Abstract
Cryptococcal meningoencephalitis is a fungal infection that predominantly affects immunocompromised patients and is uniformly fatal if left untreated. Timely diagnosis is difficult, and screening or prophylactic measures have generally not been successful. Thus, we need a better understanding of early, asymptomatic pathogenesis. Inhaled cryptococci must survive the host immune response, escape the lung, and persist within the bloodstream in order to reach and invade the brain. Here we took advantage of the zebrafish larval infection model to assess the process of cryptococcal infection and disease development sequentially in a single host. Using yeast or spores as infecting particles, we discovered that both cell types survived and replicated intracellularly and that both ultimately established a sustained, low-level fungemia. We propose that the establishment and maintenance of this sustained fungemia is an important stage of disease progression that has been difficult to study in other model systems. Our data suggest that sustained fungemia resulted from a pattern of repeated escape from, and reuptake by, macrophages, but endothelial cells were also seen to play a role as a niche for cryptococcal survival. Circulating yeast collected preferentially in the brain vasculature and eventually invaded the central nervous system (CNS). As suggested previously in a mouse model, we show here that neutrophils can play a valuable role in limiting the sustained fungemia, which can lead to meningoencephalitis. This early stage of pathogenesis—a balanced interaction between cryptococcal cells, macrophages, endothelial cells, and neutrophils—could represent a window for timely detection and intervention strategies for cryptococcal meningoencephalitis.
INTRODUCTION
Recent data from Uganda and Tanzania indicate that there are over 10,000 cases of HIV-related cryptococcosis per year in these countries alone (1, 2). Of those, roughly 5,000 to 8,000 (54 to 79%) are fatal. At a global scale, the survival rate is similarly poor, and over 600,000 deaths are attributed to Cryptococcus neoformans every year (3). The high mortality rate is in part due to difficulties in timely diagnosis; even in susceptible hosts, clinical signs of the initial pulmonary infection can be subtle or essentially absent (4). Studies in this population using serum cryptococcal antigen testing suggest that antigenemia (which may or may not represent actual fungemia) is a harbinger of meningoencephalitis but can precede it by weeks or more (5, 6). Once the infection has progressed to clinical meningoencephalitis, the prognosis is grim even with aggressive treatment. Beyond the HIV-infected population, a growing transplant and otherwise immunosuppressed population is also vulnerable, and even an immunocompetent host may be susceptible to infection and disease caused by the closely related species Cryptococcus gattii (7). An understanding of the events prior to fulminant disease is critical to better management of patients at risk for cryptococcal meningoencephalitis.
While acute new infection can cause its share of disease, it is generally thought that a large percentage of clinical cases represent remote reactivation rather than new infection (8, 9). Depending on the location, a large percentage of children already produce anticryptococcal antibody by the age of 10 (10, 11), and there is direct evidence of reactivated disease occurring years after exposure to the responsible cryptococcal strains (12). As noted above, an early harbinger of clinical disease is detectable amounts of cryptococcal antigen in the blood, although the duration of antigenemia before disease is variable, and appropriate use of this technique to guide prophylactic measures has been difficult to determine (6, 13, 14). Studies of human pathology and in the rat model suggest that one niche for long term cryptococcal survival is in granulomata (15, 16). However, the pathological changes associated with antigenemia are not known.
Cryptococcus is inhaled directly from environmental sources such as tree bark and bird guano (4). While the infectious particle(s) for humans remains formally unknown, both spores and yeast are capable of causing disseminated cryptococcosis in mouse models (17, 18). It is highly likely that humans could be exposed to both spores and yeast (either together or alone) from the environment. Also, little is known about the biological differences between cryptococcal yeast grown in rich medium and yeast germinated from the spore form after entry into the host. Therefore, we investigated both of these infectious agents in the zebrafish model to identify similarities and differences in behaviors and host responses between the two.
Once the fungus has established a foothold in the lung of a susceptible host, it is able to survive in the face of the innate immune response and gain access to the bloodstream. From there it has a specific tendency to localize to the vasculature of the central nervous system (CNS) and cross the blood-brain barrier. Many in vitro and in vivo approaches have been used to understand this process, and from these, we have made progress in understanding the interactions between Cryptococcus and specific immune cells (recently reviewed in reference 19), the intracellular life of Cryptococcus (reviewed in references 8 and 20), and the molecular mechanisms used for crossing the blood-brain barrier (21–23). Although animal models have shown prolonged fungemia to be a possible characteristic of pathogenesis (17, 24, 25), the mechanisms for initiating and maintaining fungemia have been difficult to approach. This cannot yet be examined in vitro and has been difficult to elucidate using mammalian model hosts. Here we present a model of infection that allows the direct study of this phenomenon.
A major advantage of the zebrafish model is the conserved nature of the metazoan host immune response and the relative ease with which the functions of host immune cells can be evaluated, particularly macrophages and neutrophils. Both of these cell types have been assessed in cryptococcal yeast infections both in vivo in mouse models and in vitro, allowing us to compare and contrast findings in zebrafish. Alveolar macrophages are widely held to be the initial phagocyte with which Cryptococcus interacts. Based on observations in the mouse model, cryptococcal residence inside these and other macrophages is seen to some extent throughout the course of pathogenesis (17, 19, 26). Depending upon their activation state, some macrophages are able to control and kill Cryptococcus (27), but in the progress of infection in the susceptible host, the fungus is able to survive and replicate in, and ultimately escape from, the phagosome and the macrophage (20).
Neutrophils have largely been considered either noncontributory or even detrimental to cryptococcal immunity because cryptococcosis is seen primarily in patients with dysfunctions in adaptive immunity and not patients with neutropenia. Animal studies have shown improved survival of mice transiently depleted of neutrophils at the time of pulmonary infection with Cryptococcus (28), which appears to be caused by the absence of neutrophil-mediated inflammation (29). In contrast, however, neutrophils have been implicated in Cryptococcus clearance later in disease (30, 31). Recent work has provided evidence that neutrophils are capable of clearing Cryptococcus directly from the brain vasculature of mice (32–34). In all of these cases, limitations of the mouse model made it difficult to discern the topology and kinetics of Cryptococcus-phagocyte interactions directly. Using the zebrafish, we can both evaluate and modulate this host-microbe interaction.
Two recent studies using the larval zebrafish as a model host for cryptococcal infection have demonstrated significant parallels with mammalian pathogenesis and highlighted the unique advantages this organism has to offer (35, 36). The zebrafish is genetically tractable and is now well established as a model host for several forms of infection (37–40). Perhaps most useful is the visual access the transparent larva provides, allowing for high-resolution microscopy over multiple days either serially or in continuous time-lapse applications. This access over time enables the observation of pathogenesis involving the innate immune system, a variety of tissue types, a functioning circulatory system, and a blood-brain barrier. In this study, we took advantage of the zebrafish larva to examine pathogenesis from the earliest interactions with phagocytes to the establishment of the characteristic low-level fungemia which allows for eventual brain colonization.
MATERIALS AND METHODS
Zebrafish care and maintenance.
Adult zebrafish were kept under a light/dark cycle of 14 h and 10 h, respectively. Larval zebrafish were incubated at 28.5°C in E3 buffer. For infection experiments, larvae were manually dechorionated between 24 and 30 h postinfection (hpi). Prior to microinjection or imaging, larvae were anesthetized in E3-MB containing 0.2 mg/ml Tricaine (ethyl-3-aminobenzoate; Sigma-Aldrich). For live imaging, pigment synthesis was inhibited by switching larvae to E3-MB containing 0.2 nM 1-phenyl-2-thiourea (PTU) (Sigma-Aldrich) at 18 to 24 h postfertilization (hpf). For prolonged time-lapse imaging, larvae were mounted in 1% low-melting-point agarose, 0.2 mg/ml Tricaine, and 0.2 nM PTU (final concentrations) on a custom-made glass-bottom dish. All adult and larval zebrafish procedures were in full compliance with NIH guidelines and approved by the University of Wisconsin—Madison Institutional Animal Care and Use Committee (no. M01570-0-02-13).
Transgenic zebrafish lines.
