Abstract
Enthusiasm for mining advanced biofuels from microbial hosts has increased remarkably in recent years. Isoprenoids are one of the highly diverse groups of secondary metabolites and are foreseen as an alternative to petroleum-based fuels. Most of the prokaryotes synthesize their isoprenoid backbone via the deoxyxylulose-5-phosphate pathway from glyceraldehyde-3-phosphate and pyruvate, whereas eukaryotes synthesize isoprenoids via the mevalonate pathway from acetyl coenzyme A (acetyl-CoA). Microorganisms do not accumulate isoprenoids in large quantities naturally, which restricts their application for fuel purposes. Various metabolic engineering efforts have been utilized to overcome the limitations associated with their natural and nonnatural production. The introduction of heterologous pathways/genes and overexpression of endogenous/homologous genes have shown a remarkable increase in isoprenoid yield and substrate utilization in microbial hosts. Such modifications in the hosts' genomes have enabled researchers to develop commercially competent microbial strains for isoprenoid-based biofuel production utilizing a vast array of substrates. The present minireview briefly discusses the recent advancement in metabolic engineering efforts in prokaryotic hosts for the production of isoprenoid-based biofuels, with an emphasis on endogenous, homologous, and heterologous expression strategies.
INTRODUCTION
Concerns, like climate change, diminishing fossil fuel supply, energy security, and political instability in petroleum-wealthy regions, have driven global interest toward the production of sustainable and renewable fuels from biological routes specifically from microbial origin. This directs the global interest toward the search for longer-carbon-chain-length (C ≥ 4) biomolecules as next-generation biofuels having compatibility with the existing fuel infrastructure (1–6). The field of metabolic engineering has provided a wealth of opportunities to expand the scope of biofuels from engineered microbes (1, 5, 7–9). In nature, some of the microbes are capable of decomposing complex sugars into simple sugars, while some others can convert sugars into biofuels natively. The extensive characterization of such hosts and advancement in synthetic biology tools and techniques have enabled researchers to optimize the host's pathway. Various endogenous (the host's genes and pathways), homologous (genes and pathways of other organisms distantly related to the host's), and heterologous (genes and pathways of other organisms that do not correspond in evolutionary origin to the host's) genes and pathways need to be optimized inside a host to enhance biofuel production. The plenteous genetic modifications not only improve substrate utilization, product titers, and yields but also extend the variety of biofuels from microbial hosts (10–17).
Among the vast array of secondary metabolites produced by plants and microorganisms, isoprenoids (terpenoids or terpenes) are the most abundant group of natural products, comprised of more than 40,000 known compounds (18, 19). Isoprenoids are traditionally exploited as a source of fragrances, flavors, and medicines (20). Their tremendous structural and functional diversity (7, 19, 20) provides an alluring motivation to explore biological macromolecules as next-generation biofuels for sustainable economic and industrial growth. Their low hygroscopy, high energy density, and good fluidity at low temperatures place them at the forefront of potentially good diesel and gasoline fuel alternatives (21–23). The present minireview focuses on the engineering of homologous and heterologous metabolic pathways/genes within the prokaryotic hosts for the production of isoprenoid-based advanced biofuels.
BACKGROUND
Plants are the major resources of isoprenoids, but limitations such as (i) low growth rates and seasonal production, (ii) soil and land requirements, (iii) low-level and tissue-specific biosynthesis, and (iv) difficulty in harvesting and extraction are the major barriers to the natural production of isoprenoids in large quantities from plants for fuel purposes (23, 24). During chemical synthesis, drawbacks such as stereochemical complexities of isoprenoids, use of hazardous solvents, and environmental concerns restrain their production in large amounts (23, 25). Together, these barriers have led biofuel research interests toward alternative production strategies offered by biotechnology. Biotechnological applications have enabled researchers to develop metabolically engineered microbial factories for the large-scale production of isoprenoid-based biofuels (26) (see Fig. S1 in the supplemental material). Microbes possess several advantages, such as ease in culturing and handling, high growth rates, inexpensive growth medium requirements, genetic traceability, and tractability, which bring them to the forefront of biofuel research (27). Despite the above-mentioned advantages, microbes can also be genetically modified with great success rates, and even an entire novel pathway can be introduced utilizing high-throughput synthetic biology tools and techniques (24).
Isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP) are the common precursors for isoprenoid backbone synthesis in all living organisms (28). Almost every prokaryote, with some exceptions, synthesizes its isoprenoids from the deoxyxylulose-5-phosphate (DXP) pathway (29), but some endogenous regulations of the DXP pathway restrict their large-scale production for fuel applications. On the other hand, in the eukaryotic system, the isoprenoid backbone is synthesized from the mevalonate (MVA) pathway by utilizing acetyl coenzyme A (acetyl-CoA) (30–32).
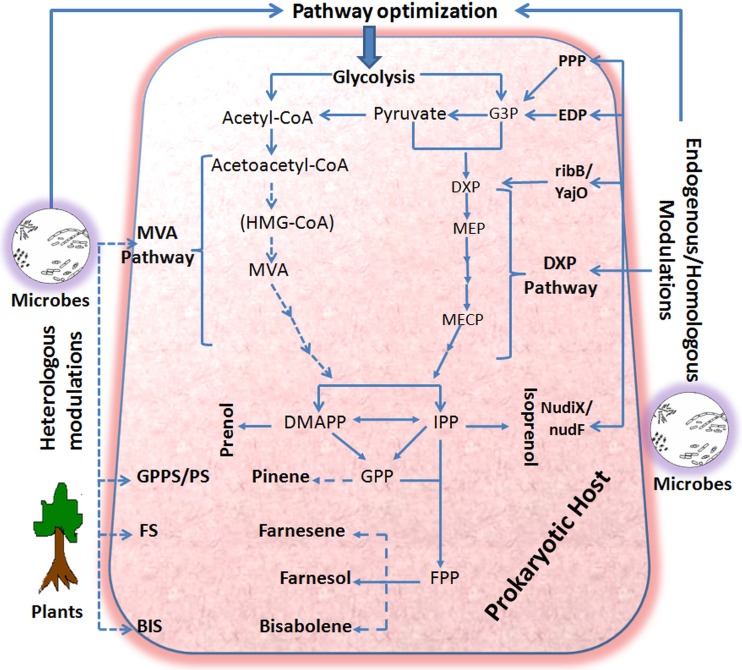
In recent years, enhanced production of isoprenoid-based biofuels has been achieved in microbial systems via tuning an endogenous pathway or engineering a heterologous pathway and genes in new hosts (5, 7, 8, 23). In the prokaryotic systems, to bypass endogenous regulations of the DXP pathway and to ensure overexpression of the isoprenoid backbone, the components of the heterologous MVA pathway have been introduced into their genomes (10–12, 33). Overexpression of key regulatory enzymes of the native DXP pathway has been also utilized to enhance IPP and DMAPP production in prokaryotes (13, 34–37). Similarly, in the eukaryotic system, efforts have been made to enhance isoprenoid production via (i) bypassing the endogenous regulation of the MVA pathway in Saccharomyces cerevisiae (38, 39) or (ii) modulating the expression of endogenous/homologous enzymes for higher production of isoprenoid-based biofuels in yeast (11, 40–42). The metabolic engineering efforts for either endogenous or heterologous gene expression have increased the production of a vast array of isoprenoid-based biofuels or fuel precursors. Figure 1 illustrates the various pathway optimization strategies for the enhanced production of isoprenoid-based biofuels in a prokaryotic host. Stoichiometric studies have shown that the DXP pathway is more competent than the MVA pathway (43); still, the production of isoprenoids by optimizing the DXP pathway cannot go beyond the levels achieved by expressing a heterologous MVA pathway in prokaryotes. In later sections of this minireview, we briefly discuss the strategies for the production of isoprenoid-based biofuels and fuel precursors in prokaryotic hosts.
FIG 1.
