ABSTRACT
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain EDL933 harbors multiple prophage-associated open reading frames (ORFs) in its genome which are highly homologous to the chromosomal nanS gene. The latter is part of the nanCMS operon, which is present in most E. coli strains and encodes an esterase which is responsible for the monodeacetylation of 5-N-acetyl-9-O-acetyl neuraminic acid (Neu5,9Ac2). Whereas one prophage-borne ORF (z1466) has been characterized in previous studies, the functions of the other nanS-homologous ORFs are unknown. In the current study, the nanS-homologous ORFs of EDL933 were initially studied in silico. Due to their homology to the chromosomal nanS gene and their location in prophage genomes, we designated them nanS-p and numbered the different nanS-p alleles consecutively from 1 to 10. The two alleles nanS-p2 and nanS-p4 were selected for production of recombinant proteins, their enzymatic activities were investigated, and differences in their temperature optima were found. Furthermore, a function of these enzymes in substrate utilization could be demonstrated using an E. coli C600ΔnanS mutant in a growth medium with Neu5,9Ac2 as the carbon source and supplementation with the different recombinant NanS-p proteins. Moreover, generation of sequential deletions of all nanS-p alleles in strain EDL933 and subsequent growth experiments demonstrated a gene dose effect on the utilization of Neu5,9Ac2. Since Neu5,9Ac2 is an important component of human and animal gut mucus and since the nutrient availability in the large intestine is limited, we hypothesize that the presence of multiple Neu5,9Ac2 esterases provides them a nutrient supply under certain conditions in the large intestine, even if particular prophages are lost.
IMPORTANCE In this study, a group of homologous prophage-borne nanS-p alleles and two of the corresponding enzymes of enterohemorrhagic E. coli (EHEC) O157:H7 strain EDL933 that may be important to provide alternative genes for substrate utilization were characterized.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) bacteria are food-borne pathogens that cause severe human gastrointestinal illness characterized by bloody diarrhea and hemolytic-uremic syndrome (HUS) (1, 2). EHEC are very heterogeneous in their genome sizes and structures as well as in their virulence gene composition (3, 4). The major pathogenicity factors of strains of classical EHEC serogroups such as O157, O26, O111, O145, and O103 are production of one or more Shiga toxins (Stx) and development of attaching and effacing lesions in the human large intestine (5). The latter effect is caused by the translocation of effector proteins into eukaryotic cells by a type III secretion system encoded by the locus of enterocyte effacement (LEE) (6). Besides LEE-positive EHEC strains, LEE-negative EHEC strains such as O113:H21 strain 98NK2 and the O104:H4 clone have caused severe human disease and outbreaks (7, 8). Stx are generally encoded in the genome of lambdoid phages. EHEC strains can carry one or more Shiga toxin-encoding prophages (Stx prophages) in their genome (9, 10). Moreover, a number of functional and cryptic non-Stx prophages have been identified in the EHEC genome. Stx phages generally carry their stx genes in a distinct DNA region downstream of the Q antiterminator and upstream of the S lysis gene, causing production of Stx during expression of the late phage genes and release of Stx during phage-mediated cell damage (11, 12).
In the same DNA region, a large open reading frame (ORF) has been described in many Stx2a-encoding phages (9). In a study by Herold et al. (13), it was shown that the corresponding ORF of phage 933W and other phages was expressed upon norfloxacin induction. Later on, the corresponding 933Wp42 protein was identified as the most prominently regulated protein, with 40-fold overexpression in simulated colonic environmental medium (SCEM) under aerobic conditions in strain EDL933 (14). Consequently, protein 933Wp42 was recombinantly produced from laboratory E. coli strain C600/933W and demonstrated cleavage of 5-N-acetyl-9-O-acetyl neuramininic acid (Neu5,9Ac2) (15). In a recent review, Vimr (16) postulated that chromosomally encoded NanS of E. coli may be responsible for the use of Neu5,9Ac2 as a carbon source. The author demonstrated by in silico analyses that pathogenic E. coli bacteria carry numerous prophage-encoded ORFs which are homologous to the chromosomal nanS (yjhS) gene, which is part of the nanCMS operon of E. coli (16–18).
Landstorfer et al. (19) compared the transcriptome of strain EDL933 grown in LB medium to the transcriptome present after growth in different growth media such as minimal media, radish sprouts, or feces or under different conditions such as the use of differing pH values or antibiotics. Although transcription of the nanS-homologous ORFs was mostly downregulated under the tested conditions, it could be shown that the nanS-homologous ORFs are transcribed under certain conditions.
The major site of infection for EHEC is the large intestine, where carbon sources are limited. Mucus, which is composed of mucins containing various carbohydrates such as galactose, hexosamines, fucose, and neuraminic (sialic) acids, is thought to be one of the most important sources of energy supply. While highly fucosylated glycans were mainly found in the small intestine of human, the content of sialic acids increases along the colon (20). Following Neu5Ac, Neu5,9Ac2 is the most frequently occurring neuraminic acid derivative in humans (16).
E. coli O157:H7 strain EDL933 was isolated from ground beef in 1982, during an EHEC outbreak in the United States (21). Since that time, EDL933 has been used as a reference EHEC strain in many laboratories worldwide. The genome sequence of EDL933 was first published in 2001 (22). Besides potential virulence factors and different metabolic pathways, 18 prophages were described. The genome of this strain has been resequenced by next-generation sequencing and has been published recently (23). The authors corrected the genome sequence by eliminating ambiguous base calls, closing a chromosomal gap, and revising the prophage regions. Therefore, a number of differences from the sequence published by Perna et al. (22) were found.
The aims of the current study were to identify, quantify, and analyze the nanS-homologous ORFs in the genome of EDL933 by in silico analyses, to clone selected nanS-homologous ORFs, and to determine their enzymatic activity. Generating isogenic and sequential deletions of the nanS-homologous ORFs demonstrated their importance for substrate utilization.
MATERIALS AND METHODS
Nomenclature of prophage-encoded nanS-homologous ORFs and use of a particular EDL933 derivative.
The genome sequence of E. coli O157:H7 strain EDL933 was annotated twice (National Center of Biological Information [http://www.ncbi.nlm.nih.gov/] accession numbers AE005174.2 and NZ_CP008957.1). The experiments of this study were initially performed using the prior sequence published by Perna et al. (22) (AE005174.2). Therefore, many of the primers used in this study (see Table S1 in the supplemental material) were named according to the locus tags used in that sequence entry. Recent work was based on this sequence and the nomenclature that we used was also applied accordingly, e.g., gene locus z1466, plasmid pET-z1466-his (13–15).
Following the naming scheme used in the more recently reported sequence (NZ_CP008957.1), we renamed the corresponding loci. Due to their homologies to the chromosomal nanS gene and their prophage locations, we termed the prophage-encoded nanS-homologous genes the nanS-p genes and numbered them in accordance with the order in which they were deleted in our study (see below) (Table 1). The nanS-p8, nanS-p9, and nanS-p10 alleles published by Latif et al. (23) were not present in the genome of our EDL933 laboratory strain derivative (see below), and the numbering of these was determined according to their appearance in the genome sequence. The nanS-p1 allele is synonymous with the ORF formerly named z1466, but when materials from former work were used in this study, we kept this name.
TABLE 1.