Tg(mpeg:H2B-mCherry) larvae express red fluorescence localized to the nucleus, specifically in macrophages. The nuclear localization was used because cytoplasmic mCherry expression from the mpeg promoter did not produce a bright enough fluorescent signal in many parts of the larva. Tg(mpx:mCherry) larvae express red fluorescence specifically in the cytoplasm of neutrophils. This line was developed in the lab of A.H. and is listed as uwm7Tg in the zfin database. The Tg(flk:mCherry) line expresses mCherry specifically in the endothelial cytoplasm and was kindly provided by Heinz-Georg Belting and Markus Affolter. Tg(mpeg:egfp) fish generated in the lab of A.H. were crossed with Tg(mpx:mCherry) fish to derive the double transgenic Tg(mpeg:egfp)/(mpx:mCherry) larvae used for simultaneous assessment of macrophage and neutrophil function. The Tg(mpx:mCherry-2A-Rac2D57N) line (zfin database name uwm3Tg; zf307Tg) was used in combination with Tg(mpx:mCherry-2A-Rac2WT) as a wild-type control as described previously (41, 42).
Cryptococcal strains, growth conditions, and spore preparation.
Strains used were of the serotype D background (Cryptococcus neoformans var. neoformans strains B-3501 and B-3502) or serotype A background (Cryptococcus neoformans var. grubii strain H99) (43, 44). All were handled using standard techniques and media as described previously (45, 46). Spores were purified from crosses using density gradient purification as described previously (47). Briefly, mixed haploid yeast cells of opposite mating types (CHY3515 and CHY3516) were spotted onto V8 solid medium at a ratio of 1:1 and a total optical density at 600 nm (OD600) of 1.0. Plates were then incubated at room temperature in the dark for 6 days on V8 agar before spore purification.
Construction of a nuclearly localized GFP expression strain pair.
A predicted nuclear localization signal (NLS) sequence was amplified from the 3′ end of the gene encoding the high-mobility group (HMG) domain-containing transcription factor Nhp6b02 (CNE04220) using primers CHO3801 (5′-CAT GGG ATCC GCC AAG AAG GAT GCC GTC G-3′) and CHO3802 (5′-CAT GAG ATC TAT CAT CAC GCC ACA CCC GG-3′). This fragment was digested with BamH1 and BglII and then ligated into pCN19 (a gift from Andrew Alspaugh) to create pCH1227. The resulting plasmid encodes a histone 3 promoter-driven enhanced green fluorescent protein (EGFP) with an 80-amino-acid NLS and a nourseothricin resistance (NATr) cassette. This integrating plasmid was then transformed via bombardment into C. neoformans B3502 to generate strain CHY3135 (47). This strain produces bright GFP that is localized to the nucleus. To generate a mating pair for spore production, CHY3135 (a) was crossed with B3501(α), and spores were isolated as described previously (70). Isolated spores were then germinated on YPD agar, and the resulting colonies were mating typed by crossing with tester strains JEC20 (a) and JEC21(α) on V8 agar (pH 7.0) and examined microscopically for the presence of filamentous growth and spores. An α NATr Gfp-NLS strain, CHY3136, was identified, which was then crossed on V8 (pH 7.0) with CHY3135 to produce GFP-expressing spores.
Microscopy.
Confocal and differential interference contrast (DIC)/epifluorescence imaging was performed on a Zeiss Z1 Observer-based system run with the Zeiss Zen software. All images from this system were obtained using a Zeiss EC Plan-Neofluar objective (40×/0.75 numerical aperture [NA]). Epifluorescence images were collected using a Photometrics Coolsnap ES2 camera. Spinning-disc confocal imaging used a Yokogawa CSU-X1 spinning disc and collection with a Photometrics Evolve 512EMCCD. Stereoscopic imaging (see Fig. 3A) was performed on a Zeiss AxioZoom.V16 microscope with a Zeiss Plan-Neofluar 1×/0.25 NA objective. Images were collected with a Zeiss AxoCam MRm using Zeiss Zen software. Live imaging of zebrafish larvae was performed with larvae anesthetized in Tricaine as previously described (50) and simply resting on the bottom of a glass-bottom dish or immobilized in 1% low-melting-point agarose.
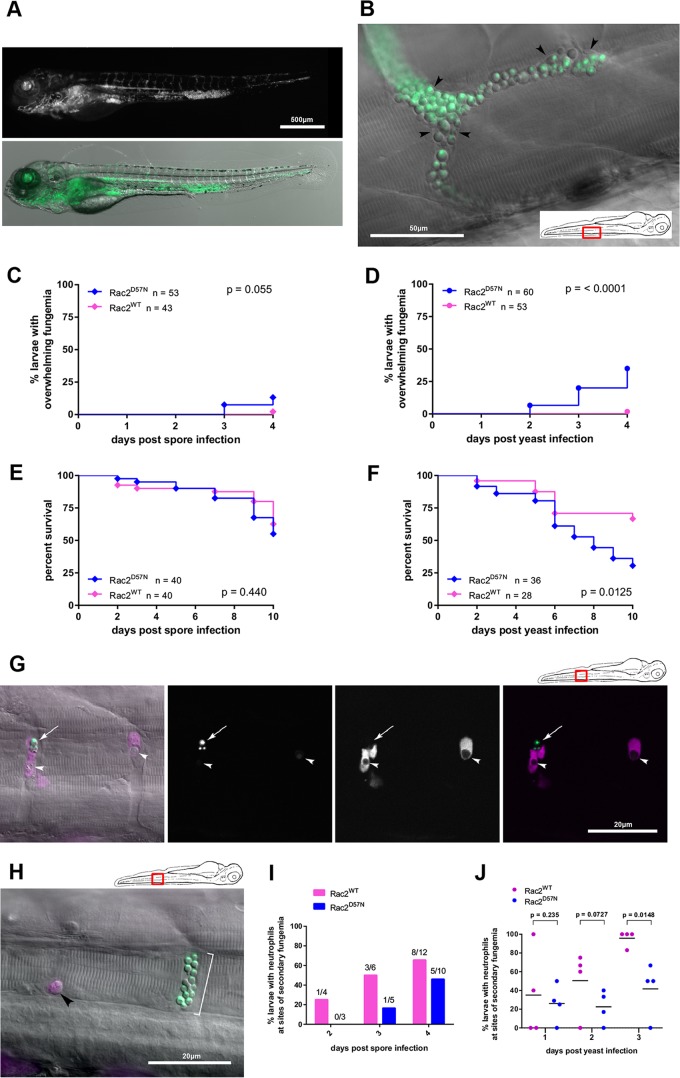
FIG 3.