Metabolic engineering for the production of isoprenoid-based biofuels in a prokaryotic host. Solid arrows indicate endogenous/homologous pathways/genes, and dashed arrows indicate heterologous pathways/genes in a prokaryotic host. EDP, Entner-Doudoroff pathway; PPP, pentose phosphate pathway; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MVA, mevalonate; G3P, glyceraldehyde-3-phosphate; DXP, deoxyxylulose-5-phosphate; MEP, 2-C-methylerythritol-4-phosphate; MECP, 2-C-methylerythritol-2,4-cyclodiphosphate; IPP, isoprenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; GPPS, geranyl pyrophosphate synthase; PS, pinene synthase; FS, farnesene synthase; BIS, bisabolene synthase.
ISOPRENOID BIOSYNTHESIS
The MVA and DXP pathways are well-known pathways for the production of a wide range of branched-chain and cyclic metabolites, collectively called isoprenoids, in living systems, e.g., quinones (ubiquinone and plastoquinone), pigments (carotenoids and the side chain of chlorophyll), hormones (gibberellins, brassinosteroids, and abscisic acid), and sterols (cholesterol, ergosterol, and campesterol), as well as other characteristic plant compounds (limonene, pinene, menthol, bacoside, camphor, etc.) (18, 19).
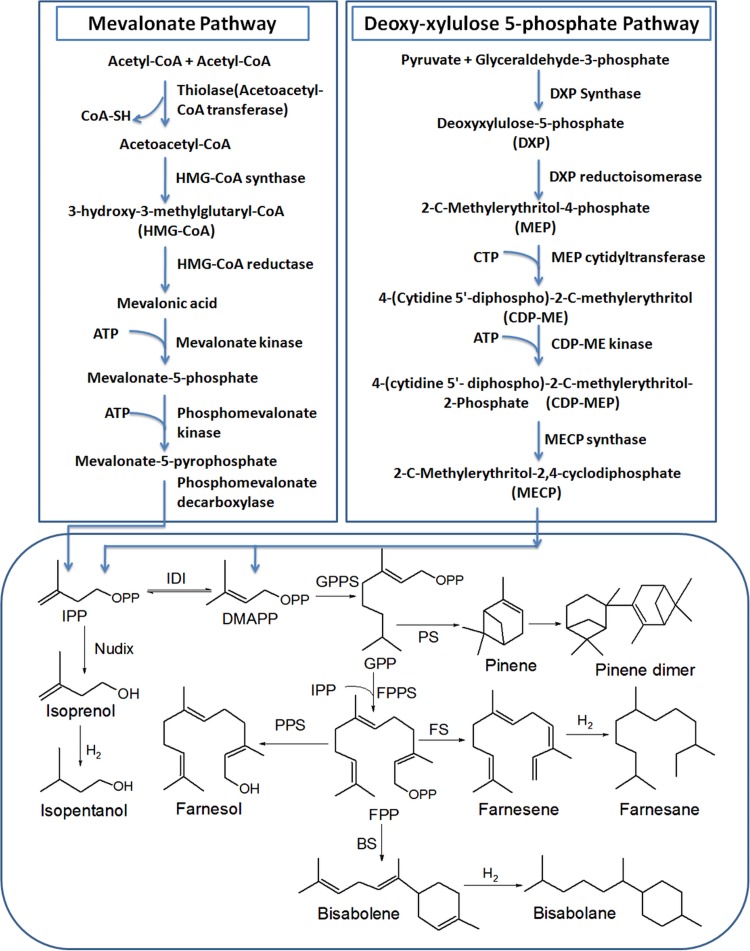
The first-recognized pathway for biosynthesis of isoprenoids is the MVA pathway (30–32), which involves a series of six enzymatic steps to convert acetyl-CoA to IPP (Fig. 2). For many decades, IPP was considered to be formed from the MVA pathway and was considered ubiquitous in all living organisms. Extensive genomic studies on plants and various eubacteria have led to the discovery of an MVA-independent pathway for IPP and DMAPP biosynthesis (44–48). This is the DXP pathway for isoprenoid production in prokaryotes and chloroplasts of higher plants. The DXP pathway consists of seven enzymatic steps that convert glyceraldehyde-3-phosphate (G3P) and pyruvic acid to IPP and DMAPP (49, 50) (Fig. 2). Though glycolysis is the primary source of the precursor supply for both pathways, there are several other known pathways which also provide precursors for isoprenoid backbone synthesis for, e.g., the pentose phosphate (PP) pathway, the Entner-Doudoroff (ED) pathway, fatty acid metabolism, and amino acid metabolism (51, 52) (Fig. 1).
FIG 2.
Metabolic routes for the production of isoprenoid-based biofuels in a living system. The mevalonate (MVA) pathway is from acetyl-CoA, and the deoxyxylulose 5-phosphate (DXP) pathway is from glyceraldehyde-3-phosphate and pyruvate. IPP, isoprenyl diphosphate; DMAPP, dimethylallyl diphosphate; IDI, isoprenyl diphosphate isomerase; nudiX, nucleoside diphosphate hydrolases; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; GPPS, geranyl pyrophosphate synthase; FPPS, farnesyl pyrophosphate synthase; PS, pinene synthase; FS, farnesene synthase; BS, bisabolene synthase; PPS, endogenous pyrophosphatase.
Downstream isoprenoid-based biofuel products.
Based on the number of carbon atoms present, isoprenoid-based biofuels can be subdivided into (i) hemiterpene-based (C5) biofuels, (ii) monoterpene-based (C10) biofuels, and (iii) sesquiterpene-based (C15) biofuels.
Hemiterpene-based biofuels are synthesized directly from IPP and DMAPP by several downstream enzymes. Various endogenous and homologous phosphatases and pyrophosphatases that can convert prenyl pyrophosphates into respective biofuel compounds have been explored in Escherichia coli (12, 16, 37, 53, 54).
Long-carbon-chain-length isoprenoid molecules, such as pinene, farnesene, and bisabolene, are not produced by prokaryotic hosts, as the enzymes responsible for their synthesis are not found in prokaryotic systems (7). Such enzymes need to be incorporated from other organisms that act on isoprenyl pyrophosphatases. Monoterpene synthesis requires geranyl pyrophosphate (GPP) as a precursor, but most prokaryotes lack endogenous GPP synthase (GPPS) (7). Prokaryotes have the farnesyl pyrophosphate synthase (FPPS or IspA) enzyme, which is responsible for farnesyl pyrophosphate (FPP) synthesis directly from IPP and DMAPP and is assumed to be a source of GPP synthesis in prokaryotes (55). Monoterpene synthase enzymes, such as pinene synthase (PS), also need to be expressed heterologously, which can convert GPP into monoterpene-based biofuel molecules (Fig. 3).
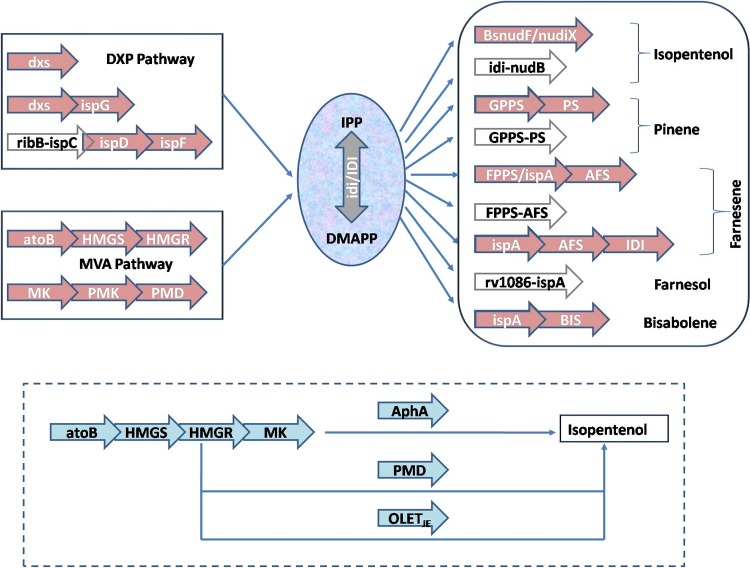
FIG 3.