Characteristics of nanS and nanS-p1-p10 and corresponding proteins in Escherichia coli O157:H7 strain EDL933 (accession no. NZ_CP008957.1)a
| Phylogroup | Gene designation | Gene length (bp) | Chromosomal position in EDL933 (NZ_CP008957) | Designation of corresponding protein | Protein length (aa) |
|---|---|---|---|---|---|
| nanS | 981 | 5440406–5441386 | NanS | 326 | |
| 1 | nanS-p1 (z1466) | 1,938 | 1356672–1358609 | NanS-p1 (933Wp42) | 645 |
| 1 | nanS-p2 | 1,947 | 3010352–3012298 | NanS-p2 | 648 |
| 3 | nanS-p3 | 1,851 | 1267489–1269339 | * | 616 |
| 3 | nanS-p4 | 1,851 | 2332008–2333858 | NanS-p4 | 616 |
| 3 | nanS-p5 | 1,854 | 3571915–3573768 | * | 617 |
| 3 | nanS-p6 | 1,854 | 1859884–1861737 | * | 617 |
| 3 | nanS-p7 | 1,851 | 2789290–2791140 | * | 616 |
| 2 | nanS-p8 | 1,938 | 1642848–1644785 | * | 645 |
| 3 | nanS-p9 | 1,851 | 1907347–1909197 | * | 616 |
| 3 | nanS-p10 | 1,854 | 2152546–2154399 | * | 617 |
*, not analyzed in this study. Italic or boldface data represent nucleotide and amino acid sequences that are altered or not present, respectively, in the EDL933 strain used in this study (accession no. CP015855).
Bioinformatic methods.
The nanS-p alleles in the E. coli O157:H7 strain EDL933 genome were analyzed by BLAST analysis (24), and the prophage regions were aligned and depicted with ClustalW using Geneious R9 software (Biomatters Ltd., Auckland, New Zealand). ln order to identify putative functional domains in the proteins, the NCBI BLASTP suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used. To analyze the phylogenetic relationships among NanS-p amino acid sequences in E. coli strain EDL933, a tree was calculated with the unweighted pair group method using average linkages (UPGMA) and Mega6 (25) from a Clustal alignment calculated with Bioedit, version 7.2.5, based on the amino acid sequences (26). The NanS sequence of EDL933 was used as an outgroup.
Bacterial strains, growth media, plasmids, and genome sequencing.
Bacterial strains and plasmids used in this study are listed in Table 2. Plasmids were prepared with a QlAprep Spin Miniprep kit (Qiagen) according to the manufacturer's recommendation. Genomic DNA of EDL933 was prepared with a GenElute bacterial genomic DNA kit from Sigma-Aldrich according to the manufacturer's instructions and was sequenced on a PacBio RS2 device (Pacific Biosciences, Menlo Park, CA, USA) with a 10-kb size-selected insertion library and P6/C4 chemistry. One SMRT cell was used. De novo assembly (HGAP3 algorithm) was performed using SMRT Analysis version 2.3 software (Pacific Biosciences). HGAP3 settings were kept at the defaults, except for the expected genome size of 5 Mbp. LB medium was used for all routine purposes and consisted of 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl. The pH was adjusted to 7.0 with 1 N NaOH. The medium was autoclaved at 121°C for 15 min. Kanamycin sulfate (Sigma-Aldrich) or ampicillin sodium salt (Roth) was added at a final concentration of 50 μg/ml or 100 μg/ml, respectively, when required.
TABLE 2.
Bacterial strains and plasmids used in this study
| Bacterial strain | Relevant characteristic(s)/plasmid | Reference or source |
|---|---|---|
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | 44 |
| E. coli BL21(DE3)/pET-z1466-his | 15 | |
| E. coli BL21(DE3)/pET-22b(+) | Donor for plasmid pET-22b(+) (Novagen, Merck) | 15 |
| E. coli BL21(DE3)/pET-nanS-p2-his | This study | |
| E. coli BL21(DE3)/pET-nanS-p4-his | This study | |
| E. coli JM 109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 λ− Δ(lac-proAB) [F− traD36 proAB lacIq lacZΔM15] | 45 |
| E. coli BW25113/pKD46 | Donor for plasmid pKD46 | 46 |
| E. coli BW25141/pKD4 | Donor for plasmid pKD4 | 46 |
| E. coli BT340 | E. coli DH5α carrying pCP20 | 47 |
| E. coli C600 | Wild type | 48 |
| E. coli C600ΔnanS | Deletion of nanS | This study |
| E. coli EDL933 | Wild type | 49 |
| E. coli EDL933ΔnanS | Deletion of nanS | This study |
| E. coli EDL933ΔnanSΔnanS-p1a | Deletion of nanS, nanS-p1a | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p2 | Deletion of nanS, nanS-p1a, nanS-p2 | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p3 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3 | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p4 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3, nanS-p4 | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p5 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3, nanS-p4, nanS-p5 | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p6 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3, nanS-p4, nanS-p5, nanS-p6 | This study |
| E. coli EDL933ΔnanSΔnanS-p1a-p7 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3, nanS-p4, nanS-p5, nanS-p6, nanS-p7 | This study |
| E. coli EDL933ΔnanSΔnanS-p6 | Deletion of nanS, nanS-p6 | This study |
M9 minimal medium was prepared basically as described previously (27). Casamino Acids (Difco) were added to reach a final concentration of 0.1% (wt/vol), and glucose, Neu5,9Ac2, or Neu5Ac was added from sterile stock solutions, when required, to reach a final concentration of 0.4% (wt/vol). For growth of E. coli C600 and derivatives, the medium was supplemented with leucine, threonine, and thiamine solutions at a final concentration of 0.0001% (wt/vol). SOC medium was prepared as described elsewhere (27).
Production of electrocompetent bacterial cells and electroporation.
For transformation experiments, overnight cultures (LB medium, 37°C, 180 rpm) of E. coli BL21(DE3), E. coli JM109, E. coli C600, or EDL933 or the corresponding deletion mutants were diluted 1:100 in 50 ml SOC medium and the cultures were incubated at 37°C and 180 rpm up to an optical density at 600 nm of 0.5 to 0.7. If strains contained temperature-sensitive plasmids, they were grown at 30°C. The cultures were cooled on ice for 30 min, and the following steps were performed at 2°C: the cells were harvested by centrifugation at 4,000 × g for 8 min and washed first with 40 ml sterile ultrapure water (Millipore), then with 20 ml sterile ultrapure water, and finally with 2 ml sterile ultrapure water containing 10% (vol/vol) glycerol. The resulting cell pellets were dissolved in 100 μl sterile ultrapure water containing 10% glycerol. Aliquots (40 μl) of competent cells were used immediately for transformation or stored as electrocompetent cells at −70°C until use.
For transformation experiments, 40-μl volumes of electrocompetent cells were thawed on ice and mixed with 30 to 300 ng DNA in an ice-cold Gene Pulser/MicroPulser cuvette (Bio-Rad). The mixture was pulsed in a Gene Pulser Xcell electroporation system (Bio-Rad) at 25 μF, 200 Ω, and 2.5 kW for 5 ms. Following this, 1 ml of prewarmed SOC medium was added immediately. The suspension was then incubated for 1 h at 37°C (or at 30°C if temperature-sensitive plasmids were present) and then spread on LB agar with or without antibiotics.
Cloning of nanS-p alleles and recombinant production of NanS-p proteins.
PCR products, which were used not only for confirmation but also for further steps, were purified using a Qiagen QlAquick PCR purification kit (Qiagen), and DNA concentrations were determined with a Nanodrop 2000 device (Thermo Scientific).