Neutrophils are required for limiting secondary fungemia. (A) Rac2D57N larvae inoculated with yeast can succumb to overwhelming fungemia. A larva at 4 days after inoculation with a visually enumerated 67 yeast cells is shown. The larva expresses no transgenic green fluorescence, and the vasculature is essentially filled with fluorescent yeast. The behavior of this larva was indistinguishable from that of an uninfected larva on the day of this image. Stereomicroscope images are shown (green channel above and overlay with contrast image below). (B) Detail of vasculature overwhelmed by yeast forms. An epifluorescence-DIC overlay image of a nontransgenic larva 4 days after inoculation with yeast is shown. The larva is different from that shown in panel A but is also behaviorally normal. Black arrowheads indicate vessel walls visible by DIC. The inset panel indicates the location in the larva. (C) Rac2D57N larvae inoculated with spores occasionally undergo overwhelming fungemia. Combined data from three replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 130 (±9) (D57N) or 128 (±9) (WT) spores i.v. and monitored daily for 4 days postinoculation. Overwhelming fungemia is defined as yeast cells flowing freely in the vasculature and too numerous to count (more than 500/larva). The P value is from the Mantel-Cox log rank test. (D) Rac2D57N larvae inoculated with spores are significantly more likely to undergo overwhelming fungemia. Combined data from three replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 69 (±5) (D57N) or 66 (±5) (WT) yeast cells i.v. and monitored daily for 4 days postinoculation. Overwhelming fungemia is defined as yeast cells flowing freely in the vasculature and too numerous to count (more than 500/larva). The P value is from the Mantel-Cox log rank test. (E) Survival is not affected by neutrophil function within 4 days of spore inoculation. Combined data from two replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 140 (±11) (D57N) or 143 (±10) (WT) spores i.v. and monitored daily for 4 days postinoculation. The P value is from the Mantel-Cox log rank test. (F) Poor neutrophil function worsens survival within 4 days of yeast inoculation. Combined data from two replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 52 (±5) (D57N) or 61 (±6) (WT) yeast cells i.v. and monitored daily for 4 days postinoculation. The P value is from the Mantel-Cox log rank test. (G) Rac2WT neutrophils engulf free yeast during secondary fungemia. A Tg(mpx:mCherry-2A-Rac2WT) larva at 48 h after yeast inoculation is shown. The cartoon at the upper right indicates the location within the larva. Two parallel intersegmental vessels are shown; the first frame is a DIC/epifluorescence overlay, and the remaining panels are confocal projections obtained immediately after. The white arrow indicates apparently extracellular yeast. White arrowheads indicate intracellular yeast, both with diminished fluorescence consistent with cell death. (H) Rac2D57N neutrophils are seen not engaged with free yeast during secondary fungemia. A Tg(mpx:mCherry-2A-Rac2D57N) larva at 48 h after yeast inoculation is shown. The cartoon at the upper right indicates the location within the larva. Two parallel intersegmental vessels are shown, with a rounded neutrophil (black arrowhead) lodged alone while a cluster of extracellular yeast cells (white bracket) is seen adjacent. (I) Rac2D57N neutrophils are less likely than WT neutrophils to associate with free germinated spores during secondary fungemia after spore inoculation. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 140 (±11) (D57N) or 143 (±10) (WT) spores i.v. and monitored at 2, 3, and 4 days after infection for neutrophil engagement with intravascular cryptococci. Because of the small number of larvae which reached the stage of secondary fungemia, we present the overall percentage of such larvae with neutrophils engaged from two replicates. (J) Rac2D57N neutrophils are less likely than WT neutrophils to associate with free yeast during secondary fungemia after yeast inoculation. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 69 (±5) (D57N) or 66 (±5) (WT) yeast cells i.v. and monitored at 2, 3, and 4 days after infection for neutrophil engagement with intravascular cryptococci. Plotted are percentages of larvae from four replicate experiments. P values represent the results of unpaired t tests of log-transformed ratios with Welch's correction.
Image processing.
For direct display, images were transferred from the Zeiss czi format to tiff and levels adjusted using Adobe Photoshop. Three-dimensional (3D) rendering work was done using Imaris rendering software (Bitplane).
MO knockdown.
PU.1 morpholino (MO) was purchased from Gene Tools (Philomath, OR) and stored at room temperature in sterile distilled water at a stock concentration of 1 mM. Three nanoliters of diluted MO was microinjected into the yolk center of embryos at the 1- to 4-cell developmental stage at 500 μM (51).
Microinjection.
Harvested spores were stored suspended in phosphate-buffered saline (PBS) at 20°C for up to 30 days. Yeast were grown from frozen stock on yeast extract-peptone-dextrose (YPD) plates overnight at 29°C. The resulting stock plate was used as a source for no more than 30 days. For microinjection, yeast cells were plated from the stock plate onto a fresh plate and grown overnight at 29°C. Just prior to injection, yeast cells were harvested off this fresh plate and suspended in PBS. For fluorescent spore and yeast injections, appropriate dilution in PBS was estimated by eye. After 3 to 5 trial injections at different volumes, the dosage was adjusted after directly assessing the inoculum in larvae. Inocula of between 150 and 250 cryptococcal cells per larva (estimated mass of 50.5 μg at 48 hpi [52]) produce an inoculum-to-mass ratio averaging 3.96, comparable to that of previously published mouse infections (1 × 105 yeast cells or spores in an ∼22-g mouse = 4.54). This dosing was also comparable to that for previously published studies in zebrafish (35, 36). For imaging and cryptococcal enumeration experiments, the inoculum for each fish was assessed by direct visual counting via epifluorescence microscopy. Consistency of counts was held by counting fish on their right sides only and at the same orientation. This accuracy of this method was confirmed by comparing the average larval inoculum in a group to a colony count from the same inoculation volume plated on YPD. Inoculation dosages with the nonfluorescent H99 strain were measured by plating the inoculation volume only. After manual dechorionation of embryos at ∼28 hpf, intravenous (i.v.) and hindbrain inoculations were performed as previously described (50), with the alteration that larvae were positioned on a 3% agarose plate formed with holding grooves as previously described (53).
Statistical analysis.
Statistical testing and graph generation were performed using GraphPad Prism version 6. Specific test modalities are indicated in the figure legends, but overall survival data and time-to-event data were generated using at least two independent replicate data sets, analyzed using Mantel-Cox proportional-hazard regression analysis, and displayed as Kaplan-Meier plots. Comparisons of normalized cryptococcal burden over time were generated by manual counting of cryptococcal numbers daily in at least 3 independent replicates. The ratios resulting from normalization to the initial inoculum were then log transformed before pairwise comparisons were made using the two-tailed Mann-Whitney test, with P values displayed in each figure. Linear regression with calculation of 95% confidence intervals was also performed using the Prism software.
RESULTS
Cryptococcal spores and yeast both establish a prolonged infection in zebrafish larvae with eventual dissemination to the brain.
For an initial comparison of spore and yeast infections, we injected 48-h-postfertilization (hpf) zebrafish larvae via the caudal vein with cryptococcal cells expressing a nuclearly localized EGFP (in the robust spore-producing B3501 [α] and B3502 [a] backgrounds). The yeast inoculum was 171 (±35) cells from a 1:1 mixture of each yeast mating type; the spore inoculum was 250 (±27) cells purified from a cross of the EGFP yeast strains. (Fig. 1A). There was no significant mortality of wild-type larvae from either spores or yeast of this background, even at the highest inoculum (Fig. 1B). In contrast, yeast cells of the hypervirulent H99 strain killed the larvae at moderate inoculum of 100 to 200 CFU (Fig. 1B), as shown previously (36). To discern differences between spore and yeast infections, we assessed survival and replication by manually counting the number of Cryptococcus cells every 24 h after inoculation.
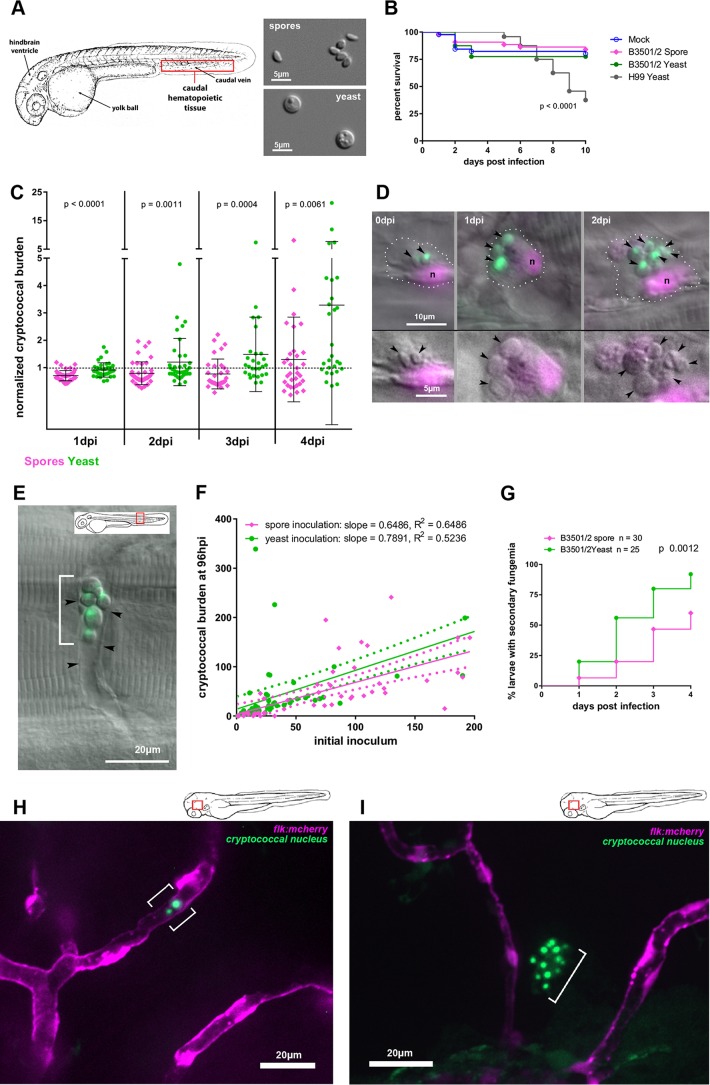
FIG 1.