Pathway optimization for the enhanced production of isoprenoid-based biofuels from prokaryotic hosts. Open and filled arrows indicate coexpression and fusion protein expression, respectively. Genes in uppercase letters are heterologous, whereas genes in lowercase letters are endogenous/homologous. The solid-line box shows conventional upstream pathways (MVA and DXP pathways), whereas the dashed-line box shows novel pathways via decoupling of the conventional MVA pathway. atoB, acetyl-CoA acetyltransferase gene; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; PMD, mevalonate diphosphate decarboxylase; dxs, deoxyxylulose-5-phosphate synthase gene; ispC, deoxyxylulose-5-phosphate reductoisomerase gene; ispD, 2-C-methylerythritol-4-phosphate cytidyltransferase gene; ispF, 2-C-methylerythritol-2,4-cyclodiphosphate synthase gene; ispG, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase gene; IPP, isoprenyl diphosphate; DMAPP, dimethylallyl diphosphate; idi/IDI, isoprenyl diphosphate isomerase; nudF, B. subtilis ADP ribose pyrophosphatase gene; nudiX, nucleoside diphosphate hydrolases; ispA, farnesyl pyrophosphate synthase (FPPS) gene; GPPS, geranyl pyrophosphate synthase; PS, pinene synthase; AFS, farnesene synthase; BIS, bisabolene synthase; OLETJE, Jeotgalicoccus sp. fatty acid decarboxylase.
Sesquiterpene (C15)-based biofuel synthesis in prokaryotes also requires heterologous enzymes, like farnesene synthase (FS) and bisabolene synthase (BIS), to convert FPP into their respective biofuels, farnesene and bisabolene (Fig. 3). For the production of farnesol (another sesquiterpene derivative) in E. coli, endogenous phosphatases and pyrophosphatases have been utilized (33, 56, 57). Later sections briefly discuss strain improvement strategies for the overproduction of isoprenoid-based biofuels and fuel precursors from prokaryotic hosts.
ENGINEERING MICROBIAL HOSTS FOR THE NONNATURAL PRODUCTION OF ISOPRENOID-BASED BIOFUELS
Most of the research concerned with the production of isoprenoid-based biofuels from prokaryotic hosts has focused on engineering the most-understood model organism, E. coli. Strain engineering efforts have provided an array of new opportunities to engineer alternate microbial strains as hosts for the nonnatural production of isoprenoid-based biofuels (13, 15, 58, 59). Engineering alternate microbial hosts also enables researchers to explore their ability to utilize a vast array of carbon sources for biofuel production. Enhanced production of isoprenoids has been achieved through a focused combinatorial approach, multivariate modular pathway engineering (MMPE), even without a full understanding of endogenous regulations (35). MMPE utilizes partitioning of the overall DXP pathway into smaller modules, and components of each module are then tuned separately in order to obtain higher yields of the desired metabolites. Codon optimization has also provided a promising platform for the optimization of nonnative genes to enhance isoprenoid-based biofuel production in microbial hosts (11, 37, 59). However, the application of codon optimization is limited by inherent low enzyme activities and specificities of the isoprenoid pathways in microbial hosts. The work of Leonard et al. has suggested a more promising way to overcome such limitations via the molecular reprogramming process (MRP) (60). In this approach, the key metabolic nodes are optimized via mutations in either transcription factors or pathway enzymes (17, 60).
In combination with pathway engineering, strategies that reduce metabolic load and improve productivity include (i) localization of pathway intermediates into protein-based synthetic compartments or bacterial microcompartments (61–63), (ii) attachment of proteins (64), RNA (65), and DNA (66) on a synthetic scaffold, (iii) tagging of proteins with localization signals (67), and (iv) harnessing of efflux pumps against product toxicity (68, 69). It is well established that overexpressed heterologous recombinant enzymes form insoluble aggregates having little or no activity (70). These aggregates are known as inclusion bodies. Zhou et al. have reported that recombinant endogenous enzymes of the DXP pathway also form inclusion bodies which impart a negative impact on metabolic flux (71). Their solubility can be increased by several means, like lowering the incubation temperature, overexpressing chaperones, mutagenizing proteins, and using chemicals such as sorbitol (71). Together, utilization of metabolic engineering with other synthetic biology approaches might maximize biofuel titers in a relatively small design space.
Tuning the endogenous DXP pathway.
It has been established that the DXP pathway is the sole source of isoprenoid supply in most prokaryotic organisms, including blue-green algae (45–48). Overexpression of DXP pathway genes has been reported for the enhanced production of isoprenoids (13, 34–37). Among the isoprenoid-based biofuels, prenol and isoprenol, collectively called isopentenols, are the simplest isoprenoid-based biofuels. The first microbial production of isopentenols was achieved by incorporating components of the heterologous MVA pathway in E. coli (53). However, not only is the hemiterpene-based biofuel research concentrated on the expression of the heterologous MVA pathway, but also the native DXP pathway has been optimized in E. coli (37). Incorporation of a codon-optimized synthetic Bacillus subtilis nudF (BsnudF) gene in E. coli W3110 conferred dephosphorylation of IPP and DMAPP into isopentenols (37). Overexpression of native DXP pathway genes (dxs and ispG) (Fig. 3) significantly increased isopentenol production 3.3-fold over that of the E. coli strain harboring NudF only. Later, the titer was improved by blocking the Embden-Meyerhof-Parnas (EMP) pathway and activating endogenous pentose phosphate (PP) and Embden-Meyerhof (ED) pathways (37). It is interesting that blocking of the EMP pathway shunts glucose metabolism to the ED and PP pathways and provides a more-balanced DXP precursor (G3P and pyruvate) pool and NADPH supply. Activation of the PP and ED pathways resulted in an additional 1.9-fold increase in the isopentenol titer (37). A final titer of 61.9 mg of isopentenols liter−1 was reported (Table 1) from 20 g liter−1 of glucose in a 5-liter-scale batch cultivation system (37).
TABLE 1.
Summary of the pathway modulations, engineered microbes, product titers, production rates, and production times of isoprenoid-based biofuels produced from engineered microbes
| Isoprenoid(s) by expression type | Engineered microbe | Titer | Rate of production | Timea | Reference |
|---|---|---|---|---|---|
| Heterologous expression | |||||
| Isopentenols | E. coli | ∼112 mg liter−1 | NAb | 43 | 53 |
| 55 mg liter−1 | NA | NA | 54 | ||
| 1.3 g liter−1 | 59.3 mg liter−1 h−1 | 48 | 12 | ||
| 2.23 g liter−1 | NA | 48 | 16 | ||
| Pinene | E. coli | 1.7 mg liter−1 | NA | NA | 10 |
| 0.97 g liter−1 | 4 mg h−1 g−1 (DCW) | 32 | 73 | ||
| 32.4 mg liter−1 | 18.1 mg liter−1 OD−1 | NA | 74 | ||
| Farnesol | E. coli | 135.5 mg liter−1 | NA | 48 | 33 |
| 143.3 mg liter−1 | NA | NA | 57 | ||
| Farnesene | E. coli | 380 mg liter−1 | NA | 48 | 83 |
| 1.1 g liter−1 | NA | 96 | 85 | ||
| Bisabolene | E. coli | ∼912 mg liter−1 | NA | 72 | 11 |
| Homologous/endogenous expression | |||||
| Isopentenols | E. coli | 61.9 mg liter−1 | NA | 48 | 37 |
| Pinene | C. glutamicum | ∼27 μg g−1 (DCW) | NA | 48 | 13 |
| Farnesene | Anabaena sp. | ∼305 μg liter−1 | ∼70 μg liter−1 OD−1 day−1 | 15 days | 59 |
| Bisabolene | S. venezuelae | ∼10 mg liter−1 | NA | NA | 15 |
| Synechococcus sp. | 0.6 mg liter−1 | 6 μg liter−1 h−1 | 96 | 58 |
In hours, unless otherwise noted.
NA, data not available.
