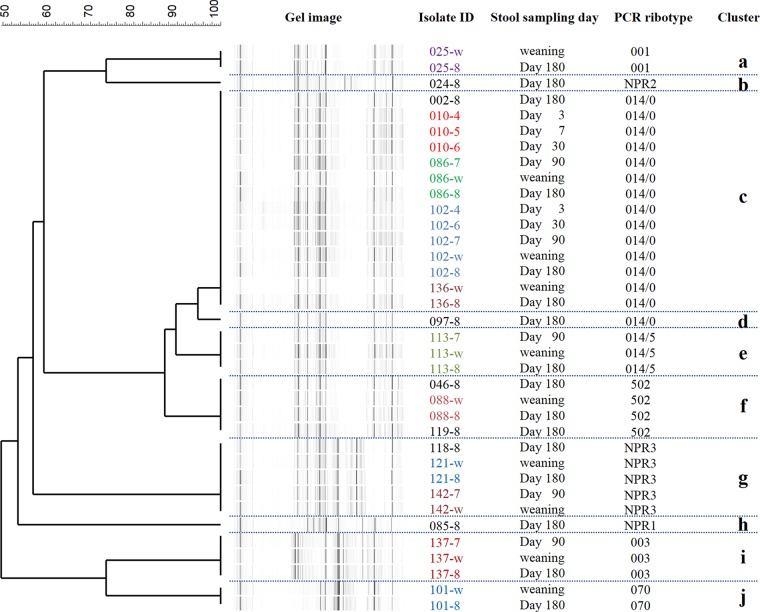
A total of 36 toxigenic C. difficile isolates from 18 infants were genotyped by CGE-PCR ribotyping. A dendrogram was generated by comparing the fragment patterns of amplicons of the 16S rRNA-to-23S rRNA intergenic spacer region. The 36 strains were classified into 10 monophyletic strains (clusters a to j) (Fig. 2). The largest cluster was cluster c, which was composed of 14 individual strains from 5 infants, followed by cluster g, which was composed of 5 individual strains from 3 infants. A Web-based database, WEBRIBO, was queried for the fragment data on each isolate, and a matched PCR ribotype (PR) was assigned to each strain. Most clusters were assigned specific registered PRs, and clusters b, g, and h, which did not match any PRs in the database, were assigned in-house PRs (NPRs 1, 2, and 3). The discrimination based on the assignment of PRs was concordant with that of clustering analysis based on the fragment patterns, except in the cases of clusters c and d. The same PR, PR 014/0 (014 subtype 0), was allocated to clusters c and d. Each of the 11 infants from whom toxigenic C. difficile was isolated at least twice retained a single monophyletic strain. In particular, infant 102 harbored the same monophyletic strain (PR 014/0) for a long period: from 3 days of age until 180 days of age (cluster c) (Fig. 2). MLST analysis showed a typing result very similar to that of CGE-PCR ribotyping (see Fig. S1 in the supplemental material) (although the resolution was slightly lower than that of CGE-PCR ribotyping), supporting our finding that each individual subject retained a particular single strain in their intestines.
FIG 2.
Dendrogram of toxigenic Clostridium difficile isolates from infants, as determined by CGE-PCR ribotyping analysis. The dendrogram was created with a multiscale setting for comparison and the unweighted pair group method with arithmetic means for clustering. Amplicons of the 16S-to-23S intergenic spacer region were analyzed by capillary gel electrophoresis with a 3130xl genetic analyzer. Genetic relatedness based on the fragment patterns was determined for fragments ranging in length from 200 to 600 nucleotides. PCR ribotypes were determined by querying fragment files (.fsa) against the Web-based database WEBRIBO (http://webribo.ages.at/). If no matched PCR ribotype was obtained, a new PCR ribotype (NPR plus one digit) specific to this study was allocated to the strain.
Comparison of characteristics of C. difficile-positive and -negative infants at 6 months of age.