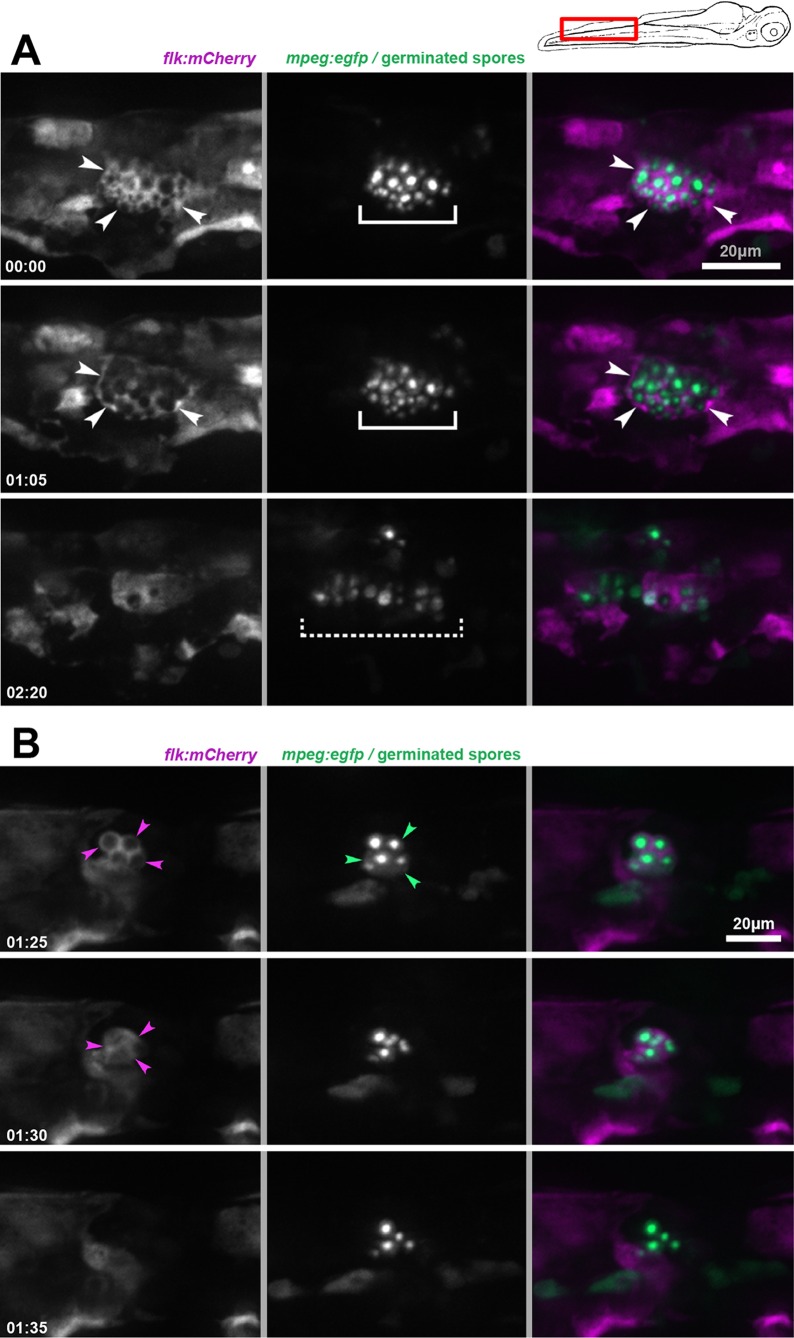
Cryptococcal spores and yeast establish a prolonged infection in zebrafish larvae, with differing chronologies of similar basic events. (A) Schematic view of the zebrafish larva at 32 hpf with anatomical landmarks. The insets show DIC images of spores and yeast cells, representing their relative sizes. (B) No significant difference is found in survival of zebrafish larvae inoculated i.v. with PBS (mock), 250 (±27) spores, or 171 (±35) yeast cells (3-way curve comparison; P = 0.75, Mantel-Cox log rank test). Data are from two independent experiments with 22 larvae per group per experiment. As previously shown, 100 to 200 CFU of H99 yeast does induce decreased survival (P value shown from Mantel-Cox log-rank test). Combined data of two replicates are shown. Mock, n = 46; spore, n = 44; yeast, n = 40, H99 yeast, n = 24. (C) Inoculated spores and yeast both establish stable infection with subsequent expansion, although inoculated yeast cells do so sooner. The cryptococcal burden normalized to the initial inoculum of individual larvae is shown, log transformed for analysis with the two-tailed Mann-Whitney test. (D) i.v. inoculated spores germinate within macrophages. Spores expressing EGFP inoculated into Tg(mpeg:H2B-mCherry) larvae, which express the red fluorescent protein mCherry specifically in macrophages, localized to the nucleus. DIC/fluorescence overlays at 0, 1, and 2 dpi are shown. Black arrowhead, fluorescent nuclei. Bottom row, enlarged image with green signal removed for clarity. Note the smaller size and angular shape of spores at 0 dpi (see panel A for spore and yeast shapes). Note the increasing diameter of yeast by 1 dpi and the capsule layer (not fluorescent but edge clearly seen by DIC) by 2 dpi. The scale bar applies to all 3 panels in each row. (E) Germinated spores lodged temporarily in an intersegmental vessel during secondary fungemia (cryptococci released back into the bloodstream after initial phagocytosis). The inset shows the location within larva. The white bracket indicates germinated spores, now with plentiful capsule material, extracellular and in the bloodstream. Black arrowheads indicate endothelial walls visible by DIC. (F) Cryptococcal expansion after spore and yeast inoculation is dose dependent and occurs at different rates. Counts at 96 hpi versus initial inoculum of spores are shown with dose-response curves. Linear regression data are shown; dashed lines represent 95% confidence intervals. (G) Likelihood of signs of secondary fungemia increases over time and begins earlier after yeast inoculation. Combined data from two replicates are shown. Larvae were inoculated with 143 (±16) spores or 72 (±8) yeast cells i.v. and the presence of intravascular yeast scored by DIC and fluorescence microscopy. The P value is from the Mantel-Cox log rank test. (H) Germinated spores and yeast lodge in brain vasculature during secondary fungemia. Confocal imaging of Tg(flk:mCherry) larvae, which express the red fluorescent mCherry protein specifically in endothelial cells, at 3 days postinoculation with yeast is shown. The bracket emphasizes the location of yeast cells, which are contained within the vessel. The inset shows the location within the larva. (I) Some yeast cells lodged in the brain vasculature invade the brain parenchyma after secondary fungemia. Confocal imaging of Tg(flk:mCherry) larvae at 5 days postinoculation with yeast is shown. The bracket emphasizes a cluster of yeast cells clearly outside the vasculature. The inset shows the location within the larva.
In the case of spore inoculation, the cryptococcal burden most often decreased over the first 3 days, with a few larvae experiencing mild expansion (Fig. 1C). By the fourth day there was a broad variety in fungal burden: a few larvae experienced large increases, while most remained near or just below the initial inoculum. To investigate the fate of spores after inoculation we used direct microscopy of Tg(mpeg:H2B-egfp) transgenic larvae, which express the enhanced green fluorescent protein specifically in macrophages and localized to the nucleus, and Tg(mpx:mCherry) larvae, which express the red fluorescent mCherry protein specifically in the cytoplasm of neutrophils. By 2 hpi we found that there were no visible spores still free in the bloodstream (data not shown). All visible spores had been phagocytosed and were residing in macrophages and, to a lesser extent, neutrophils. Over the first 2 days postinfection (dpi), inoculated spores within macrophages showed morphological changes consistent with germination and subsequent replication (as evidenced by the appearance of daughter buds on yeast cells) (Fig. 1D). These changes were not uniform, however, as some subsets of spores germinated but remained small and did not replicate. Although most germinated spores (for clarity, we will use the term “germinated spores” for yeast cells seen after spore inoculation) remained intracellular, by 48 to 72 hpi we began to see occasional cells free in the bloodstream. These cells were often lodged in characteristic vascular locations such as the intersegmental tail vessels (Fig. 1G), gill vasculature (see Fig. S1A in the supplemental material), and brain vasculature (see Fig. S1B in the supplemental material). We termed this appearance of previously intracellular Cryptococcus in the bloodstream “secondary fungemia,” considering the “primary” fungemia in this model to have taken place immediately after inoculation and prior to initial phagocytosis.