Monoterpene-based (C10) biofuels, such as α-pinene, camphene, and limonene, have viscosities, densities, and a net heat of combustion similar to those of the widely used missile fuel J-10 (21, 72). Microbes do not synthesize pinene naturally, and they require a heterologous PS enzyme to convert GPP into pinene. Most prokaryotes also lack endogenous GPPS and depend on IspA for the condensation of IPP and DMAPP into GPP. Therefore, a heterologous GPPS is required to incorporate into their genomes to increase the GPP supply. GPPS and PS enzymes from various sources, such as Abies grandis, Picea abies, and Pinus taeda, have been studied to enhance pinene yield in microbial hosts (13, 73, 74). Both enzymes have been introduced in microbes either with a heterologous MVA pathway (73, 74) or with an optimized endogenous DXP pathway (13). Modulation of the DXP pathway in alternate microbial hosts, such as cyanobacteria, Corynebacterium glutamicum, and Streptomyces sp., has improved biofuel yields in such hosts (13, 15, 58, 59). Combinatorial gene expression of heterologous PS and GPPS (from P. taeda and A. grandis) with native deoxyxylulose-5-phosphate synthase (DXS) and isoprenyl diphosphate isomerase (IDI) improved pinene titers in C. glutamicum (13). Twelve strains of C. glutamicum were constructed using various combinations of PS and GPPS with either DXS or IDI or both. Coexpression of P. taeda PS and A. grandis GPPS with both endogenous DXP pathway enzymes (DXS and IDI) resulted in ∼27 μg α-pinene g−1 (dry weight of cells [DCW]) (13) (Table 1). Coexpression of PS and GPPS increased pinene production up to a detectable level; however, the fusion protein product of PS and GPPS can also be utilized to enhance pinene titer in alternate hosts.
As with pinene, microbes do not synthesize sesquiterpene-based biofuels naturally. Sesquiterpenes have been proposed as diesel fuels, and their estimated cetane value, cloud point, and energy density are also in the range of those of diesel fuels (2, 7, 15, 75). Recently, metabolic engineering approaches have been utilized in cyanobacteria to exploit their ability to produce isoprenoids from H2O and CO2 (simplest carbon source) (14, 22, 58, 59). The advantages associated with cyanobacteria, such as fast growth compared to that of plants and yeast, high photosynthetic rate, genetic tractability, and sequenced genomes bring them to the forefront of biofuel research. Incorporation of the codon-optimized fs gene from P. abies in the filamentous cyanobacterium Anabaena sp. strain PCC 7120 enabled photosynthetic production of farnesene through an endogenous DXP pathway (59). The total photosynthetic accumulation of farnesene was ∼305 μg liter−1 during 15 days of growth, with specific farnesene productivity at ∼20 μg liter−1 optical density (OD)−1 day−1 (59). Further, optimization of the DXP pathway and expression of protein fusion products may improve farnesene titers in cyanobacteria. Biofuel production from cyanobacteria is a slow process, and the yield obtained is also not appropriate for biofuel purposes. However, it requires the least input in terms of growth and production. Insertion of sugar transporter systems in cyanobacteria has overcome diurnal running conditions and has provided an excellent alternative for 24-h chemical production (76). Other constraints, such as obtaining maximum photosynthetic efficiency and minimizing heat generation for the optimization of a photo-bioreactor, can be addressed by (i) providing a saturating amount of light (77) and maintaining culture depth and rate of cell mixing to avoid shelf shading (78), (ii) utilizing hyperthermophilic bacteria (79), or (iii) converting nonphotosynthetically active radiation into usable wavelengths (80).
Bisabolene, a monocyclic sesquiterpene, is a candidate biodiesel fuel and is produced by microbial hosts through the incorporation of a heterologous BIS gene from various plant sources (11, 15, 17, 58). Recently, attempts have been made to produce bisabolene by modulating the DXP pathway in different microbial hosts capable of utilizing alternate carbon sources (15, 17, 58). In Streptomyces venezuelae, FPP flux was redirected toward bisabolene synthesis via deletion of the geosmin synthase and 2-methylisoborneol (2-MIB) synthase genes in the downstream DXP pathway, with simultaneous overexpression of endogenous FPP synthase 1 (FPPS1) (15). Incorporation of a heterologous AgBIS gene facilitated the conversion of accumulated FPP into bisabolene in an engineered host. In order to produce bisabolene from alternate carbon sources, the maximum-bisabolene-producing strain, S. venezuelae SZ04, was subjected to different carbon sources, like cellobiose, cellulose, and ionic liquid (IL)-treated switch grass. Bacteria supplemented with cellobiose yielded a bisabolene titer similar to that of glucose, while low-level production was obtained with IL-treated switch grass, and it was in the order of magnitude of pinene (10). Bacteria supplemented with cellulose failed to improve production over that of the control (15). The euryhaline cyanobacterium Synechococcus sp. PCC 7002 also produces its isoprenoids through the DXP pathway from CO2 and H2O. Incorporation of the AgBIS gene at a neutral site (termed NSI) between two open reading frames encoding hypothetical proteins in the Synechococcus sp. genome produced 0.6 mg of bisabolene liter−1 (0.3 mg g−1 [DCW]) (58) (Table 1). The production of biofuels might be optimized in such alternate hosts via consolidated bioprocessing (CBP) approaches and utilization of protein fusion products.
Expressing the heterologous MVA pathway.
Most studies utilizing the MVA pathway to bypass endogenous DXP pathway regulations and to increase the precursor supply for isoprenoid-based biofuel production are focused on engineering well-established prokaryotic host E. coli strains. Expression of the MVA pathway has enhanced isoprenoid titers severalfold over titers produced by DXP pathway modulations. Introduction of the MVA pathway in E. coli produced toxicity due to the accumulation of isoprenoid precursors (53). To overcome the toxicity associated with IPP and DMAPP accumulation, a library of 19,000 clones harboring B. subtilis genomic DNA fragments was screened in E. coli. The toxicity was overcome by the expression of BsnudF, which encodes an enzyme capable of dephosphorylating prenyl diphosphates into prenyl alcohols. After 43 h of incubation, a final titer at 112 mg of isopentenol liter−1 was observed from engineered E. coli (53) (Table 1). This study opens up a gateway to explore the heterologous MVA pathway with simultaneous overexpression of bacterial homologous phosphatase and pyrophosphatase superfamilies for the overproduction of isopentenols and other high-yield isoprenoid alcohols in engineered microbes.
Encouraged by the results of the above-mentioned study, Chou and Keasling (54) screened E. coli-endogenous nucleoside diphosphate linked to X (NudiX) hydrolase superfamily genes in an engineered E. coli strain incorporated with a heterologous MVA pathway. This screening revealed that overexpression of five NudiX hydrolase superfamily genes (nudB, nudF, nudI, nudJ, and nudM) resulted in a >2-fold increase in the production of isopentenol compared to the production level in a control that expressed only the MVA pathway genes. Overexpression of E. coli NudB (EcNudB) resulted in the highest isoprenol production, at 55 mg of isoprenol liter−1 from 2 g liter−1 of glucose (54). Later, Zheng et al. (12) improved isopentenol titers by introducing components of the entire MVA pathway, including idi from S. cerevisiae (idi1), into E. coli. They also overexpressed different phosphatases and pyrophosphatases from various sources in engineered E. coli strains to remove pyrophosphate from IPP and DMAPP. Among all enzymes, B. subtilis NudF (BsNudF) removed pyrophosphate most efficiently and produced the highest yield of isoprenol and prenol in E. coli (YY158). To obtain single-isoprenol production, the conversion of IPP into DMAPP was blocked by inactivating the idi1 gene in E. coli (YY158). The resultant strain, E. coli YY159, produced 1.3 g of a single isoprenol liter−1, whereas overexpressing E. coli NudF (EcNudF) with the entire MVA pathway and idi1 gene in E. coli (YY168) produced 0.2 g of isopentenols liter−1, with ∼80% of the total production as prenol (12).
The studies on hemiterpene-based biofuels suggested that a consortium of homologous/endogenous genes with heterologous pathways or genes could enhance the nonnatural production of isoprenoid-based biofuels at significant levels. In a recent study, the production of hemiterpene-based biofuels was found to be increased by several modifications in endogenous and heterologous enzymes in an engineered E. coli strain (16). First, the production level of isoprenol was enhanced by 60% from background level by engineering the Shine-Dalgarno sequence (the ribosomal binding site in the mRNA) of an endogenous nudB gene. Engineering the Shine-Dalgarno sequence also elevated protein levels and reduced intracellular IPP accumulation by 4-fold. Further, expression of a heterologous mevalonate kinase (MK) enzyme was optimized by investigating various MK enzymes from different microbes. These modifications enhanced isoprene production to a significant level, and finally, a titer of 2.23 g of isoprenol/liter was obtained (which was ∼70% of the pathway-dependent theoretical yield) by using an oleyl alcohol overlay (16). To the best of our knowledge, this is the highest yield achieved for isoprenoid-based biofuel from an engineered strain. The above-mentioned studies have also established NudiX hydrolase superfamily genes as a key player having an important role in the conversion of isoprenoid precursors into biofuels.