The nanS-p4 allele of E. coli O157:H7 strain EDL933 was amplified by PCR with primers z6054-NdeI-for and z6054-XhoI-rev (see Table S1 in the supplemental material).
The nanS-p2 allele showed high sequence similarity to nanS-p1. Therefore, it was amplified in a nested PCR approach using primers z3342-for and z3342-rev (see Table S1 in the supplemental material). Since nanS-p2 contains a NdeI restriction site, this site had to be removed to facilitate cloning. Therefore, the nucleotide sequence of the restriction site was modified by site-directed mutagenesis (28). Briefly, A-tailing with Taq polymerase modified the first-round nested PCR product of nanS-p2 according to the specifications of the manufacturer (New England BioLabs). It was then ligated to vector pGEM-T Easy (Promega) and transformed (see above) in E. coli JM109 according to the manufacturer's specifications. Correct insertion was verified by PCR with oligonucleotides M13-f and M13-r (see Table S1). A nucleotide exchange of adenine to cytosine was performed by PCR with primers z3342-18A/C-for and z3342-13T/G-rev at position 312 of nanS-p2, using Phusion High-Fidelity DNA polymerase (Thermo Scientific) (see Table S1). This procedure was modified from that described in a QuikChange II site-directed mutagenesis kit instruction manual (Agilent Technologies). The former NdeI restriction site changed to 5′-CCTATG-3′, which did not result in an amino acid change. A digestion with NdeI was performed subsequently (by the method described in a Thermo Scientific manual) to cut the unchanged template plasmid. The resulting target plasmid was used as the template for amplification of nanS-p2 with primers z3342-NdeI-for and z3342-XhoI-rev (see Table S1).
The PCR products of each approach were ligated into expression vector pET22b(+) as described elsewhere (17) and transformed into E. coli BL21(DE3) (see above). Correct insertion of the genes into pET22b(+) and the corresponding nucleotide sequences were confirmed by sequencing both DNA strands.
The resulting recombinant E. coli BL21(DE3)/pET-nanS-p2-his and BL21(DE3)/pET-nanS-p4-his strains, as well as the previously described BL21(DE3)/pET-z1466-his strain and the negative-control BL21(DE3)/pET-22b(+) strain (15), were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), harvested, and lysed, and the resulting protein solutions were purified and confirmed by Western blotting with an anti-His-tag antibody as described previously (15). The protein solutions were dialyzed in Slide-A-Lyzer dialysis cassettes (Thermo Scientific) (3.5-K-molecular-weight cutoff) in sodium-potassium buffer (10 mM Na2HPO4, 10 mM KH2PO4, pH 7.0) at 4°C in a 2.5-liter volume. The buffer was changed after 16 h, and the dialysis process continued for a further 4 h. The resulting protein preparations were designated NanS-p2-His and NanS-p4-His in addition to 933Wp42-His and the no-insertion control (NIC). The determination of protein concentrations by Bradford protein assay was carried out as previously described (15) using bovine serum albumin as a standard. The protein solutions were stored at −70°C.
Determination of the esterase activity of recombinant NanS-p proteins using Neu5,9Ac2 and mucin.
5-N-Acetyl-9-O-acetyl neuraminic acid (Neu5,9Ac2) was provided by Wolfgang Fessner and Ning He, Technical University Darmstadt, Darmstadt, Germany. It was synthesized as described previously (29) and analyzed by nuclear magnetic resonance (NMR) spectroscopy. The purity was >95%. 5-N-Acetyl neuraminic acid (Neu5Ac) was purchased from Applied BioTech, Austria. Neu5,9Ac2 and Neu5Ac were used at a concentration of 6.25 mg/ml, corresponding to molar concentrations of 17.79 and 20.21 mmol, respectively, in 50 mM Tris-HCl (pH 8.0). Mucin from porcine stomach (Sigma-Aldrich) and mucin from bovine submaxillary gland (Merck Millipore), both dissolved in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2HPO4, pH 7.4), were used at a final concentration of 12.5 mg/ml. NanS-p2-His and NanS-p4-His as well as the 933Wp42-His positive control (15) and the NIC (see above) were diluted in 10 mM sodium potassium buffer (pH 7.0) to the concentrations used. The final protein concentrations used were 0.18 mg/liter for deacetylation of Neu5,9Ac2 and 6.7 mg/liter for deacetylation of mucin. In this case, 1 katal (kat) esterase activity was defined as the release of 1 mol acetic acid per second.
Mixtures of 100 μl Neu5,9Ac2 or the mucin solutions, respectively, were preincubated at 37°C. The reactions were started by adding 5 μl of the diluted protein solutions of the recombinant proteins or the positive control, respectively, with shaking at 300 rpm for 20 min. All assays were performed in triplicate. The reactions were stopped at 95°C and 500 rpm for 10 min. The released acetic acid concentrations were detected with a commercial kit (acetic acid; Boehringer Mannheim/R-Biopharm) according to the manufacturer's instructions. Neu5Ac and the NIC were used as negative controls.
The measurement range was 999 to 4,995 μmol acetic acid according to the instructions of the manufacturer. Amounts of substances below those values were assessed as representing negative conditions.
Biochemical characterization of recombinant NanS-p esterases.
Esterase activity was determined by measuring hydrolysis of 4-methylumbelliferyl acetate (4-MUF-Ac) (Sigma-Aldrich) for investigation of the pH and temperature optima as described by Nübling et al. (15) with minor modifications as described below. The absorption measurements were carried out in an Infinite M200 microplate reader (Tecan). The 4-MUF-Ac stock solution was 5 mM. The buffers used had a molar concentration of 50 mM. Briefly, 80 μl of the corresponding recombinant esterase solution was preincubated for 4 min in 352 μl of the corresponding buffer system at the corresponding temperature. The reaction was started by adding 48 μl substrate stock solution and stopped by adding 100 μl 50% (vol/vol) acetic acid solution. A pH range of 4.0 to 9.0 in acetate buffer (pH 4.0 to 5.0), potassium phosphate buffer (pH 5.0 to 7.0), or Tris-HCl (pH 7.0 to 9.0), and a temperature range of 10 to 70°C were tested. The measurement of pH optimum was performed at 20°C or 40°C for NanS-p4-His or 933Wp42-His and NanS-p2-His, respectively. For detection of the temperature optima, a Tris-HCl (pH 7.0) buffer system was used. For the investigation of substrate specificity, 4-methylumbelliferyl caprylate (Santa Cruz Biotechnology), 4-methylumbelliferyl butyrate (Sigma-Aldrich), and 4-MUF-Ac were used as potential substrates. A commercial lipase (from Candida rugosa; Sigma-Aldrich) served as a positive control.
All measurements were performed in triplicate. Percentages of relative activity were calculated by assigning the highest level of enzymatic activity a value of 100%. For calculation of the enzymatic activity, determinations of calibration curves between 0 and 90 μmol 4-methylumbelliferone (Sigma-Aldrich) in each buffer system at each pH were performed. One katal (kat) of esteraseMuf-Ac activity was defined as the release of 1 mol 4-methylumbelliferone within 1 s. For determinations of specific esteraseMuf-Ac activity (measured as nanokatals per milligram), the activity (in nanokatals) was quantified in reference to 1 mg protein. The limit of quantification (LOQ) was calculated by multiplying the standard deviation of the blank values by a factor of 9.
Construction of nanS- and isogenic sequential nanS-p deletion mutants and growth experiments.