Events after yeast inoculation were only slightly different. Yeast inoculation also resulted in a 24-h burden below the initial number, but the drop was not as pronounced as it was with spores (Fig. 1C). While both cell types were capable of establishing progressive infection, inoculated yeast began replicating earlier, often progressing to a heavy fungal burden during the four-day observation period (Fig. 1C). Although individual instances of heavy proliferation were more common after yeast inoculation, linear regression analysis of the inoculum-normalized fungal burden at 96 hpi showed that the two forms were comparable in their abilities to replicate and establish prolonged infection (Fig. 1F). Secondary fungemia was also observed after yeast inoculation, usually earlier, averaging 2.38 (±0.18) days for yeast and 2.84 (±0.22) days for spores (two-tailed t test, P = 0.12), but still after an interval during which all yeast cells were intracellular (Fig. 1G). In both spore and yeast infections, only yeast forms are seen in secondary fungemia.
From these results we conclude that spores must undergo germination before establishing secondary fungemia and that inoculated yeast cells require 24 to 48 h to acquire this ability. The appearance of secondary fungemia was typically low-level: only one or two germinated spores or yeast cells could be seen lodged in the vasculature in a given larva. On several occasions single larvae showed secondary fungemia on one day but not the next, but in no instance did secondary fungemia precede a drop in overall cryptococcal numbers (see Tables S1 and S2 in the supplemental material). Based on these observations, we conclude that a sustained, sporadic release of small numbers of yeast cells into the circulation was occurring after both spore and yeast inoculations.
The transparent zebrafish larvae offered an ideal opportunity to follow the fate of these newly extracellular yeast cells or germinated spores. By inoculating Tg(flk:mCherry) larvae (54), which express mCherry in all endothelial cells, we specifically visualized the lodging of Cryptococcus inside the brain vasculature (Fig. 1H), which was followed by invasion out of the vessels and into the brain parenchyma (Fig. 1I). These results are consistent with previous reports using the H99 strain in zebrafish larvae (36). While inoculated yeast could lodge temporarily in the brain vasculature in the very first hours after inoculation (data not shown), invasion into the parenchyma was not seen until after the establishment of secondary fungemia. All together, these observations suggest that both spore and yeast inoculations produced sustained, low-level fungemia after intravenous infection, which eventually led to CNS invasion. Both cell types require time to acquire the ability to cause this secondary fungemia, and that refractory time is spent inside host phagocytes.
During primary infection, inoculated spores and yeast attract phagocytes at different rates, but both are more susceptible to killing by neutrophils than by macrophages.
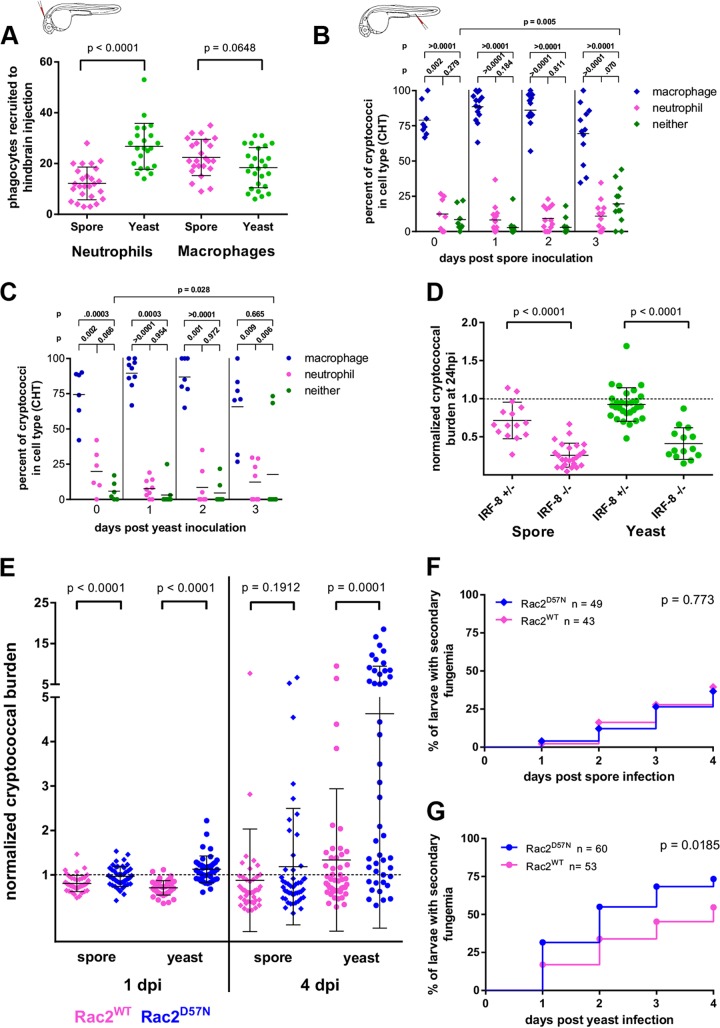
To examine the establishment of secondary fungemia by spores or yeast in a stepwise fashion, we undertook an analysis of early pathogenesis in chronological order. To assess phagocyte recruitment, we used the hindbrain recruitment assay (37, 55), in which yeast or spores were inoculated into the hindbrain ventricle (Fig. 1A) of transgenic larvae expressing the red fluorescent protein mCherry from either the mpeg promoter (for macrophages) [Tg(mpeg1:mCherry)] (42) or the mpx promoter (for neutrophils) [Tg(mpx:mCherry)]. We then counted the number of phagocytes recruited at 6 hpi. The rate of recruitment of macrophages was the same for both cell types, but yeast attracted significantly more neutrophils than spores did (Fig. 2A). To determine the fate of spores and yeast after they became intracellular, we inoculated Tg(mpeg1:mCherry)/Tg(mpx:mCherry) double transgenic larvae via the caudal vein and assessed the percentages of cryptococcal cells associated with either macrophages or neutrophils at multiple time points within the caudal hematopoietic tissue (CHT). Although in this experiment both Cryptococcus and transgenic macrophages expressed green fluorescence, the nuclear localization of fluorescence in our engineered cryptococcal strain produced a bright, punctate signal easily distinguished from that of transgenic macrophages. Spores inoculated intravenously into zebrafish larvae were found primarily within macrophages at 24 h (Fig. 2B), as were yeast cells (Fig. 2C) (reported previously for yeast [36]). A small percentage of cryptococcal cells was not associated with either macrophages or neutrophils. From these data we concluded that most spores and yeast cells are within macrophages, although a small percentage was found inside neutrophils. This was somewhat surprising given our previous findings of a strong attraction of neutrophils by yeast.
FIG 2.