Recently, novel MVA-mediated pathways have been successfully assembled in E. coli via decoupling of the MVA pathway (81, 82). Kang et al. (81), in their study, recruited phosphomevalonate decarboxylase (PMD) and an E. coli-endogenous phosphatase (AphA) to bypass the MVA pathway. PMD produced isopentenol directly from MVA, whereas ApaH synthesized isopentenol via an intermediate, isopentenyl monophosphate (IP), from MVA monophosphate (MVAP) (81) (Fig. 3). In another study, a fatty acid decarboxylase from Jeotgalicoccus species (OleTJE) was introduced into E. coli for direct conversion of MVA into isoprenol (82) (Fig. 3). The pathway bypass reduced cellular energetic requirements and escaped the host from IPP-related toxicity. It also eliminated the cellular dependency on NudiX hydrolase superfamily proteins to convert excess IPP and IMAPP into their respective biofuels.
In an attempt to produce pinene, Bokinsky et al. (10) engineered an E. coli (MG1655) strain with a heterologous MVA pathway and a downstream pinene-specific enzyme, PS (Fig. 3). Since the authors used IL-treated biomass as a carbon source, the strains of E. coli were further engineered separately with heterologous β-glucosidase (cel3A) and β-xylosidase (gly43F) genes from Cellvibrio japonicas to metabolize celluloses and hemicellulose, respectively. A coculture of both the engineered strains produced 1.7 mg of pinene liter−1 in MOPS-M9 medium containing 3.9% IL-treated switch grass (10). The findings of this study unlock the gateway to explore a consortium of microbes capable of digesting cellulose and hemicellulose, with the engineered microbes capable of converting simple sugar to biofuels.
As IspA quickly takes up endogenous GPP for FPP synthesis in E. coli, the excess GPP can also be supplied by expressing heterologous GPPS enzymes from various plant sources. Expression of PS and GPPS (from A. grandis, P. abies, and P. taeda) in various combinations in E. coli with a heterologous MVA pathway has been reported to improve pinene titers (73, 74). Coexpression of GPPS from A. grandis and α-PS from P. taeda with a heterologous MVA pathway improved the pinene titer in E. coli BL21(DE3) (73) (Fig. 3). Later, the culture conditions were optimized to improve the pinene titer in a shake flask (73). A pinene titer at 5.44 mg liter−1 was obtained in a shake flask containing 0.25 mmol isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer and MD beef extract as an organic nitrogen source at 30°C. Further, in a scale-up study in a fed-batch fermentor, a maximum concentration of 0.97 g of pinene liter−1 was achieved at 32 h postinduction (Table 1) while a glucose concentration of less than 0.5 g liter−1 was maintained. The specific productivity of pinene was recorded at 4 mg h−1 g−1 (DCW), with a conversion efficiency of glucose to α-pinene as 2.61% (73).
Despite many attempts to produce pinene from a microbial origin, the titers obtained do not match the orders of magnitude of those of hemiterpenes (12, 16, 37) and sesquiterpenes (11, 33, 83). The possible reasons may be (i) the toxicity associated with the monoterpenes (23), (ii) inhibition of GPPS and PS by GPP, and (iii) the use of magnesium as a cofactor for catalysis (74). Improvement of efflux pumps has shown a potential to overcome product-related toxicity (69, 84) and might be utilized for monoterpene-based biofuels also. Fusion products of two successive enzymes of the isoprenoid pathway have overcome the enzyme inhibition and improved isoprenoid production in E. coli (74, 83). To explore the effect of fusion proteins, several GPPS-PS fusion products from A. grandis, P. abies, and P. taeda have been studied in E. coli expressing a heterologous MVA pathway (74). The GPPS-PS fusion product (Fig. 3) improved pinene titers compared to those of coexpression systems used in the same study. The highest pinene titer at 32.4 mg liter−1 with a specific productivity of 18.1 mg liter−1 OD−1, was achieved by expressing the A. grandis GPPS-PS fusion product (74). The titer was ∼6-fold higher than that in the previous report of Yang et al. (73). Redirecting metabolic flux toward the DXP pathway (15, 37) and expressing the GPPS-PS fusion product (74, 83) can improve the nonnatural production of monoterpene-based biofuels in alternate microbes.
In a study, α-farnesene (i) was synthesized by incorporating the apple α-farnesene synthase (afs) gene into E. coli overexpressing DXS and IDI, (ii) was incorporated with the bottom portion of the MVA pathway (cells were not able to synthesize mevalonate, and mevalonate was supplied externally), or, in a third case, (iii) was incorporated with the entire MVA pathway (cells were able to synthesize mevalonate). Overexpression of DXS marginally increased the background production of α-farnesene, while an ∼5-fold increase was recorded in E. coli overexpressing IDI. Later, expression of the bottom portion of a heterologous MVA pathway showed an increase in the yield by ∼15-fold, which was enhanced to ∼72-fold by the protein fusion product FPPS (IspA)-AFS. Lastly, an approximately 317-fold increase (∼380 mg of α-farnesene liter−1) over the initial production was achieved via incorporation of the complete MVA pathway by integration with the FPPS (IspA)-AFS fusion protein (Fig. 3) (83).
Zhu et al. (85) revealed that endogenous IDI plays a key role in α-farnesene synthesis in vitro as well as in vivo. Overexpression of IDI with IspA and AFS enhanced the farnesene yield up to ∼2,000-fold over that produced by the control strain in E. coli incorporated with the MVA pathway (85). A maximum output at 731 mg liter−1 was obtained 48 h postinduction in a shake flask, which was twice that previously reported (83) under the same conditions. After 96 h postinduction, farnesene production reached ∼1.1 g liter−1 in a shake flask (85). The overexpression of endogenous genes to direct metabolic flux toward FPP production not only enhanced biofuel production (83, 85) but also restored the growth of engineered strains (83). This suggests that a consortium of endogenous/homologous genes with heterologous genes/pathways can be utilized to overcome the bottlenecks in the production of biofuels more efficiently.
Farnesol (FOH) is another FPP derivative, and its production was first reported in culture broth of squalene synthase-deficient S. cerevisiae (86). Early research on FOH production was focused solely on production from S. cerevisiae (41, 42, 87, 88). The extracellular fraction and cell lysate of E. coli overexpressing endogenous ispA and idi produce 389 μg of FOH liter−1 by the action of potato acid phosphatase (56). Incorporation of the heterologous MVA pathway and overexpression of IspA improved the FOH titer by ∼350-fold over that previously reported (56) from an engineered E. coli strain (33). The engineered strain produced a farnesol titer at 135 mg liter−1, and endogenous phosphatases were believed to hydrolyze accumulated FPP to FOH nonspecifically. Identification and optimization of endogenous phosphatases and pyrophosphatases having specificity toward FPP might be utilized to increase the FOH yield in E. coli.