Deletion mutagenesis was performed according to the method described by Datsenko and Wanner (28) using plasmids pKD46, pKD4, and pCP20 (Table 2). Primer pairs and PCR programs for deletion of nanS in E. coli C600 and EDL933 and sequential deletion of nanS-p1a to nanS-p7 in strain EDL933 are listed in Table S1 in the supplemental material. PCR products were purified and the DNA concentration was measured as described above. Deletions were confirmed by PCR with primer pairs surrounding the position where the deletion had taken place (see Table S1).
The resulting E. coli C600ΔnanS mutant strain was grown at an initial optical density at 600 nm of 0.1 in 5 ml M9 minimal medium with Neu5,9Ac2 as a carbon source (see above) and a supplementation of 4 μg/ml NanS-p2-His, NanS-p4-His, or 933Wp42-His at 37°C and 180 rpm and compared to controls without supplement. Similarly, EDL933 and its mutant derivatives (Table 2) were cultivated with Neu5,9Ac2, Neu5Ac, or glucose or without a carbohydrate source. The mutant strain EDL933ΔnanSΔnanS-p1a-p7, with eight deletions, was cultivated with Neu5,9Ac2 and with supplemental recombinant protein. Growth experiments were generally performed in triplicate with two measurements of optical density each per measurement point. Complementation experiments were performed in duplicate for the EDL933 deletion mutants.
Accession number(s).
The sequence determined in this work was deposited in the NCBI database under accession no. CP015855 and organism designation Escherichia coli EDL933-1.
RESULTS
Bioinformatic analyses of the nanS-p alleles.
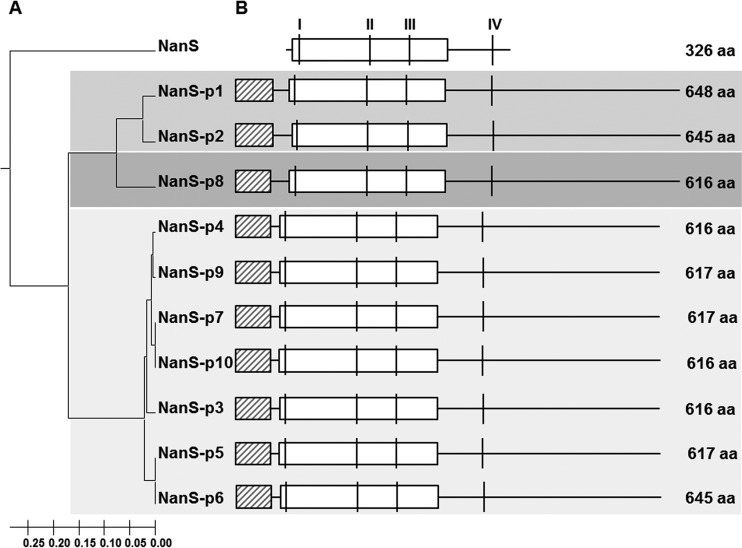
The E. coli O157:H7 strain EDL933 genome as published by Latif et al. (23) contains a single chromosomal nanS gene and 10 nanS-p alleles (nanS-p1 to nanS-p10) (Fig. 1), the latter with high nucleotide sequence similarities to nanS. All nanS-p alleles in E. coli EDL933 are located in the genome of functional or cryptic lambdoid prophages. They are located between the respective late antiterminator Q gene and the lysis S gene, and the DNA region surrounding nanS-p is conserved in E. coli EDL933 (Fig. 2).
FIG 1.
Phylogenetic analysis and structure of NanS and NanS-p protein sequences in E. coli O157:H7 strain EDL933. (A) The UPGMA tree of NanS and the NanS-p family was calculated with Mega6 (25) based on the amino acid (aa) sequences. (B) Structural organization of NanS and the NanS-p family. Potential functional domains (DUF, domain of unknown function) were predicted using BLAST. Horizontal boxes with black diagonal stripes depict DUF1737; white horizontal boxes indicate DUF303. Amino acid sequence blocks typical of the SGNH enzyme family of hydrolases as described previously (18) are depicted as vertical lines I, II, III, and IV. Thin horizontal continuous lines illustrate continuous amino acid chains.
FIG 2.
Comparison of the prophage regions between genes Q and S in E. coli O157:H7 strain EDL933. ClustalW alignment of prophage regions was performed using Geneious R9 software. Blank and pink arrows depict different prophage genes and transfer ribonucleic acids (tRNA), respectively. The compared prophage regions are flanked by an antiterminator Q gene and a lysis S gene. The stx1A and stx1B genes as well as the stx2A and stx2B genes are labeled.
Sequence and phylogenetic analyses revealed that the corresponding NanS-p proteins of E. coli EDL933 cluster into three phylogroups (Fig. 1A and Table 1). NanS-p1 and NanS-p2, belonging to group I, are encoded by nanS-p1 and nanS-p2, located in close proximity to and downstream of stx2a and stx1a in the corresponding prophage genomes, and are 645 and 648 amino acids in length, respectively. They share overall amino acid similarity of 91.5%. Differences occur mainly between amino acids 34 and 78. NanS-p8 belongs to group II, with a size of 645 amino acids. The corresponding proteins of the seven nanS-p alleles nanS-p3 to nanS-p7, nanS-p9, and nanS-p10 belong to group III. In this group, the proteins consist of 616 or 617 amino acids and share sequence similarity of 95.0% to 99.7%.
The chromosomally encoded NanS is 326 amino acids in length and carries domain of unknown function 303 (DUF303), which has been previously linked to its esterase activity (18). In addition, NanS harbors four sequence blocks (blocks I to IV) (Fig. 1B) which have been described for the SGNH superfamily of enzymes (18). All NanS-p proteins also contain a DUF303 superfamily domain and possess those conserved sequence blocks which are present in enzymes of the SGNH superfamily. Moreover, the NanS-p proteins harbor a conserved DUF1737 domain of unknown function at their N-terminal ends. The DUF1737 sequence is similar to the CIII protein sequence of phage lambda from amino acid 14 to amino acid 37 which is the essential region for the activity of λCIII (30).
Since the genome sequence reported by Latif et al. (23) was generated with an EDL933 derivative deposited in the American Type Culture Collection (ATCC) and since we used a derivative that has been propagated over 30 years in German laboratories, we found some differences and decided to determine the genome sequence of our EDL933 derivative. A total of 63,172 reads with 651 Mbp were produced. The HGAP3 analysis produced a complete de novo genome sequence with 100-fold coverage over the entire molecules. Although we found the sequence to be mostly in accordance to the recently described EDL933 genome sequence (NZ_CP008957.1), there are some differences concerning the nanS-p alleles. Besides nanS, we discovered only 7 nanS-p alleles (nanS-p1 to nanS-p7) instead of the 10 present in the Latif genome. Alleles nanS-p8 to nanS-p10 were not present. The nanS-p1 allele (formerly z1466) which is located on the stx2a prophage is slightly altered in its sequence in comparison with the z1466 sequence present in 933W. We designated it nanS-p1a. Alleles nanS-p2, nanS-p3, nanS-p4, nanS-p5, nanS-p6, and nanS-p7 showed the same nucleotide sequences as those previously published (23).
Functional characterization of recombinant NanS-p2 and NanS-p4 proteins.