During primary infection, inoculated spores and yeast attract phagocytes at different rates, but both are more susceptible to killing by neutrophils than by macrophages. (A) Phagocyte recruitment by spores and yeast as measured by a hindbrain recruitment assay using Tg(mpeg:H2B-mCherry) and Tg(mpx:mCherry) (a line which expresses cytoplasmic mCherry specifically in neutrophils) larvae to enumerate recruitment of macrophages and neutrophils, respectively. Yeast cells attract significantly more neutrophils than spores do. Data are pooled from two experiments with at least 7 larvae per group, and the P values shown result from a 2-tailed, unpaired t test. The inset shows the location of inoculation (this is the only experiment using hindbrain inoculation that we present). (B) After i.v. spore inoculation, cryptococci within the CHT are found predominantly in macrophages, although the numbers seen in neutrophils or not associated with phagocytes increases over time. Spores were inoculated into double transgenic [Tg(mpeg:egfp, mpx:mCherry)] larvae and analyzed by 3D confocal microscopy at 6 hpi (0 dpi) and daily thereafter. The inset image to underscore i.v. inoculation applies to this and all subsequent experiments. P values represent the results of unpaired t tests of log-transformed percent values obtained in three independent replicate experiments. Except for the comparison of cryptococci found associated with neither phagocyte type at 0 and 3 dpi, no other longitudinal percentages of the same category (macrophages each day or neutrophils each day) were found to be significantly different. (C) Same analysis as for panel B but after yeast inoculation. (D) In IRF-8 mutant larvae, an absence of macrophages and surfeit of neutrophils result in significantly more cryptococcal killing over the first 24 h. Data are pooled from two independent experiments for each condition, with at least 3 larvae per genotype. P values are the results of a 2-tailed unpaired t test. (E) Control of cryptococcal burden is impaired in larvae with poor neutrophil function. Spores or yeast cells were inoculated into either Tg(mpx:mCherry-2A-Rac2D57N) larvae, which express a dominant negative allele of Rac2 specifically in neutrophils, or Tg(mpx:mCherry-2A-Rac2WT) larvae as a control. The cryptococcal burden was normalized to the initial inoculum of individual larvae, log transformed for analysis with a two-tailed Mann-Whitney test. (F) The likelihood of secondary fungemia after spore inoculation is not affected by neutrophil function. Combined data from three replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 130 (±9) (D57N) or 128 (±9) (WT) spores i.v. and the presence of intravascular yeast scored by DIC and fluorescence microscopy. The P value is from the Mantel-Cox log rank test. (G) Neutrophil dysfunction increases the likelihood of secondary fungemia after yeast inoculation. Combined data from three replicates are shown. Tg(mpx:mCherry-2A-Rac2D57N) or Tg(mpx:mCherry-2A-Rac2WT) larvae were inoculated with 69 (±5) (D57N) or 66 (±5) (WT) yeast cells i.v. and the presence of intravascular yeast scored by DIC and fluorescence microscopy. The P value is from the Mantel-Cox log rank test.
To evaluate the nature of Cryptococcus-phagocyte interactions during infection, we inoculated an IRF-8 mutant strain of zebrafish, which lacks macrophages for the first few days of life but has an increased number of neutrophils during this time (57). By again counting the number of fluorescent cryptococcal cells at 2 hpi and on subsequent days, we discovered that there were reduced numbers of cryptococcal cells at 24 hpi in mutant zebrafish compared to heterozygote clutchmates, and this effect was greater in the case of yeast-infected larvae (Fig. 2D). The difference between IRF-8 mutant larvae and their heterozygous clutchmates decreased with each successive time point (data not shown), perhaps reflecting the effect of returning macrophage numbers after 24 hpi (72 h post fertilization). These findings suggest that neutrophils are more likely to kill the cryptococcal cells they ingest than are macrophages, particularly in the case of yeast. However, it is also possible that the loss of IRF-8 function affects cells other than macrophages. To address further the function of neutrophils, we used a transgenic line of larvae with a human dominant negative allele of Rac2 expressed specifically in neutrophils [Tg(mpx:mCherry-2A-Rac2D57N), referred to here as Rac2D57N larvae]. Rac2 is a small Rho GTPase which is required for normal neutrophil function, and patients with the Rac2D57N mutation have impaired immune function resulting in severe recurrent infections (71). While neutrophils are present in the circulation of these transgenic larvae, their adhesion and migration are impaired, and the neutrophils also likely have deficits in intracellular killing (41, 42, 58). When spores or yeast were inoculated into the bloodstream of Rac2D57N larvae, we found that the larvae did not control the fungal burden as well as Rac2WT controls [Tg(mpx:mCherry-2A-Rac2WT), a neutrophil-specific transgenic line expressing the same construct as Rac2D57N larvae without the D57N mutation] (Fig. 2E). Once again, this effect was observed after spore inoculation, but it was much more marked in yeast infections. This again supports the conclusion that in this model neutrophils are more efficient than macrophages at killing cryptococcal cells, particularly yeast. Given this finding, it was curious that some neutrophils harboring cryptococcal cells could be observed throughout infection (Fig. 2B and C). One possible explanation for this observation is that secondary fungemia provided neutrophils with a small, continuous supply of cryptococcal cells to phagocytose. To test this possibility, we compared the rates of secondary fungemia after spore and yeast inoculation in Rac2D57N larvae. If neutrophils counter secondary fungemia by phagocytosing free cryptococcal cells, we would expect to see more evidence of secondary fungemia in Rac2D57N larvae. We observed no differences in fungemia between Rac2WT and Rac2D57N larvae infected with spores (Fig. 2F). In contrast, there was significantly more secondary fungemia after yeast inoculation in the Rac2D57N larvae than in the wild type (Fig. 2G). These data support the idea that secondary fungemia can contribute to the small population of yeast within the neutrophils of yeast-infected larvae. The difference between yeast- and spore-infected larvae seems most likely due to the relative delay by spores in producing secondary fungemia. As seen in previous experiments, the kinetics of spore interactions with the host differed from that for yeast, and the resultant delay in secondary fungemia did not allow us to detect it in the time frame used here.
Functional neutrophils are required for limiting secondary fungemia.
To test the direct effects of neutrophil function during secondary fungemia, we examined the fate of infected Rac2D57N larvae after secondary fungemia was established. Rac2WT and Rac2D57N larvae were infected with yeast or spores and evaluated after 4 dpi. We observed that many larvae developed overwhelming fungemia in which their entire vasculature became filled with fluorescent yeast (Fig. 3A and B). In these experiments only one out of 96 (1%) Rac2WT larvae developed such massive fungemia, compared to 27 out of 113 (19%) Rac2D57N larvae. As expected based on prior results, overwhelming fungemia was much more likely after yeast inoculation than after spore inoculation (Fig. 3C and D), again likely due to the difference in kinetics after spore inoculation. The incidence of overwhelming fungemia preceded but corresponded to the incidence of larval death (Fig. 3E and F), which is in accordance with our previous observation that larvae dying of H99 infections were overwhelmed with intravascular yeast. On closer inspection of the neutrophils in Rac2WT larvae, we observed that they were often engaged with and ingesting intravascular yeast during secondary fungemia (Fig. 3G), whereas neutrophils of Rac2D57N larvae were seldom associated with the yeast (Fig. 3H to J). From these data we conclude that neutrophils not only restrain secondary fungemia but are also crucial for control of the cryptococcal burden during secondary fungemia in this model.
Secondary fungemia is preceded by cryptococcal residency within both macrophages and endothelial cells.
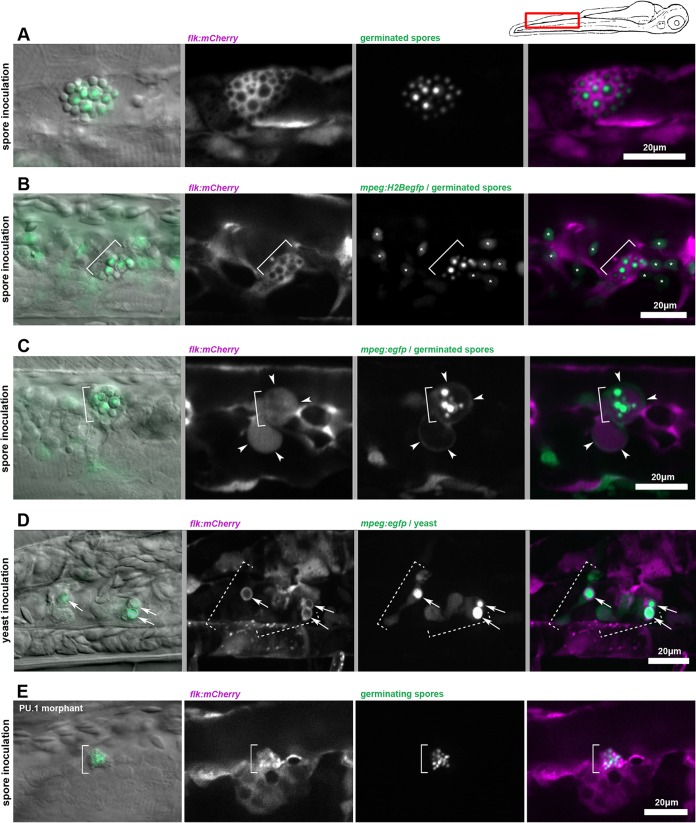
To determine the mechanisms governing the development of secondary fungemia, we focused our attention specifically on the caudal hematopoietic tissue and vein, where the great majority of infected cells are found after initial inoculation, while we noted previously that as infection proceeded, spores and yeast cells remained primarily inside macrophages and neutrophils, but a small number were associated with neither phagocyte population (Fig. 2B and C). Based on our observations that cryptococcal cells not associated with phagocytes were perivascular but stationary, we first hypothesized that these cryptococcal cells were associated with endothelial cells. To test this hypothesis, we infected an endothelial cell-specific [Tg(flk:mCherry)] transgenic line. We observed that after spore inoculation clusters of yeast could be found inside endothelial cells of the CHT (Fig. 4A). In a representative experiment, such clusters were found in 2 of 18 (11%) of larvae at 48 hpi, in 6 of 17 (35%) at 72 hpi, and in 11 of 16 (69%) at 96 hpi. To ensure that this apparent finding was not misexpression of mCherry by infected macrophages, we infected Tg(flk:mCherry)/Tg(mpeg:H2B-egfp) double-transgenic larvae to allow positive identification of both endothelial cytoplasm and macrophage nuclei. We found that labeled macrophage nuclei were not found within the cytoplasmic signal containing the germinated yeast (Fig. 4B; see Movie S1 in the supplemental material), indicating that the germinated spores were indeed residing within endothelial cells.