Later, Wang and coworkers produced a Z,E isoform of FOH (Z,E-FOH), via expression of a heterologous Z,E-FPP synthase (Rv1086) from Mycobacterium tuberculosis in E. coli (57). In nature, the chain elongation of the newly formed double bonds can proceed in two possible stereospecific configurations of E and Z. The FPPS catalyzes the condensation of GPP and IPP into FPP in four different potential configurations (E,E, E,Z, Z,E, and Z,Z) (89). E. coli-endogenous FPPS (IspA) catalyzes the formation of all E configurations of FPP (E,E-FPP) (55), while Rv1086 catalyzes Z condensation of GPP and IPP into Z,E-FOH in M. tuberculosis (90). The expression of Rv1086 in engineered E. coli produced 75.3 mg of FOH liter−1 (E,E-FOH and Z,E-FOH), which was lower than the titer of FOH produced by the overexpression of IspA only. Coexpression of IspA in Rv1086-expressing E. coli enhances total FOH production but reduced Z,E-FOH due to the consumption of GPP by IspA. It is well known that prokaryotes lack endogenous GPP synthase; hence, another enzyme, IspA, is responsible for GPP synthesis, which it simultaneously takes up to form FPP (55). To increase GPP availability to Rv1068 the fusion protein IspA-Rv1068 was constructed. This allowed Rv1068 to effectively compete with IspA for GPP and IPP. Z,E-FOH at 115 mg liter−1 was produced by the engineered strain harboring the IspA-Rv1068 fusion product, which was 80.7% of the total FOH production (57). The strategy of using the fusion protein might also be utilized for the production of monoterpenes in microbial hosts, where efficient GPP synthase is not available. The fusion product of monoterpene synthase with IspA may be an interesting platform for excess GPP supply to overproduce monoterpene-based biofuels. It was also reported that the Z,E- isoform of FPP did not restore the growth of the ispA deletion mutant and that it was not metabolized by E. coli for the synthesis of its essential isoprenoids (57). Utilization of isoprenoid precursor isomers also reduced the precursors' flux toward other endogenous isoprenoids. This encourages the recruitment of the precursors' isomers to minimize metabolic wastes during biofuel production.
In order to produce bisabolene, BIS genes from Arabidopsis thaliana, Pseudotsuga menziesii, A. grandis, and P. abies have been screened in the E. coli strain carrying the entire MVA pathway (11). A. grandis α-bisabolene synthase (AgBIS) was found to be the highest producer of bisabolene (78 mg liter−1), and its codon optimization improved bisabolene yield by ∼5-fold in E. coli. For further improvement in bisabolene synthesis, the MVA pathway was divided into (i) an upper module to convert acetyl-CoA into MVA and (ii) a lower module to convert MVA into FPP in E. coli. In addition to pathway modulations, codon optimization of hydroxymethylglutaryl-CoA synthase (HMGS), hydroxymethylglutaryl-CoA reductase (HMGR), MK, and phosphomevalonate kinase (PMK) enhanced bisabolene yield in E. coli. Lastly, the expression of enzymes of the bottom portion of the MVA pathway was optimized by placing the genes under the control of a strong promoter. The final bisabolene titer was measured at 912 mg liter−1, and bisabolene was converted to bisabolane by chemical hydrogenation (11).
The yields obtained through an optimized DXP pathway are not as high as those from the heterologous MVA pathway, as is quite evident from Table 1. However, the efforts to increase the DXP precursors' pool size and bypassing the endogenous regulatory mechanism may contribute to high-level production of isoprenoid-based biofuels. Recently, a novel route was uncovered to enhance the isoprenoid precursors' pool via direct conversion of C5 sugars to DXP (17). Overexpression of E. coli-endogenous ribB mutant ribB(G108S) and yajO genes enabled the direct conversion of ribulose-5-phosphate (Ru5P) to DXP. Expression of the RibB-DXP reductoisomerase (RibB-DXR) fusion product and AgBIS with overexpressed DXP pathway genes (ispA, ispD, ispF, and idi) (Fig. 3) produced a maximum bisabolene titer at ∼9 mg g−1 (DCW) (17). The alternate route from pentose to DXP synthesis may be utilized further to circumvent endogenous regulation of the DXP pathway for enhanced production of isoprenoid-based biofuel and fuel precursors. Most of the biofuels need to be hydrogenated before becoming practical biodiesel alternatives. Identification of crystal structures of geranylgeranyl reductase (GGR) from Sulfolobus acidocaldarius and its engineering to generate sequence variants (91) opened an innovative approach to in vivo reduction of isoprenoid-based biofuels. Enzymatic reduction of double bonds would also eliminate the capital cost associated with the chemical hydrogenation of isoprenoid-based biofuels.
CHALLENGES AND FUTURE PROSPECTS
Over the past few decades, intensive research has been done to synthesize isoprenoids from biological routes (Table 1). Microbial engineering was done via (i) overexpressing endogenous/homologous genes of the DXP pathway, (ii) introducing components of the MVA pathway or the entire MVA pathway, and (iii) utilizing a consortium of homologous/endogenous and heterologous genes to achieve nonnative production as well as improve titers of isoprenoid-based biofuels (10–12, 15, 83). Despite the commercialization of isoprenoids (75, 92, 93), challenges still persist for achieving higher titers of isoprenoid-based biofuels from biological routes, which can be addressed by utilizing various computational, rational, and combinatorial approaches (63, 94).
While biomass is used for biofuel production, the yield of biofuels critically depends upon the conversion of total sugars released from biomass hydrolysis into the final product. Hemicellulosic biomass hydrolysate generally comprises up to 30% of the pentose sugars of the total carbohydrates; hence, strategies to engineer alternate pathways for xylose catabolism in biofuel-producing microbial strains might enhance the precursors' pool inside the cell (17, 95). Strain engineering efforts with balanced enzyme expression and effective recovery strategies can be utilized to achieve higher yields of the biofuels from microbial hosts. The utilization of cyanobacteria has gained considerable popularity in recent years due to their ability to produce liquid fuels directly from the least-complex carbon source, i.e., CO2, and solar energy (14, 96, 97). However, their nutrient requirements (nitrogen and phosphorus) may place an additional strain on the already heavily loaded fertilizer industry. The utilization of cyanobacteria to generate isoprenoid-based biofuels and fuel precursors is still debatable and in the primary stage of investigation. To date, significant improvements have been made in the titers and yields of isoprenoid-based biofuels in engineered microbes. However, there is a long way to go for their commercialization, as the current energy policy relies upon volume rather than the energy content of the fuel. This requires robust fluxomics and metabolomic tools in coordination with bioprocess techniques and needs integration of other engineering disciplines for the conversion of available feedstock into desired biofuels and fuel precursors.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01192-16.
REFERENCES
- 1.Atsumi S, Liao JC. 2008. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19:414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buijs NA, Siewers V, Nielsen J. 2013. Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol 17:480–488. doi: 10.1016/j.cbpa.2013.03.036. [DOI] [PubMed] [Google Scholar]
- 3.Gronenberg LS, Marcheschi RJ, Liao JC. 2013. Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol 17:462–471. doi: 10.1016/j.cbpa.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nozzi NE, Desai SH, Case AE, Atsumi S. 2014. Metabolic engineering for higher alcohol production. Metab Eng 25:174–182. doi: 10.1016/j.ymben.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. 2012. Microbial engineering for the production of advanced biofuels. Nature 488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 6.Wen M, Bond-Watts BB, Chang MCY. 2013. Production of advanced biofuels in engineered E. coli. Curr Opin Chem Biol 17:472–479. doi: 10.1016/j.cbpa.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 7.George KW, Alonso-Gutierrez J, Keasling JD, Lee TS. 2015. Isoprenoid drugs, biofuels, and chemicals-artemisinin, farnesene, and beyond. Adv Biochem Eng Biotechnol 148:355–389. doi: 10.1007/10_2014_288. [DOI] [PubMed] [Google Scholar]