In order to characterize some NanS-p proteins in E. coli O157:H7 strain EDL933 enzymatically, we selected nanS-p2 and nanS-p4 for cloning. The nanS-p2 allele, which is located in the genome of the Stx1a-encoding phage (15), is highly homologous to the already described z1466 (nanS-p1) and belongs to group I. NanS-p4 belong to the more distantly related group III (Fig. 1). Since nanS-p8 was not present in our EDL933 laboratory strain, we did not analyze the group II protein. The selected nanS-p alleles were cloned into vector pET-22b(+), transformed into E. coli BL21(DE3), expressed as His-tagged proteins, and purified. The resulting proteins were designated NanS-p2-His and NanS-p4-His. For comparison purposes, 933Wp42-His (15) was used as a positive control in all experiments. Polyacrylamide gel electrophoresis of all preparations resulted in large (approximately 70-kDa) bands for all recombinant proteins. Western blotting with an anti-His-tag antibody demonstrated that all homologous proteins could be detected with high specificity (data not shown).
Characterization of the esterase activity of NanS-p2-His and NanS-p4-His.
To demonstrate the esterase activity of NanS-p2-His and NanS-p4-His, cleavage of acetic acid from Neu5,9Ac2 and from two different mucins was investigated with a commercial acetic acid detection assay. Release of acetic acid from Neu5,9Ac2 demonstrated enzymatic activities of 7,578 ± 637 nkat/mg, 9,231 ± 300 nkat/mg, and 7,882 ± 294 nkat/mg for NanS-p2-His, NanS-p4-His, and the 933Wp42-His control, respectively, after incubation with the putative recombinant esterases. This result demonstrated slight differences in the esterase activities of the recombinant enzymes tested. Using the NIC, the free acetic acid molar concentration was below the recommended measurement range of the assay and was therefore considered a negative result. Moreover, acetic acid release from Neu5Ac was also considered a negative result. These results confirmed the assumption that NanS-p2-His and NanS-p4-His as well as 933Wp42-His (15) possess an O-acetyl esterase activity and cleave the acetyl group from C9 of Neu5,9Ac2.
However, differences in the amounts of released acetic acid were observed when porcine stomach mucin and mucin from bovine submaxillary glands were used as substrates. Release of acetic acid from mucin from bovine submaxillary gland demonstrated specific enzymatic activities of 193 ± 12 nkat/mg, 196 ± 5 nkat/mg, and 200 ± 8 nkat/mg for NanS-p2-His, NanS-p4-His, and 933Wp42-His, respectively. Negative-control results were below the recommended measurement range.
In contrast, acetic acid release could not be detected after incubation of mucin from porcine stomach with the Neu5,9Ac2 esterases. Kit controls were tested and gave positive results; thus, the possibility of any disturbing interaction between kit components and samples could be excluded.
Determination of characteristic enzymatic properties of the selected recombinant Neu5,9Ac2 esterases.
Due to the amounts of the different nanS-p alleles that were present in E. coli EDL933 and their proved Neu5,9Ac2 esterase activity, we hypothesize that the different Neu5,9Ac2 esterases of E. coli EDL933 may act under different environmental conditions. Initially, standard enzymatic tests were performed as described earlier (15). Using 4-methylumbelliferyl acetate, 4-methylumbelliferyl butyrate, and 4-methylumbelliferyl caprylate as synthetic esterase substrates, it was shown that NanS-p2-His, NanS-p4-His, and the positive control 933Wp42-His could cleave the acetic acid residues from 4-methylumbelliferyl acetate but not from 4-methylumbelliferyl butyrate and 4-methylumbelliferyl caprylate. Lipase activity could not be detected for any of the enzymes (data not shown).
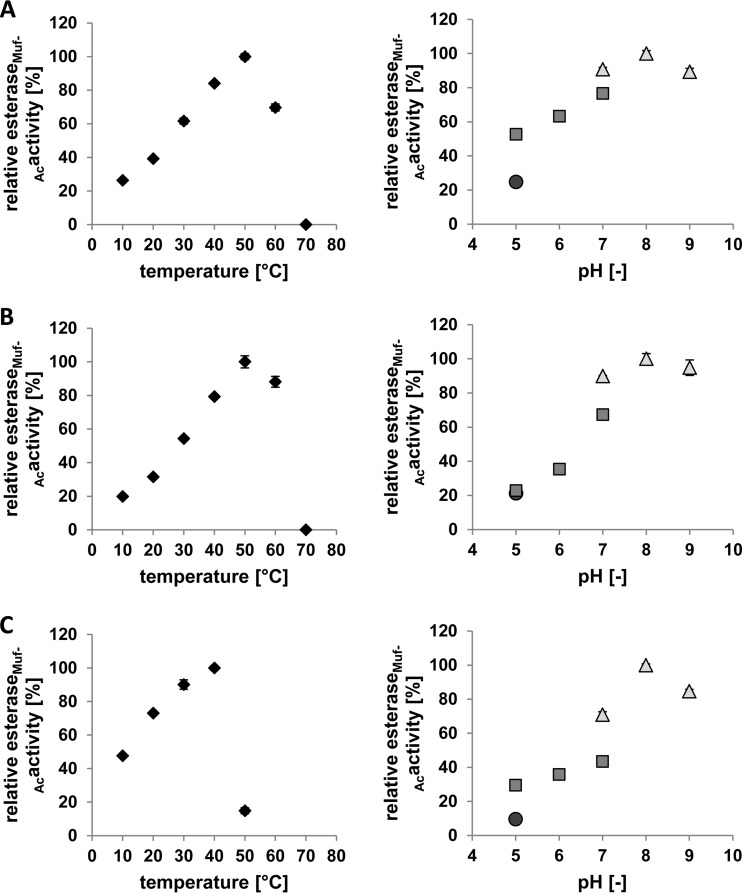
In a next step, the temperature optima of the 3 Neu5,9Ac2 esterases were determined in a range between 10°C to 70°C and the pH optimum was measured in a range between 4.0 and 9.0 in different buffer systems (Fig. 3). For NanS-p2-His and 933Wp42-His, a temperature of 50°C was determined to be optimal (Fig. 3A and B). Moreover, for NanS-p4-His, the temperature optimum was 40°C (Fig. 3C). The optimal pH range determined for all Neu5,9Ac2 esterases in Tris-HCl buffer was between 7 and 9 (Fig. 3).
FIG 3.
Measurement of temperature and pH optima of NanS-p2-His (A), 933Wp42-His (B), and NanS-p4-His (C). The substrate was 4-methylumbelliferyl acetate. Temperature conditions for measurement of pH optimum and buffer system (dot, acetate; square, potassium phosphate; triangle, Tris-HCl) were 40°C or 20°C for panels A and B or panel C, respectively. The temperature optimum was detected in Tris-HCl buffer (pH 7.0). In the case of temperature optima, 100% relative esteraseMuf-Ac activity data represent 50.9 ± 0.9 nkat/mg (A), 35.3 ± 1.3 nkat/mg (B), and 53.1 ± 0.7 nkat/mg (C). In the case of pH optima, 100% relative esteraseMuf-Ac activity data represent 41.5 ± 0.7 nkat/mg (A), 30.3 ± 0.9 nkat/mg (B), and 52.7 ± 0.4 nkat/mg (C).
Supplementation of growth medium with recombinant Neu5,9Ac2 esterases facilitates growth of E. coli C600ΔnanS.