FIG 4.
Secondary fungemia is preceded by cryptococcal residency within both macrophages and endothelial cells. In all panels of this figure, the first frame is a DIC and epifluorescence overlay, while the remaining frames are confocal projections obtained immediately after. The cartoon of a larva above indicates the approximate location for all panels. (A) By 96 hpi, germinated yeast forms can be seen growing inside the endothelial cytoplasm. EGFP-expressing spores were inoculated into Tg(flk:mCherry) larvae. (B) Observation in double-transgenic larvae demonstrates that endothelial occupancy is not an artifact of transgene misexpression. EGFP-expressing spores were inoculated into Tg(flk:mCherry, mpeg:H2B-egfp) larvae and observed at 96 hpi. The bracket indicates location of a cluster of germinated spores in all frames. The asterisks in frames 3 and 4 indicate locations of macrophage nuclei. A focus-through animation of this image is shown in Movie S1 in the supplemental material. (C) Germinated spores are found within pools of endothelial cytoplasm, which is partially or completely engulfed by macrophages. Spores were inoculated into Tg(flk:mCherry, mpeg:dendra) larvae and imaged at 96 hpi. The bracket in all panels marks the location of the cluster of germinated spores. White arrowheads indicate a thin rim of green fluorescent signal representing macrophage cytoplasm, which surrounds areas of endothelial cytoplasm. An animation of a similar example is shown in Movie S2 in the supplemental material. (D) Endothelial interaction after yeast inoculation is present but not as extensive. Some yeasts inside macrophages are surrounded by a thin ring of endothelial cytoplasm. EGFP-expressing yeast cells were inoculated into Tg(flk:mCherry, mpeg:egfp) larvae and imaged at 24 hpi. Dashed brackets indicate the locations of two adjacent macrophages. White arrows indicate the location of a thin ring of endothelial cytoplasmic signal. 3D animation of this image is shown in Movie S3 in the supplemental material. (E) Endothelial cells are capable of engulfing cryptococcal spores in the absence of phagocytes. Spores were inoculated into PU.1 morphant Tg(flk:mCherry) larvae and imaged at 6 to 8 h after injection. The bracket indicates the location of a cluster of inoculated spores.
To examine more closely the relationship between phagocytes and endothelium at this stage, we injected both spores and yeast into Tg(flk:mCherry)/Tg(mpeg:egfp) double-transgenic larvae to identify the cytoplasms of both endothelial cells and macrophages. Serial observations of these larvae over days after inoculation with spores again revealed germinated spores inside endothelial cells. Over time these infected endothelial cells became entangled with and even partially engulfed by neighboring macrophages (Fig. 4C; for a different example, see Movie S2 in the supplemental material).
These findings contrasted with those for yeast-infected larvae, in which inoculated yeast cells were seen in small clusters surrounded by thin layers of endothelial signal (Fig. 4D; see Movie S3 in the supplemental material). Movie S4 in the supplemental material, taken at ∼3 hpi, shows inoculated yeast cells just prior to uptake by a macrophage. One of these is surrounded by endothelial signal both before and after uptake. Whether this is due to a vesicle containing endothelial cytoplasm or to adherence of fluorescent protein to the cryptococcal cells is uncertain. These data suggest that while inoculated yeast cells interact physically with endothelium, they are not able to colonize it to the extent of germinated spores. To test whether phagocytes are required for endothelial cells to take up Cryptococcus, we inoculated EGFP-expressing spores into Tg(flk:mCherry) larvae which had been injected with the PU.1 morpholino at the 1-cell stage. These morphant larvae contain no phagocytes of any kind. In these larvae, the endothelium appeared to absorb the spores gradually, with intraendothelial residence found commonly (Fig. 4E). From these findings we conclude that inoculated spores are able to occupy endothelium prior to secondary fungemia. Although inoculated yeast cells also have significant interactions with the endothelium, they are unable to colonize it in the same way.
Secondary fungemia is initiated by both lytic and nonlytic release of yeast into the bloodstream.
To determine how the transition from intracellular life back into the bloodstream occurs and whether or not endothelial occupancy affects this step, we used time-lapse confocal microscopy of infected double-transgenic [Tg(flk:mCherry, mpeg:egfp)] larvae from 72 to 96 hpi. After spore infection, we found evidence for two distinct mechanisms of escape into the bloodstream. In the first, the cytoplasmic fluorescence of both endothelium and macrophages appeared to surround a cluster of yeast cells, and the subsequent release appeared to occur without lysis of the host cell(s) (Fig. 5A; see Movie S5 in the supplemental material). This is reminiscent of the exocytic process previously observed in cryptococcal infections in vitro (48, 49, 59) and in vivo in mice (60). Although many of the yeast cells adhered to nearby macrophages after release, a subset of them entered the bloodstream. In the second mechanism, we observed abrupt disappearance of the host cytoplasmic signal at the time of release. Figure 5B shows an example, in which a green fluorescent (macrophage) cytoplasmic pocket bursts immediately before the subsequent lysis of a red fluorescent (endothelial) compartment containing yeast (Fig. 5B; see Movie S6 in the supplemental material). This finding is consistent with the phagocytosis or partial phagocytosis of an endothelial cell containing cryptococcal cells by a macrophage, which subsequently lyses. Thus, our evidence suggests that residence within, or at least in close association with, endothelial cells occurs near the time of establishment of secondary fungemia and subsequent dissemination after spore inoculation. We were unable to find significant involvement of endothelial cells during release of inoculated yeast. Thus, while both spores and yeast lead to secondary fungemia, at least during the time period we have observed, only after spore inoculation do endothelial cells appear to have a direct role.
FIG 5.
Secondary fungemia is initiated by both lytic and nonlytic release of yeast into the bloodstream. (A) Cryptococci within a peripheral endothelial cell after spore inoculation are released into the bloodstream without host cell lysis. The red channel signal representing endothelial cytoplasm rearranges abruptly but does not dissolve, consistent with release without cell lysis. The image was taken 5 days after spore inoculation into Tg(flk:mCherry, mpeg:H2B-egfp) larvae. These are still images from Movie S5 in the supplemental material; the time elapsed from the beginning of the movie (in hours and minutes) is shown in the first panel of each row. White arrowheads indicate the extent of the containing cell. Solid brackets indicate the extent of an intracellular cluster of germinated spores. The dashed bracket indicates the extent of spreading by germinated spores immediately after release. (B) Cryptococci within a complex of endothelial and macrophage cytoplasm are released into the bloodstream with apparent host cell lysis. The green signal representing macrophage cytoplasm dissolves abruptly, consistent with lysis of the host cell. Minutes later, the red channel signal representing endothelial cytoplasm also dissolves. The image was taken 5 days after spore inoculation into Tg(flk:mCherry, mpeg:dendra) larvae. These are still images from Movie S6 in the supplemental material; the time elapsed from the beginning of the movie (in hours and minutes) is shown in the first panel of each row. Magenta arrowheads indicate the edge of the endothelial compartment. Green arrowheads indicate the edge of the enclosing macrophage.