- 8.Gupta P, Phulara SC. 2015. Metabolic engineering for isoprenoid-based biofuel production. J Appl Microbiol 119:605–619. doi: 10.1111/jam.12871. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Rodriguez S, Keasling JD. 2011. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opin Biotechnol 22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD. 2011. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U S A 108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS. 2011. Identification and microbial production of a terpene-based advanced biofuel. Nat Commun 2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Liu Q, Li L, Qin W, Yang J, Zhang H, Jiang X, Cheng T, Liu W, Xu X, Xian M. 2013. Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol Biofuels 6:57. doi: 10.1186/1754-6834-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang M-K, Eom J-H, Kim Y, Um Y, Woo HM. 2014. Biosynthesis of pinene from glucose using metabolically-engineered Corynebacterium glutamicum. Biotechnol Lett 36:2069–2077. doi: 10.1007/s10529-014-1578-2. [DOI] [PubMed] [Google Scholar]
- 14.Halfmann C, Gu L, Zhou R. 2014. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chem 16:3175–3185. doi: 10.1039/C3GC42591F. [DOI] [Google Scholar]
- 15.Phelan RM, Sekurova ON, Keasling JD, Zotchev SB. 2014. Engineering terpene biosynthesis in Streptomyces for production of the advanced biofuel precursor bisabolene. ACS Synth Biol 4:393–399. doi: 10.1021/sb5002517. [DOI] [PubMed] [Google Scholar]
- 16.George KW, Thompson MG, Kang A, Baidoo E, Wang G, Chan LJG, Adams PD, Petzold CJ, Keasling JD, Lee TS. 2015. Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E. coli. Sci Rep 5:11128. doi: 10.1038/srep11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby J, Nishimoto M, Chow RWN, Baidoo EEK, Wang G, Martin J, Schackwitz W, Chan R, Fortman JL, Keasling D. 2015. Enhancing terpene yield from sugars via novel routes to 1-deoxy-d-xylulose 5-phosphate. Appl Environ Microbiol 81:130–138. doi: 10.1128/AEM.02920-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange BM, Rujan T, Martin W, Croteau R. 2000. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A 97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. 2008. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm 5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 20.Breitmaier E. 2006. Terpenes: flavors, fragrances, pharmaca, pheromones. John Wiley & Sons, Weinheim, Germany. [Google Scholar]
- 21.Meylemans HA, Quintana RL, Harvey BG. 2012. Efficient conversion of pure and mixed terpene feedstocks to high density fuels. Fuel 97:560–568. doi: 10.1016/j.fuel.2012.01.062. [DOI] [Google Scholar]
- 22.Hellier P, Al-Haj L, Talibi M, Purton S, Ladommatos N. 2013. Combustion and emissions characterization of terpenes with a view to their biological production in cyanobacteria. Fuel 111:670–688. doi: 10.1016/j.fuel.2013.04.042. [DOI] [Google Scholar]
- 23.Tippmann S, Chen Y, Siewers V, Nielsen J. 2013. From flavors and pharmaceuticals to advanced biofuels: production of isoprenoids in Saccharomyces cerevisiae. Biotechnol J 8:1435–1444. doi: 10.1002/biot.201300028. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Pfeifer BA. 2014. Heterologous production of plant-derived isoprenoid products in microbes and the application of metabolic engineering and synthetic biology. Curr Opin Plant Biol 19:8–13. doi: 10.1016/j.pbi.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 25.McCaskill D, Croteau R. 1997. Prospects for the bioengineering of isoprenoid biosynthesis. Adv Biochem Eng Biotechnol 55:107–146. [DOI] [PubMed] [Google Scholar]
- 26.Chandran SS, Kealey JT, Reeves CD. 2011. Microbial production of isoprenoids. Process Biochem 46:1703–1710. doi: 10.1016/j.procbio.2011.05.012. [DOI] [Google Scholar]
- 27.Keasling JD. 2008. Synthetic biology for synthetic chemistry. ACS Chem Biol 3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Concepción M. 2006. Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev 5:1–15. doi: 10.1007/s11101-005-3130-4. [DOI] [Google Scholar]
- 29.Boucher Y, Doolittle WF. 2000. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol Microbiol 37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 30.Katsuki H, Bloch K. 1967. Studies on the biosynthesis of ergosterol in yeast. Formation of methylated intermediates. J Biol Chem 242:222–227. [PubMed] [Google Scholar]
- 31.Lynen F. 1967. Biosynthetic pathways from acetate to natural products. Pure Appl Chem 14:137–168. [DOI] [PubMed] [Google Scholar]
- 32.Miziorko HM. 2011. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys 505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Yoon S-H, Shah AA, Chung Y-R, Kim J-Y, Choi E-S, Keasling JD, Kim S-W. 2010. Farnesol production from Escherichia coli by harnessing the exogenous mevalonate pathway. Biotechnol Bioeng 107:421–429. doi: 10.1002/bit.22831. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Keasling JD. 2001. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72:408–415. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Ajikumar PK, Xiao W-H, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. 2010. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue J, Ahring BK. 2011. Enhancing isoprene production by genetic modification of the 1-deoxy-d-xylulose-5-phosphate pathway in Bacillus subtilis. Appl Environ Microbiol 77:2399–2405. doi: 10.1128/AEM.02341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Wang Y, Tang Q, Kong W, Chung W-J, Lu T. 2014. MEP pathway-mediated isopentenol production in metabolically engineered Escherichia coli. Microb Cell Fact 13:135. doi: 10.1186/s12934-014-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partow S, Siewers V, Daviet L, Schalk M, Nielsen J. 2012. Reconstruction and evaluation of the synthetic bacterial MEP pathway in Saccharomyces cerevisiae. PLoS One 7:e52498. doi: 10.1371/journal.pone.0052498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlsen S, Ajikumar PK, Formenti LR, Zhou K, Phon TH, Nielsen ML, Lantz AE, Kielland-Brandt MC, Stephanopoulos G. 2013. Heterologous expression and characterization of bacterial 2-C-methyl-d-erythritol-4-phosphate pathway in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:5753–5769. doi: 10.1007/s00253-013-4877-y. [DOI] [PubMed] [Google Scholar]
- 40.Millis J, Maurina-Brunker J, McMullin TW. February 2004. Production of farnesol and geranylgeraniol. US patent US6689593 B2.
- 41.Muramatsu M, Ohto C, Obata S, Sakuradani E, Shimizu S. 2009. Alkaline pH enhances farnesol production by Saccharomyces cerevisiae. J Biosci Bioeng 108:52–55. doi: 10.1016/j.jbiosc.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Ohto C, Muramatsu M, Obata S, Sakuradani E, Shimizu S. 2009. Overexpression of the gene encoding HMG-CoA reductase in Saccharomyces cerevisiae for production of prenyl alcohols. Appl Microbiol Biotechnol 82:837–845. doi: 10.1007/s00253-008-1807-5. [DOI] [PubMed] [Google Scholar]
- 43.Dugar D, Stephanopoulos G. 2011. Relative potential of biosynthetic pathways for biofuels and bio-based products. Nat Biotechnol 29:1074–1078. doi: 10.1038/nbt.2055. [DOI] [PubMed] [Google Scholar]
- 44.Rohmer M, Knani TM, Simonin P, Sutter B, Sahm H. 1993. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz MK. 1994. Terpen-Biosynthese in Ginkgo biloba: eine überraschende Geschichte. Thesis no. 10951 ETH Zürich, Zürich, Switzerland. [Google Scholar]
- 46.Eisenreich W, Menhard B, Hylands PJ, Zenk MH, Bacher A. 1996. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc Natl Acad Sci U S A 93:6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. 1996. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J Am Chem Soc 118:2564–2566. doi: 10.1021/ja9538344. [DOI] [Google Scholar]
- 48.Schwender R, Seemann M, Lichtenthaler HK, Rohmer M. 1996. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem J 316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohdich F, Kis K, Bacher A, Eisenreich W. 2001. The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr Opin Chem Biol 5:535–540. doi: 10.1016/S1367-5931(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 50.Hunter WN. 2007. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J Biol Chem 282:21573–21577. doi: 10.1074/jbc.R700005200. [DOI] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Withers ST, Gottlieb SS, Lieu B, Newman JD, Keasling JD. 2007. Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl Environ Microbiol 73:6277–6283. doi: 10.1128/AEM.00861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou HH, Keasling JD. 2012. Synthetic pathway for production of five-carbon alcohols from isopentenyl diphosphate. Appl Environ Microbiol 78:7849–7855. doi: 10.1128/AEM.01175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujisaki S, Hara H, Nishimura Y, Horiuchi K, Nishino T. 1990. Cloning and nucleotide sequence of the ispA gene responsible for farnesyl diphosphate synthase activity in Escherichia coli. J Biochem 108:995–1000. [DOI] [PubMed] [Google Scholar]