To investigate factors that could be related to colonization with C. difficile, we compared a number of characteristics of C. difficile-positive (n = 47) and -negative (n = 58) infants at 6 months of age (Table 5). Although there was no significant difference in the male-to-female ratio and mode of delivery between the two groups, the proportion of infants who had continued to receive breast milk until 6 months of age was significantly higher in the C. difficile-negative group (P < 0.001). At 6 months of age, the percentage of infants who had begun ingesting solids was higher in the C. difficile-positive group than in the negative group (P < 0.05).
TABLE 5.
Comparison of characteristics of Clostridium difficile-positive infants and C. difficile-negative infants at 6 months of agea
| Characteristic | Value for group |
|||
|---|---|---|---|---|
|
C. difficile-positive infants infected with: |
C. difficile-negative infants (n = 58) | |||
| Toxigenic strains (n = 16) | Nontoxigenic strains (n = 31) | Total (n = 47) | ||
| No. of males/no. of females | 9/7 | 19/12 | 28/19 | 29/29 |
| No. of infants with natural delivery/no. of infants with cesarean delivery | 14/2 | 24/7 | 38/9 | 40/18 |
| No. (%) of infants with continual intake of breast milk for 6 mo | 2 (13) | 8 (26) | 10 (21)*** | 32 (55) |
| No. (%) of infants with initiation of solid food | 15 (94) | 30 (97) | 45 (96)* | 47 (81) |
Comparison between C. difficile-positive infants (total) and C. difficile-negative infants was done by using Fisher's exact test or a χ2 test. *, P < 0.05; ***, P < 0.001.
DISCUSSION
To our knowledge, this is the largest study to have investigated patterns of colonization by C. difficile in the infant gut. Previous studies were limited to reports of the bacterium's prevalence at a single time point or in relatively small numbers of infants (22, 23). We examined both the carriage rates and bacterial counts for total and toxigenic C. difficile strains in 111 infants over time, from immediately after birth until 180 days of age. We observed that carriage of C. difficile increased dramatically between 90 days of age and weaning (at an average of 143 days of age) (Fig. 1). In France, Rousseau et al. (12) examined stool samples of 10 healthy infants for C. difficile monthly during the first year of life; they reported that the C. difficile carriage rate rose sharply from 30% at 3 months of age to 80% at 4 months of age. This sharp rise was similar to what we detected, although their carriage rates were much higher than those reported here. In our study, regardless of the age of infants, carriers of toxigenic C. difficile accounted for approximately one-third of the total C. difficile carriers (Fig. 1). This finding was also consistent with those of a single-point survey of 85 infants ranging in age from 1 month to 3 years by Rousseau et al. (12): they found that 38 infants (45%) carried C. difficile and that 11 infants (13%) carried toxigenic C. difficile. An advantage of our study was the quantitative analysis of both total and toxigenic C. difficile bacteria. Although there have been several reports of the quantification of C. difficile bacteria in infants (9, 24–26), we report here a longitudinal quantitative analysis of both total C. difficile and toxigenic strains in healthy infants. A total of 18 asymptomatic infants carried toxigenic C. difficile, with an average count of ∼107 cells/g of stool (see Table S1 in the supplemental material). Eleven of the 18 infants retained toxigenic C. difficile, some at levels as high as 108 cells/g of stool (Table 3). In our previous study, the mean C. difficile count in asymptomatic elderly individuals was 104 cells/g of stool (16); thus, C. difficile levels as a proportion of the total microbiota population were relatively high in infants. Moreover, Naaber et al. (27) reported an average C. difficile count in patients ranging in age from 3 to 89 years (median, 72 years of age) with antibiotic-associated diarrhea of 107.9 cells/g of stool. Thus, our asymptomatic infants carried quite high numbers of C. difficile bacteria, comparable to those in patients with antibiotic-associated diarrhea.