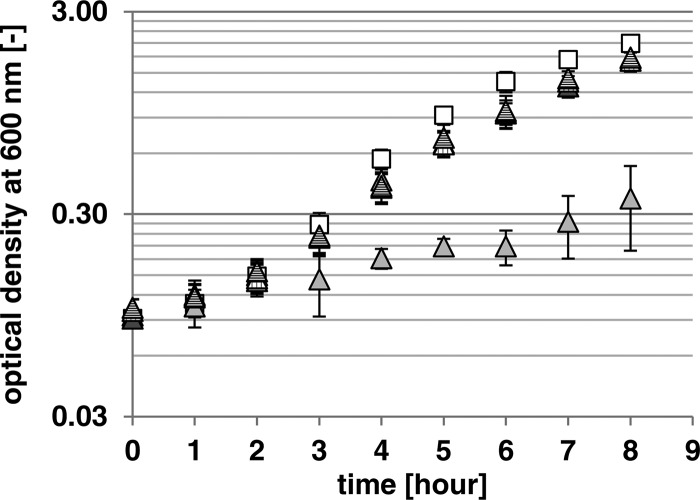
To test the hypothesis that the enzymatic activity of the prophage-encoded Neu5,9Ac2 esterases allows growth of E. coli on Neu5,9Ac2 as a carbon source, an E. coli C600 derivative lacking the chromosomal nanS gene was constructed and designated mutant strain C600ΔnanS. The wild-type E. coli C600 strain grew well in M9 minimal medium containing Neu5,9Ac2 as a sole carbon source until an optical density at 600 nm of 2.1 was reached (Fig. 4).
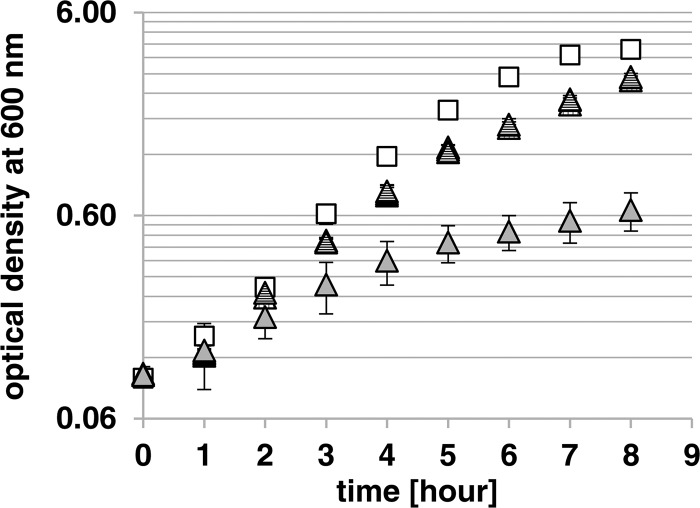
FIG 4.
Supplementation of the growth medium of E. coli C600ΔnanS with the different recombinant NanS-p proteins. Data represent the results of growth of E. coli C600 (white squares) and the C600ΔnanS mutant (light gray triangles) on 0.4% Neu5,9Ac2 in M9 minimal medium at 37°C and 180 rpm and of growth of the C600ΔnanS mutant with 4 μg/ml supplemented 933Wp42-His, NanS-p2-His, and NanS-p4-His (triangles with vertical stripes, dark gray triangles, and triangles with horizontal stripes, respectively).
In contrast, the C600ΔnanS deletion mutant grew only minimally in the medium (Fig. 4). Given that there should have been no growth, since nanS was deleted, the weak growth was probably due to the presence of a low concentration of Casamino Acids in the medium. Furthermore, the possibility cannot be excluded that there were side activities of other enzymes present which were produced from the strain. After addition of each of the Neu5,9Ac2 esterases to the growth medium, the C600ΔnanS mutant regained the ability to grow on Neu5,9Ac2. The growth rate and the obtained optical density were nearly identical to those seen with the E. coli C600 wild-type strain, suggesting that the Neu5,9Ac2 esterases can complement the chromosomal NanS function.
Sequential deletion of nanS-p alleles induces loss of the ability of EDL933 to grown on Neu5,9Ac2.
Deletion mutagenesis in EDL933 was performed sequentially starting with the deletion of the chromosomal nanS gene. Subsequently, nanS-p1a, nanS-p2, nanS-p3, nanS-p4, nanS-p5, nanS-p6, and nanS-p7 were deleted in the same strain. This resulted in a mutant of EDL933, designated mutant strain EDL933ΔnanSΔnanS-p1a-p7, which had eight deletions and which did not contain any nanS-p allele. Each deletion step was confirmed by PCR analysis. Furthermore, after deletion of all nanS-p alleles, we double-checked the number of homologs by PCR using primers which bind in all nanS-p alleles that are present in the EDL933 genome sequence (NZ_CP008957.1). We confirmed that there were no further nanS-p alleles in the EDL933ΔnanSΔnanS-p1a-p7 mutant strain.
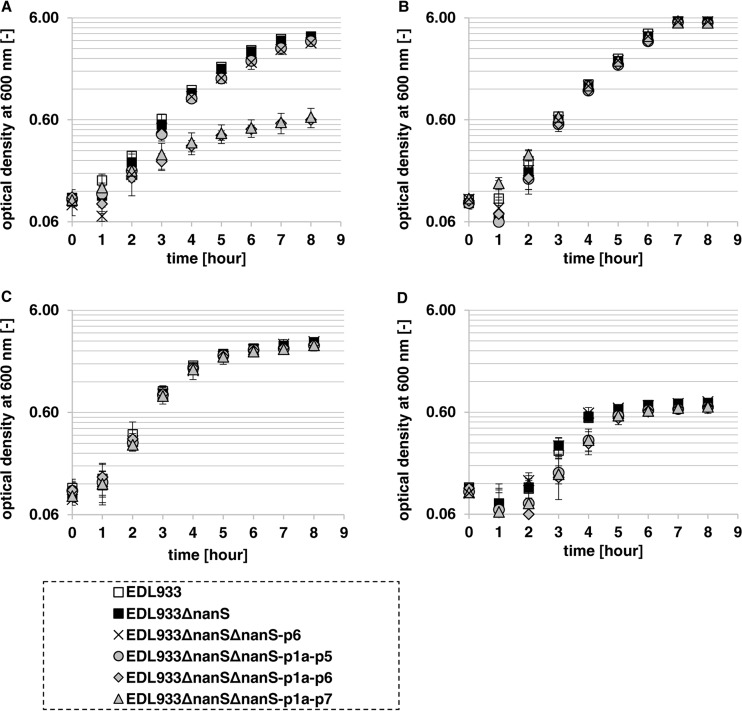
Preliminary tests did not show noteworthy growth defects on Neu5,9Ac2 of mutant strains EDL933ΔnanSΔnanS-p1a-p2, EDL933ΔnanSΔnanS-p1a-p3, EDL933ΔnanSΔnanS-p1a-p4, and EDL933ΔnanSΔnanS-p1a-p5 compared to the wild type (data not shown). Therefore, we performed an experiment in which we grew mutant strains EDL933, EDL933ΔnanS, EDL933ΔnanSΔnanS-p1a-p5, EDL933ΔnanSΔnanS-p1a-p6, and EDL933ΔnanSΔnanS-p1a-p7 in M9 minimal medium with Neu5,9Ac2 as a carbon source (Fig. 5A). Mutant strains EDL933ΔnanS and EDL933ΔnanSΔnanS-p1a-p5 grew just as well as the EDL933 wild-type strain. Interestingly, starting with the 7-fold deletion mutant, EDL933ΔnanSΔnanS-p1a-p6, the strain lost its ability to grow on Neu5,9Ac2, although nanS-p7 was still present. To exclude any undesired defects in the nan operons or in growth ability in general, we let this strain and the other mutant strains grow on Neu5Ac and glucose in parallel (Fig. 5B and C). To test whether the growth defect of mutant strain EDL933ΔnanSΔnanS-p1a-p6 was attributable to the deletion of nanS-p6 or whether it was a consequence of the number of nanS-p alleles that had been deleted, we constructed a double mutant with deletion of nanS and nanS-p6 (mutant strain EDL933ΔnanSΔnanS-p6) and let it grow as before. Indeed, mutant strain EDL933ΔnanSΔnanS-p6 grew well (Fig. 5A), and we reasoned that the growth defect of mutant strain EDL933ΔnanSΔnanS-p1a-p6 on Neu5,9Ac2 was attributable to the quantity of deletions and thus that a gene dose effect exists. Mutant strain EDL933ΔnanSΔnanS-p1a-p7 could not grow on Neu5,9Ac2, as expected, and a basal level of growth could be measured in all cases (Fig. 5D) that was attributable to the supplemented Casamino Acids.