- 56.Ohto C, Muramatsu M, Obata S, Sakuradani E, Shimizu S. 2009. Prenyl alcohol production by expression of exogenous isopentenyl diphosphate isomerase and farnesyl diphosphate synthase genes in Escherichia coli. Biosci Biotechnol Biochem 73:186–188. doi: 10.1271/bbb.80446. [DOI] [PubMed] [Google Scholar]
- 57.Wang C, Zhou J, Jang H, Yoon S, Kim J, Lee G, Choi E, Kim S. 2013. Engineered heterologous FPP synthases-mediated Z,E-FPP synthesis in E. coli. Metab Eng 18:53–59. doi: 10.1016/j.ymben.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Davies FK, Work VH, Beliaev AS, Posewitz MC. 2014. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front Bioeng Biotechnol 2:21. doi: 10.3389/fbioe.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halfmann C, Gu L, Gibbons W, Zhou R. 2014. Genetically engineering cyanobacteria to convert CO2, water, and light into the long-chain hydrocarbon farnesene. Appl Microbiol Biotechnol 98:9869–9877. doi: 10.1007/s00253-014-6118-4. [DOI] [PubMed] [Google Scholar]
- 60.Leonard E, Ajikumar KP, Thayer K, Xiao W, Mo JD, Tidor B, Stephanopoulos GA, Prather KLJ. 2010. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci U S A 107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H, DeLoache WC, Dueber JE. 2012. Spatial organization of enzymes for metabolic engineering. Metab Eng 14:242–251. doi: 10.1016/j.ymben.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Chen AH, Silver PA. 2012. Designing biological compartmentalization. Trends Cell Biol 22:662–670. doi: 10.1016/j.tcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Woolston BM, Edgar S, Stephanopoulos G. 2013. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng 4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 64.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD. 2009. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 65.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. 2011. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 66.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnek J, Tomšič N, Avbelj M, Gaber R, Koprivnjak T, Mori J, Glavnik V, Vovk I, Beninča M, Hodnik V, Anderluh G, Dueber JE, Jerala R, DeLisa MP. 2012. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res 40:1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin W. 2010. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci 365:847–855. doi: 10.1098/rstb.2009.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. 2011. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol 7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foo JL, Leong SSJ. 2013. Directed evolution of an E. coli inner membrane transporter for improved efflux of biofuel molecules. Biotechnol Biofuels 6:81. doi: 10.1186/1754-6834-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee KK, Jang CS, Yoon JY, Kim SY, Kim TH, Ryu KH, Kim W. 2008. Abnormal cell division caused by inclusion bodies in E. coli; increased resistance against external stress. Microbiol Res 163:394–402. doi: 10.1016/j.micres.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Zhou K, Zou R, Stephanopoulos G, Too H-P. 2012. Enhancing solubility of deoxyxylulose phosphate pathway enzymes for microbial isoprenoid production. Microb Cell Fact 11:148. doi: 10.1186/1475-2859-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvey BG, Wright ME, Quintana RL. 2010. High-density renewable fuels based on the selective dimerization of pinenes. Energy Fuels 24:267–273. doi: 10.1021/ef900799c. [DOI] [Google Scholar]
- 73.Yang J, Nie Q, Ren M, Feng H, Jiang X, Zheng Y, Liu M, Zhang H, Xian M. 2013. Metabolic engineering of Escherichia coli for the biosynthesis of alpha-pinene. Biotechnol Biofuels 6:60. doi: 10.1186/1754-6834-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarria S, Wong B, Martín HG, Keasling JD, Peralta-Yahya P. 2014. Microbial synthesis of pinene. ACS Synth Biol 3:466–475. doi: 10.1021/sb4001382. [DOI] [PubMed] [Google Scholar]
- 75.Renninger NS, McPhee DJ. December 2010. Fuel compositions comprising farnesane and farnesane derivatives and method. US patent US7846222 B2.
- 76.McEwen JT, Machado IMP, Connor MR, Atsumi S. 2013. Engineering Synechococcus elongatus PCC 7942 for continuous growth under diurnal conditions. Appl Environ Microbiol 79:1668–1675. doi: 10.1128/AEM.03326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iwaki T, Haranoh K, Inoue N, Kojima K, Satoh R, Nishino T, Wada S, Ihara H, Tsuyama S, Kobayashi H, Wadano A. 2006. Expression of foreign type I ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) stimulates photosynthesis in cyanobacterium Synechococcus PCC7942 cells. Photosynth Res 88:287–297. doi: 10.1007/s11120-006-9048-x. [DOI] [PubMed] [Google Scholar]
- 78.Qiang H, Zarmi Y, Richmond A. 1998. Combined effects of light intensity, light-path and culture density on output rate of Spirulina platensis (Cyanobacteria). Eur J Phycol 33:165–171. doi: 10.1080/09670269810001736663. [DOI] [Google Scholar]
- 79.Onai K, Morishita M, Kaneko T, Tabata S, Ishiura M. 2004. Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: a simple and efficient method for gene transfer. Mol Genet Genomics 271:50–59. doi: 10.1007/s00438-003-0953-9. [DOI] [PubMed] [Google Scholar]
- 80.Wondraczek L, Batentschuk M, Schmidt MA, Borchardt R, Scheiner S, Seemann B, Schweizer P, Brabec CJ. 2013. Solar spectral conversion for improving the photosynthetic activity in algae reactors. Nat Commun 4:2047. doi: 10.1038/ncomms3047. [DOI] [PubMed] [Google Scholar]
- 81.Kang A, George KW, Wang G, Baidoo E, Keasling JD, Lee TS. 2016. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab Eng 34:25–35. doi: 10.1016/j.ymben.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Yang J, Nie Q, Liu H, Xian M, Liu H. 2016. A novel MVA-mediated pathway for isoprene production in engineered E. coli. BMC Biotechnol 16:1. doi: 10.1186/s12896-016-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang C, Yoon S-H, Jang H-J, Chung Y-R, Kim J-Y, Choi E-S, Kim S-W. 2011. Metabolic engineering of Escherichia coli for α-farnesene production. Metab Eng 13:648–655. doi: 10.1016/j.ymben.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 84.Dunlop MJ. 2011. Engineering microbes for tolerance to next-generation biofuels. Biotechnol Biofuels 4:32. doi: 10.1186/1754-6834-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu F, Zhong X, Hu M, Lu L, Deng Z, Liu T. 2014. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol Bioeng 111:1396–1405. doi: 10.1002/bit.25198. [DOI] [PubMed] [Google Scholar]
- 86.Chambon C, Ladeveze V, Oulmouden A, Servouse M, Karst F. 1990. Isolation and properties of yeast mutants affected in farnesyl diphosphate synthetase. Curr Genet 18:41–46. doi: 10.1007/BF00321113. [DOI] [PubMed] [Google Scholar]
- 87.Muramatsu M, Ohto C, Obata S, Sakuradani E, Shimizu S. 2008. Various oils and detergents enhance the microbial production of farnesol and related prenyl alcohols. J Biosci Bioeng 106:263–267. doi: 10.1263/jbb.106.263. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi S, Yeo Y, Greenhagen BT, McMullin T, Song L, Maurina-Brunker J, Rosson R, Noel JP, Chappell J. 2007. Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol Bioeng 97:170–181. doi: 10.1002/bit.21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thulasiram HV, Erickson HK, Poulter CD. 2007. Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science 316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 90.Schulbach MC, Brennan PJ, Crick DC. 2000. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J Biol Chem 275:22876–22881. doi: 10.1074/jbc.M003194200. [DOI] [PubMed] [Google Scholar]
- 91.Kung Y, McAndrew RP, Xie X, Liu CC, Pereira JH, Adams PD, Keasling JD. 2014. Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase. Structure 22:1028–1036. doi: 10.1016/j.str.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 92.Ubersax JA, Platt DM. December 2010. Genetically modified microbes producing isoprenoids. US patent 20100311065A1.
- 93.Marliere P, Anissimova M, Chayot R, Delcourt M. July 2013. Process for the production of isoprenol from mevalonate employing a diphosphomevalonate decarboxylase. US patent 20130052706 A1.
- 94.Lo TM, Teo WS, Ling H, Chen B, Kang A, Chang MW. 2013. Microbial engineering strategies to improve cell viability for biochemical production. Biotechnol Adv 31:903–914. doi: 10.1016/j.biotechadv.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Stephanopoulos G. 2007. Challenges in engineering microbes for biofuels production. Science 315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 96.Machado IMP, Atsumi S. 2012. Cyanobacterial biofuel production. J Biotechnol 162:50–56. doi: 10.1016/j.jbiotec.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 97.Nozzi NE, Oliver JWK, Atsumi S. 2013. Cyanobacteria as a platform for biofuel production. Front Bioeng Biotechnol 1:7. doi: 10.3389/fbioe.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.