Even though C. difficile seems to be an early colonizer of the infant gut, its source remains unknown. We did not detect C. difficile in any meconium specimens (Fig. 1), and it was detected in only 1 of 109 maternal stool specimens collected before delivery (data not shown), in agreement with data from previous studies (28–32). Therefore, it is quite unlikely that C. difficile is transmitted from the mother to the infant gut. Acquisition of C. difficile from the hospital environment at birth is generally considered a possible transmission route (11). We considered that this may have been the case in infants in whom we detected C. difficile at 1 month of age. Interestingly, the rate of acquisition of C. difficile within the first month of life in infants delivered by cesarean section (20.7%; 6/29) was higher than that in naturally delivered infants (4.9%; 4/82) (P < 0.05); this suggests that infants delivered by cesarean section are less resistant to C. difficile colonization. On the other hand, the carriage rate of C. difficile rose dramatically between 90 days of age and weaning (at an average of 143 days of age) (Fig. 1). Taking into account the fact that infants of these ages spend most of their time at home, or in day nurseries, it is unlikely that they acquired C. difficile in the hospital. Therefore, we speculated on two potential sources of C. difficile: solid foods and the surrounding living environment. Unfortunately, we did not collect samples immediately before the initiation of solids, and this limited our ability to determine whether food diversification was the reason for C. difficile acquisition. In the previous study by Rousseau et al. (12), the proportion of C. difficile-positive infants jumped to 80% at 4 months of age, but only 25% of these positive infants had started food diversification. In contrast, Fallani et al. (33) reported that the proportion of infants carrying C. difficile significantly decreased after weaning. Accordingly, solid foods could not have been the main source of C. difficile in infants. Generally, infants become increasingly physically mobile at around 3 months of age or older and are inclined to put various objects into their mouths; therefore, we considered that oral intake of C. difficile from surfaces in living environments was reasonable. Although it is unclear whether individual houses are contaminated with C. difficile, one study demonstrated the contamination of day nurseries with C. difficile and also the transmission of certain C. difficile strains between these facilities and the children attending them (28). We therefore suggest that infants acquire C. difficile bacteria that are ubiquitous in their living environments, although further investigations are needed.
Clostridium difficile was detected at least twice in 29 of the 55 C. difficile-positive infants, and 27 of these 29 infants retained the same toxin types of C. difficile (Table 3). Moreover, both of our typing analyses (CGE-PCR ribotyping and MLST) revealed that the same monophyletic strain was retained in each infant (Fig. 2; see also Fig. S1 in the supplemental material). Figure 3 summarizes the longitudinal changes of C. difficile bacteria in the 18 toxigenic C. difficile-positive infants. In agreement with findings of previous studies (12, 23), specific single strains of the same genotype were retained over time in each of the 11 infants who yielded toxigenic C. difficile isolates on at least two occasions. These results suggested that C. difficile in a majority of infants was not transient, but rather was a colonizer, in their intestines. The reason why C. difficile is more commonly found in the infant gut than in adults has not been thoroughly elucidated. It was recently suggested that certain bacterial species belonging to Clostridium cluster XIVa can prevent colonization by C. difficile (2, 3). Intestinal microbiota that are of low complexity and immature and that lack such bacteria possessing possible C. difficile-inhibitory effects may be responsible for colonization (34). As an extrinsic factor, intake of breast milk decreased carriage rates of C. difficile (Table 5) and was an inhibitor of colonization by C. difficile, as reported previously in other studies (24–26, 35).
FIG 3.