FIG 5.
Growth curves of E. coli O157:H7 strain EDL933 and selected EDL933 deletion mutants with different numbers of deleted nanS-p alleles. Data represent the results of cultivation in M9 minimal medium with 0.4% Neu5,9Ac2 (A), Neu5Ac (B), or glucose (C) or without a special added carbohydrate source (D) at 37°C and 180 rpm over 8 h. (A and D) The EDL933 wild-type strain and the EDL933ΔnanS, EDL933ΔnanSΔnanS-p6, and EDL933ΔnanSΔnanS-p1a-p5 mutant strains grew well to approximately the same optical density (A), while the EDL933ΔnanSΔnanS-p1a-p6 and EDL933ΔnanSΔnanS-p1a-p7 mutant strains grew just up to the basal optical density (A and D). (B and C) All mutants grew as well as the wild-type strain on the positive controls with Neu5Ac (B) or glucose (C) as the carbon source. (D) Basal growth was measured in the negative control, where no carbon source had been added.
In order to analyze whether the ability of mutant strain EDL933ΔnanSΔnanS-p1a-p7 to grow could be restored, M9 minimal medium was supplemented with recombinant 933Wp42-His, NanS-p2-His, or NanS-p4-His. It was found that the growth of the complemented strain could be restored to approximately the wild-type level in all three cases (Fig. 6).
FIG 6.
Supplementation of the growth medium of the EDL933ΔnanSΔnanS-p1a-p7 mutant with different recombinant NanS-p proteins. Data represent the results of growth of the EDL933 wild-type strain (white squares) and the EDL933ΔnanSΔnanS-p1a-p7 mutant strain (light gray triangles) on 0.4% Neu5,9Ac2 in M9 minimal medium at 37°C and 180 rpm and of growth of the EDL933ΔnanSΔnanS-p1a-p7 mutant strain with 4 μg/ml supplemented 933Wp42-His, NanS-p2-His, and NanS-p4-His (triangles with vertical stripes, dark gray triangles, and triangles with horizontal stripes, respectively).
DISCUSSION
E. coli O157:H7 strain EDL933 has been used as an EHEC reference strain worldwide for more than 30 years. Its genome sequence was analyzed and published twice (NCBI accession numbers AE005174.2 and NZ_CP008957.1), with the most notable sequence differences located in the repeated prophage regions (22, 23). In this context, Vimr (16) described 7 complete and 5 partial nanS-homologous alleles in the EDL933 genome, while we found 10 complete nanS-p alleles using the recently determined nucleotide sequence (23). Moreover, analyzing our EDL933 laboratory strain by whole-genome sequencing, only seven nanS-p alleles were detected. We hypothesize, in accordance with a recent publication (31), that the EDL933 genomes of different isolates were different from that of the original isolate due to evolutionary events that took place over the period of time since the strain was first isolated (21). Different storage and inoculation techniques could probably trigger such mutations. Such events have been demonstrated earlier by Papadopoulos et al. (32), who have shown the dynamic nature of the E. coli genome. Moreover, the specific genetic structures of the phages encoding NanS-p6 and NanS-p9, localized in tandem and separated only by transposases, would probably lead to instability in this region (data not shown).
In silico analysis of EDL933 identified the location of all nanS-p alleles in the region of late gene expression of prophages. Moreover, the positions of nanS-p1 and nanS-p2 are directly downstream of the positions of stx2a and stx1a, respectively, in the genome of Stx phages. In fact, nanS-p1 and stx2a or nanS-p2 and stx1a are cotranscribed after phage induction (13). The other nanS-p alleles are located in the genome of non-Stx phages but positioned in the corresponding region. Detailed investigation within the nanS-p alleles showed differences from the nanS gene. The additional nucleotide sequences which are present downstream of the typical sequence blocks for the SGNH enzyme family may be relevant for a further enzymatic function or for structural purposes, but the details are currently not known. A further region (DUF1737) which shows similarities to a prophage repressor gene was identified at the N terminus, but its function is currently unknown (see Fig. 1).
The presence of multiple nanS-p alleles in the EHEC genomes raises the issue of the functionalities and activities of the corresponding gene products. Based on the bioinformatic analyses and molecular experiments performed during this study, there is strong body of evidence that derivatives of Neu5Ac may serve as substrates for carbon nutrition in the large intestine. The presence of multiple nanS-p genes suggests that their gene products may give an advantage for growth under certain environmental conditions. By performing deletion experiments in EDL933, we were able to show that it did not lose the ability to grow on Neu5,9Ac2 if at least 2 nanS-p alleles were still present. That indicated that large mutations or other genetic events could happen without loss of carbohydrate utilization in the large intestine.
To evade naturally occurring colonization resistance in the gut, EHEC bacteria have to outcompete the gut microbiome for at least one nutrient (33). Furthermore, E. coli O157:H7 needs mechanisms to avoid host defense and to penetrate the mucus layer to reach the gut epithelium (34). Human gastrointestinal mucus consists of two layers, a thick loosely mucus layer and a thinner layer attached to the mucosa (35). While the inner layer is nearly sterile, the outer mucus layer forms a microbial niche that also includes nonmucolytic bacteria (36, 37). One of those bacterial species is E. coli (38). E. coli bacteria lack the ability to degrade the complex sugar chains present in the MUC2 mucus glycoprotein. However, they are actually in equilibrium with the turnover rate of mucus. One possibility is explained in the “Restaurant” hypothesis, which implies that E. coli bacteria grow in synergy with gut anaerobes which can degrade large polysaccharide chains to mono- or disaccharides (34). Indeed, it was shown that commensal E. coli strain MG1655 as well as E. coli O157:H7 strain EDL933 can grow well on mucus-derived saccharides such as N-acetylglucosamine, gluconate, galactose, N-acetyl neuraminic acids, and others (39). Interestingly, the same authors showed that deletion defects in one of the nan operons, responsible for growth on N-acetyl neuraminic acid, led to a growth defect in E. coli MG1655 but not in EDL933.