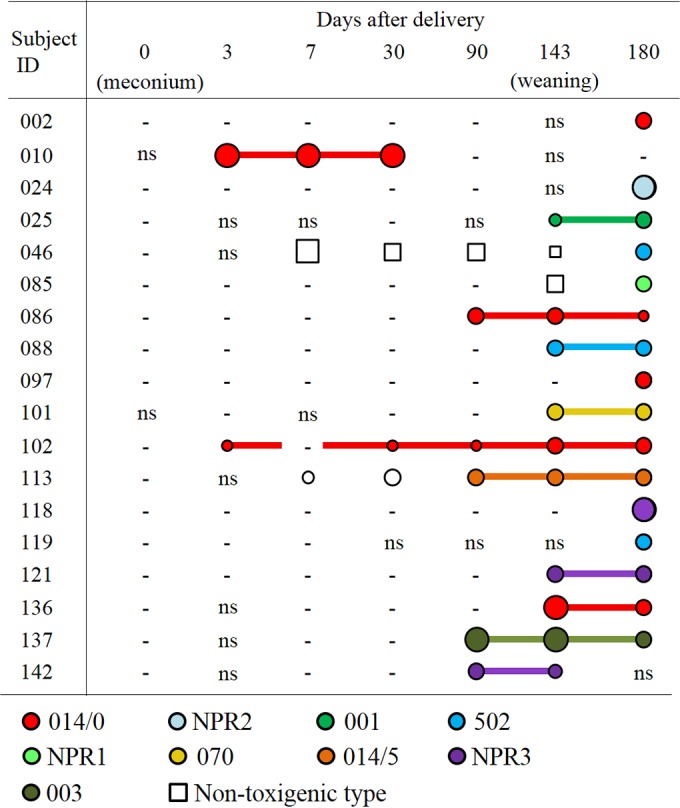
Longitudinal changes in bacterial counts and genotypes of toxigenic Clostridium difficile isolates in 18 infants. Circles and squares indicate the detection of toxigenic C. difficile and nontoxigenic C. difficile by qPCR, respectively. The sizes of the symbols reflect bacterial numbers as determined by qPCR, where small symbols indicate <6.0 log10 cells/g stool, medium symbols indicate 6.0 to 8.0 log10 cells/g stool, and large symbols indicate >8.0 log10 cells/g stool. A minus symbol means that no C. difficile was detected by qPCR. “ns,” no samples. Circles filled with color indicate the isolation of a C. difficile strain by stool culture, and each color corresponds to a genotype of toxigenic C. difficile identified by PCR ribotyping. For each infant, strains that were confirmed to be monophyletic are linked with a line.
We report here an in-depth survey of genotypes of C. difficile isolates in Belgian infants. The most frequently detected toxigenic C. difficile strain in the CGE-PCR ribotyping analysis was of PR 014/0 (Fig. 2); in the MLST analysis, it was of ST33 (see Fig. S1 in the supplemental material). Fifteen of the 36 toxigenic C. difficile isolates (42%) were identified as being both ST33 and PR 014/0 isolates. A previous study showed that ST33 was the most frequently identified strain type of toxigenic C. difficile in French infants (13). Many PCR ribotyping-based studies have revealed that PR 014 is the most frequent strain type of toxigenic C. difficile in French infants (12, 13) and the second most frequent one after PR 001 in Swedish infants (23). PR 014 strains are also frequently isolated from adult CDI patients (36). Interestingly, a recent survey of CDI patients in Belgium revealed that PR 014 had become one of the most prevalent strain types identified in hospitals (37). It is probable that PR 014 is the most common strain type, not only in healthy infants but also in adult CDI patients, in Belgium, and this may support the recently proposed hypothesis that asymptomatic infants are a reservoir of pathogenic strains and contribute to the mediation of CDI in adults (12, 14, 23). The results of a literature search support the associations of specific PRs between infants and adult patients in other countries, namely, PR 014 in France (12, 38) and PR 001 in Sweden (23, 39). The possibility of direct transmission of specific strains from asymptomatic babies to their mothers, who then suffer recurrent CDI, has been reported (14), although this form of transmission remains speculative. Furthermore, it was recently reported that close contact with children less than 2 years of age is a risk factor for community-associated CDI in adults (40). We consider it highly credible that CDI in adults can be mediated by asymptomatic infant carriers of toxigenic C. difficile.
In conclusion, we investigated longitudinal changes in the carriage rates, counts, and genotypes of toxigenic C. difficile bacteria in infants from after birth until the age of 6 months. We found that considerable numbers of toxigenic C. difficile bacteria colonized the intestines of healthy infants. The toxigenic strains were of the same genotype as that capable of causing CDI in adults, supporting the hypothesis that asymptomatic infants can mediate CDI in adults by transmitting pathogenic C. difficile. These findings are informative not only for ecological studies but also for efforts to prevent or control the spread of CDI in adults.
Supplementary Material
Funding Statement
This work was supported by Yakult Honsha European Research Center for Microbiology ESV and Danone Research (Centre for Specialised Nutrition, Wageningen, Netherlands, and Centre Daniel Carasso, Palaiseau, France). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01540-16.
REFERENCES
- 1.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. 2012. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, Brassard P, Dendukuri N, Beliveau C, Oughton M, Brukner I, Dascal A. 2011. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med 365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 5.Garey KW, Dao-Tran TK, Jiang ZD, Price MP, Gentry LO, Dupont HL. 2008. A clinical risk index for Clostridium difficile infection in hospitalised patients receiving broad-spectrum antibiotics. J Hosp Infect 70:142–147. doi: 10.1016/j.jhin.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Asha NJ, Tompkins D, Wilcox MH. 2006. Comparative analysis of prevalence, risk factors, and molecular epidemiology of antibiotic-associated diarrhea due to Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus. J Clin Microbiol 44:2785–2791. doi: 10.1128/JCM.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, Takakuwa H, Saikai T, Kobayashi K, Yamagishi T, Nakamura S. 2001. Colonisation and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J Med Microbiol 50:720–727. doi: 10.1099/0022-1317-50-8-720. [DOI] [PubMed] [Google Scholar]
- 8.Miyajima F, Roberts P, Swale A, Price V, Jones M, Horan M, Beeching N, Brazier J, Parry C, Pendleton N, Pirmohamed M. 2011. Characterisation and carriage ratio of Clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. PLoS One 6:e22804. doi: 10.1371/journal.pone.0022804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark PL, Lee A, Parsonage BD. 1982. Colonization of the large bowel by Clostridium difficile in healthy infants: quantitative study. Infect Immun 35:895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton RP, Tait SK, Dear PR, Losowsky MS. 1984. Asymptomatic neonatal colonisation by Clostridium difficile. Arch Dis Child 59:466–472. doi: 10.1136/adc.59.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFarland LV, Brandmarker SA, Guandalini S. 2000. Pediatric Clostridium difficile: a phantom menace or clinical reality? J Pediatr Gastroenterol Nutr 31:220–231. doi: 10.1097/00005176-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A. 2012. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 55:1209–1215. doi: 10.1093/cid/cis637. [DOI] [PubMed] [Google Scholar]
- 13.Rousseau C, Lemee L, Le Monnier A, Poilane I, Pons JL, Collignon A. 2011. Prevalence and diversity of Clostridium difficile strains in infants. J Med Microbiol 60:1112–1118. doi: 10.1099/jmm.0.029736-0. [DOI] [PubMed] [Google Scholar]
- 14.Hecker MT, Riggs MM, Hoyen CK, Lancioni C, Donskey CJ. 2008. Recurrent infection with epidemic Clostridium difficile in a peripartum woman whose infant was asymptomatically colonized with the same strain. Clin Infect Dis 46:956–957. doi: 10.1086/527568. [DOI] [PubMed] [Google Scholar]
- 15.Jangi S, Lamont JT. 2010. Asymptomatic colonization by Clostridium difficile in infants: implications for disease in later life. J Pediatr Gastroenterol Nutr 51:2–7. doi: 10.1097/MPG.0b013e3181d29767. [DOI] [PubMed] [Google Scholar]
- 16.Kubota H, Sakai T, Gawad A, Makino H, Akiyama T, Ishikawa E, Oishi K. 2014. Development of TaqMan-based quantitative PCR for sensitive and selective detection of toxigenic Clostridium difficile in human stools. PLoS One 9:e111684. doi: 10.1371/journal.pone.0111684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, Tanaka R. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One 8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, Pons JL. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. 2009. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol 75:1961–1969. doi: 10.1128/AEM.01843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57:1377–1382. doi: 10.1099/jmm.0.47714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tullus K, Aronsson B, Marcus S, Mollby R. 1989. Intestinal colonization with Clostridium difficile in infants up to 18 months of age. Eur J Clin Microbiol Infect Dis 8:390–393. doi: 10.1007/BF01964052. [DOI] [PubMed] [Google Scholar]