Regarding the distribution of sialic acids in the human gut, the monoacetylated Neu5Ac and the diacetylated Neu5,9Ac2 are represented in large amounts (16). In the mucin, they play a role in protection of the core protein against microbes. Nevertheless, there are some anaerobes which can remove the sialic acids from the glycoprotein by sialidases (40–42). Mostly, sialidases are specialized to cleave only a few bonds of sialic acids to the glycoprotein or to have narrow substrate specificity. Thus, Tannerella forsythia, a pathogenic member of the phylum Bacteroidetes, uses NanS to deacetylate glycolytically bound Neu5,9Ac2 and Neu5Gc,9Ac (43). The residual Neu5Ac or Neu5Gc can then be separated from the glycoprotein with NanH, which is the sialidase corresponding to Bacteroidetes, and the neuraminic acids can then be used as the carbon source. Although E. coli does not express any sialidases, it still possesses the nanS gene (17, 18).
We hypothesize that the multiple Neu5,9Ac2 esterases could provide E. coli O157:H7 with a colonization advantage by enabling it to outcompete the commensal E. coli strains in the gut for Neu5,9Ac2.
While NanS is located in the periplasm of the bacterial cells (16), it is probable that, similarly to Stx, additional Neu5,9Ac2 esterases may be released by phage lysis into the environment. This hypothesis is justified by the localization of the genes in the late regulated prophage regions. Therefore, we added different Neu5,9Ac2 esterases to the medium containing Neu5,9Ac2 and grew laboratory E. coli strain C600ΔnanS and EDL933 mutant strain EDL933ΔnanSΔnanS-p1a-p7 under this set of conditions. As expected, both grew to almost the same optical density as the wild-type strain without deletions in the nanS gene and the nanS-p gene, respectively.
The different Neu5,9Ac2 esterases could be part of a larger network of substrate utilization enzymes in the gut. This could be a reason for the high number of homologous genes with similar functions present in EHEC strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ning He and Wolfgang Fessner, University of Darmstadt, Germany, for providing 5-N-acetyl-9-O-acetyl neuraminic acid. We also thank Melanie Schneider and Markus Kranz for skillful technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01671-16.
REFERENCES
- 1.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Dobrindt U, Gartner J, Gallitz I, Hacker J, Karch H, Muller D, Schubert S, Alexander SM, Sorsa LJ, Zdziarski J. 2007. Aspects of genome plasticity in pathogenic Escherichia coli. Int J Med Microbiol 297:625–639. doi: 10.1016/j.ijmm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen Y, Sperandio V. 2012. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol 2:90. doi: 10.3389/fcimb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 7.Paton AW, Srimanote P, Talbot UM, Wang H, Paton JC. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J Exp Med 200:35–46. doi: 10.1084/jem.20040392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karch H, Denamur E, Dobrindt U, Finlay BB, Hengge R, Johannes L, Ron EZ, Tonjum T, Sansonetti PJ, Vicente M. 2012. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol Med 4:841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unkmeir A, Schmidt H. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun 68:4856–4864. doi: 10.1128/IAI.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogg PC, Saunders JR, McCarthy AJ, Allison HE. 2012. Cumulative effect of prophage burden on Shiga toxin production in Escherichia coli. Microbiology 158:488–497. doi: 10.1099/mic.0.054981-0. [DOI] [PubMed] [Google Scholar]
- 11.Wagner PL, Waldor MK. 2002. Bacteriophage control of bacterial virulence. Infect Immun 70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt H. 2001. Shiga-toxin-converting bacteriophages. Res Microbiol 152:687–695. doi: 10.1016/S0923-2508(01)01249-9. [DOI] [PubMed] [Google Scholar]
- 13.Herold S, Siebert J, Huber A, Schmidt H. 2005. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother 49:931–944. doi: 10.1128/AAC.49.3.931-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polzin S, Huber C, Eylert E, Elsenhans I, Eisenreich W, Schmidt H. 2013. Growth media simulating ileal and colonic environments affect the intracellular proteome and carbon fluxes of enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Appl Environ Microbiol 79:3703–3715. doi: 10.1128/AEM.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nübling S, Eisele T, Stober H, Funk J, Polzin S, Fischer L, Schmidt H. 2014. Bacteriophage 933W encodes a functional esterase downstream of the Shiga toxin 2a operon. Int J Med Microbiol 304:269–274. doi: 10.1016/j.ijmm.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Vimr ER. 2013. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol 2013:816713. doi: 10.1155/2013/816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steenbergen SM, Jirik JL, Vimr ER. 2009. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J Bacteriol 191:7134–7139. doi: 10.1128/JB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangarajan ES, Ruane KM, Proteau A, Schrag JD, Valladares R, Gonzalez CF, Gilbert M, Yakunin AF, Cygler M. 2011. Structural and enzymatic characterization of NanS (YjhS), a 9-O-acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7. Protein Sci 20:1208–1219. doi: 10.1002/pro.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landstorfer R, Simon S, Schober S, Keim D, Scherer S, Neuhaus K. 2014. Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15:353. doi: 10.1186/1471-2164-15-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbe C, Capon C, Coddeville B, Michalski JC. 2004. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J 384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 22.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 23.Latif H, Li HJ, Charusanti P, Palsson BO, Aziz RK. 14 August 2014. A gapless, unambiguous genome sequence of the enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Genome Announc doi: 10.1128/genomeA.00821-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall TA. 2016. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/N. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 27.Maniatis T, Sambrook J, Fritsch EF. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Hutchison CA III, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M. 1978. Mutagenesis at a specific position in a DNA sequence. J Biol Chem 253:6551–6560. [PubMed] [Google Scholar]
- 29.Ogura H, Furuhata K, Sato S, Anazawa K, Itoh M, Shitori Y. 1987. Synthesis of 9-O-acyl- and 4-O-acetyl-sialic acids. Carbohydr Res 167:77–86. doi: 10.1016/0008-6215(87)80269-0. [DOI] [PubMed] [Google Scholar]
- 30.Kobiler O, Rokney A, Oppenheim AB. 2007. Phage lambda CIII: a protease inhibitor regulating the lysis-lysogeny decision. PLoS One 2:e363. doi: 10.1371/journal.pone.0000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fellner L, Huptas C, Simon S, Muhlig A, Scherer S, Neuhaus K. 7 April 2016. Draft genome sequences of three European laboratory derivatives from enterohemorrhagic Escherichia coli O157:H7 strain EDL933, including two plasmids. Genome Announc doi: 10.1128/genomeA.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci U S A 96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freter R, Brickner H, Botney M, Cleven D, Aranki A. 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun 39:676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway T, Cohen PS. June 2015. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr doi: 10.1128/microbiolspec.MBP-0006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atuma C, Strugala V, Allen A, Holm L. 2001. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest. Liver Physiol 280:G922–G929. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, Krogfelt KA. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun 62:5191–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le GG, de Vos WM, Taylor GL, Juge N. 2015. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun 6:7624. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crost EH, Tailford LE, Le GG, Fons M, Henrissat B, Juge N. 2013. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nees S, Veh RW, Schauer R. 1975. Purification and characterization of neuraminidase from Clostridium perfringens. Hoppe Seylers Z Physiol Chem 356:1027–1042. doi: 10.1515/bchm2.1975.356.s1.1027. [DOI] [PubMed] [Google Scholar]
- 43.Phansopa C, Kozak RP, Liew LP, Frey AM, Farmilo T, Parker JL, Kelly DJ, Emery RJ, Thomson RI, Royle L, Gardner RA, Spencer DI, Stafford GP. 2015. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem J 472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 44.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.O'Brien AO, Lively TA, Chen ME, Rothman SW, Formal SB. 1983. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet i:702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.