- 23.Adlerberth I, Huang H, Lindberg E, Aberg N, Hesselmar B, Saalman R, Nord CE, Wold AE, Weintraub A. 2014. Toxin-producing Clostridium difficile strains as long-term gut colonizers in healthy infants. J Clin Microbiol 52:173–179. doi: 10.1128/JCM.01701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. 2005. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett 243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 25.Tonooka T, Sakata S, Kitahara M, Hanai M, Ishizeki S, Takada M, Sakamoto M, Benno Y. 2005. Detection and quantification of four species of the genus Clostridium in infant feces. Microbiol Immunol 49:987–992. doi: 10.1111/j.1348-0421.2005.tb03694.x. [DOI] [PubMed] [Google Scholar]
- 26.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 27.Naaber P, Stsepetova J, Smidt I, Ratsep M, Koljalg S, Loivukene K, Jaanimae L, Lohr IH, Natas OB, Truusalu K, Sepp E. 2011. Quantification of Clostridium difficile in antibiotic-associated-diarrhea patients. J Clin Microbiol 49:3656–3658. doi: 10.1128/JCM.05115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuki S, Ozaki E, Shozu M, Inoue M, Shimizu S, Yamaguchi N, Karasawa T, Yamagishi T, Nakamura S. 2005. Colonization by Clostridium difficile of neonates in a hospital, and infants and children in three day-care facilities of Kanazawa, Japan. Int Microbiol 8:43–48. [PubMed] [Google Scholar]
- 29.Larson HE, Barclay FE, Honour P, Hill ID. 1982. Epidemiology of Clostridium difficile in infants. J Infect Dis 146:727–733. doi: 10.1093/infdis/146.6.727. [DOI] [PubMed] [Google Scholar]
- 30.Martirosian G, Kuipers S, Verbrugh H, van Belkum A, Meisel-Mikolajczyk F. 1995. PCR ribotyping and arbitrarily primed PCR for typing strains of Clostridium difficile from a Polish maternity hospital. J Clin Microbiol 33:2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Jumaili IJ, Shibley M, Lishman AH, Record CO. 1984. Incidence and origin of Clostridium difficile in neonates. J Clin Microbiol 19:77–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherkasskaia RS, Dzhamali N, Marina M, Makarova NV, Samsygina GA, Semina NA, Komarovskaia TP. 1992. Clostridium difficile and diarrhea in infants in the first half-year of life. Pediatriia 1992:15–20. [PubMed] [Google Scholar]
- 33.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Dore J, Edwards CA. 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto-Osaki T, Kamiya S, Sawamura S, Kai M, Ozawa A. 1994. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J Med Microbiol 40:179–187. doi: 10.1099/00222615-40-3-179. [DOI] [PubMed] [Google Scholar]
- 35.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL. 2013. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 37.Viseur N, Lambert M, Delmee M, Van Broeck J, Catry B. 2011. Nosocomial and non-nosocomial Clostridium difficile infections in hospitalised patients in Belgium: compulsory surveillance data from 2008 to 2010. Euro Surveill 16(43):pii=20000 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20000. [PubMed] [Google Scholar]
- 38.Eckert C, Coignard B, Hebert M, Tarnaud C, Tessier C, Lemire A, Burghoffer B, Noel D, Barbut F. 2013. Clinical and microbiological features of Clostridium difficile infections in France: the ICD-RAISIN 2009 national survey. Med Mal Infect 43:67–74. doi: 10.1016/j.medmal.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Magnusson C, Wullt M, Lofgren S, Iveroth P, Akerlund T, Matussek A. 2013. Ribotyping of Clostridium difficile strains associated with nosocomial transmission and relapses in a Swedish county. APMIS 121:153–157. doi: 10.1111/j.1600-0463.2012.02950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. 2008. A case-control study of community-associated Clostridium difficile infection. J Antimicrob Chemother 62:388–396. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.