DISCUSSION
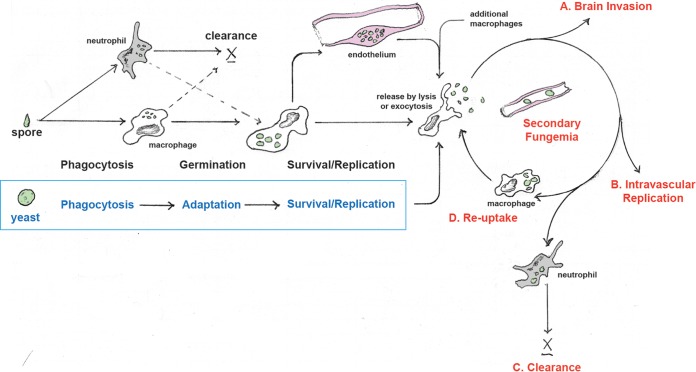
In 2000, Marta Feldmesser and colleagues wrote: “Persistence of infection in lung tissue may involve repeating cycles of phagocytosis, intracellular residence, and phagocytic cell destruction” (26). Taken together, our findings in the larval zebrafish model suggest expansion of this concept to persistence of infection in the entire host, as summarized in Fig. 6. Our findings show that shortly after inoculation, phagocytosis, predominantly by macrophages, was universal for both spores and yeast. For both cell types there was a compulsory period of residency in phagocytes, during which spores underwent germination and yeast, we propose, adapted to this intracellular niche. Based on the time measured before secondary fungemia, spores required 24 to 48 h or more within phagocytes, while inoculated yeast required as little as 12 h. It seems likely that spores need the extra time to germinate, but during this time they were capable of colonizing both macrophages and endothelial cells. There were also extensive interactions between macrophages and endothelium after yeast inoculation, but we show that these did not result in endothelial colonization. In either case, when sufficient time had passed, cryptococcal cells began making intermittent, brief tours through the bloodstream, and based on our observations were usually taken back up by macrophages. Thereafter, we propose that an equilibrium develops between intracellular cryptococcal expansion and intravascular clearance (for which neutrophils are required), with sustained, low-level fungemia lasting for most larvae throughout the observation period. Given the proclivity for yeast to lodge in the brain vasculature, eventually this low level of fungemia gave rise to CNS invasion. The sustained, low-level, and asymptomatic fungemia at the center of this model has been described in other models (17, 24) and represents a perhaps underappreciated stage in pathogenesis. Given the limits of sensitivity in clinical blood culture, asymptomatic, very low-level fungemia during human infection is plausible. There is evidence in human disease for asymptomatic fungemia or at least cryptococcal antigenemia well ahead of the development of CNS disease (6, 14), although how and when to screen for this, prevent it, or act on it remains uncertain (6, 13). It does appear that cryptococcemia can persist for long periods before invasive disease is manifest both in humans (5) and in animal studies (17). The events initiating and controlling this fungemia, whether during reactivation from a latent state or during initial infection, represent potential targets for new diagnostic or treatment approaches.
FIG 6.
Spore and yeast pathways to secondary fungemia. Inoculated spores are readily phagocytosed by macrophages and neutrophils, but survival is more likely within macrophages. Spores germinate and survive within macrophages, endothelial cells, or a combination of these. Secondary fungemia represents a cycle of release from the intracellular compartment and brief extracellular periods in the bloodstream. Possible fates of circulating cryptococci include brain invasion (A), replication within the circulation (B), clearance (especially by neutrophils) (C), or reuptake by macrophages and eventual repetition of the cycle. Inoculated yeast cells undergo a similar process, but their residency within the endothelium is either much more limited or more rapidly interrupted by macrophages.
Here we report evidence of germinated spores using peripheral endothelial cells as a temporary niche for survival. Interactions between Cryptococcus and endothelial cells have been observed both in vitro and in vivo in prior studies (17, 61–63). Subsequent investigations have focused mainly on how cryptococcal-endothelial interactions may play a part in CNS invasion (64, 65), but our results suggest that endothelial cells in other tissues may also absorb or contain the fungus. It has generally been assumed that Cryptococcus resides mainly in the lung in early infection, although human clinical disease has been found in practically every organ (4). The idea of the endothelium as a niche for cryptococcal survival and replication has been entertained in the past (66, 67), but the question of where Cryptococcus may reside between initial infection and brain invasion has been difficult to study. The zebrafish model provides a way to appreciate such a process in the context of the entire arc of pathogenesis. Formation of cryptococcomas in the zebrafish has been described already (36), and we were able to observe this phenomenon as well (data not shown), so future studies will allow the assessment of multiple niches for long-term persistence and/or initiation of reactivated disease. While we observed endothelium in particular as a nonphagocyte host cell type, there could well be other cell types hosting the fungus. Our observations of cell residency over time (Fig. 2B) revealed a gradually rising percentage of yeast cells associated with neither macrophages nor neutrophils. Whether these other yeast cells are all accounted for by endothelial residency or if there is another niche available remains unknown.
Of course, macrophages are considered to be the primary host cell harboring Cryptococcus, and our findings in the zebrafish support these conclusions. At every time point we examined, macrophages were consistently the cell type containing most intracellular cryptococcal cells. We used the IFR-8 knockout zebrafish to investigate the role of macrophages and found enhanced fungal killing at initial infection in the mutant, supporting the idea that neutrophils are better killers of Cryptococcus than macrophages are. The fact that IRF-8 mutant larvae eventually produce macrophages at 4 to 5 days postfertilization (57) is the likely reason that cryptococcal killing faded in subsequent days during this experiment. Previous work using the zebrafish to study cryptococcal infection showed markedly rapid mortality in the absence of macrophages (36). That study used the more virulent H99 strain, which belongs to the serotype A group. The difference in our results could be due to our use of the less virulent B3501 and B3502 (serotype D) strains. While there is a good argument that the studies of H99 may have more clinical relevance, many serotype D strains are also found to be pathogenic, and the excellent production of spores by the B3501 and B3502 mating pair made it a suitable background strain for this work. Also, the relatively slow development of what appear to be patterns of pathogenesis very similar to those with H99 allowed ample time for us to observe the events prior to mortality. Another possible reason for our differing results could be differences in phagocyte behavior in the context of titrated morpholino knockdown versus lack of IRF-8 function. Our finding that only a subset of germinated cryptococcal spores developed into robust, replicating yeast is likely underpinned by the known heterogeneity in zebrafish macrophage subsets, which is still under investigation (68, 69). Future work with this model will allow investigation of the impact of the different subsets of macrophages on cryptococcal spore germination and survival.
Consistent with previous findings, our results support a function for neutrophils less in early infection and more in controlling or eliminating secondary fungemia. This finding is very much in line with recent findings using the mouse model (32–34) and with clinical evidence for the efficacy of granulocyte colony-stimulating factor (G-CSF) as an adjuvant treatment for cryptococcal meningoencephalitis in AIDS patients (30, 31). Thus, impaired neutrophil function does not necessarily predispose to cryptococcal infection or meningoencephalitis but rather plays a role in host defense and could still be the target of new forms of intervention. One shortcoming of our model of spore pathogenesis is direct inoculation into the bloodstream. Whether in spore or yeast form, it is accepted that the majority of cryptococcal infections begin via inhalation. While effective models of infection involving a distinct anatomical compartment in the zebrafish have been established using the hindbrain ventricle (37) and the swim bladder (38), these approaches were problematic in this case. Inoculation into the hindbrain makes study of dissemination and CNS invasion problematic at best. As for the swim bladder, the late appearance of this structure during development reduces the time of observation possible before larvae require much more intensive husbandry. Nonetheless, our data suggest that even spores inoculated into the bloodstream interact differently with the host than their yeast parent strains, at least in the early stages of infection. Germinated spores were capable of occupying endothelial cells, and at least some of the time they initiated secondary fungemia from that reservoir. Although inoculated yeast cells interacted extensively with endothelial cells, we found no evidence that they reside inside intact endothelium in significant numbers. This difference could reflect a difference in the biology of freshly germinated yeast compared with inoculated yeast or perhaps could be a result of differing phagocyte responses to these two cell types. On the other hand, endothelial colonization might simply require more time than is allowed by the relatively short time that inoculated yeast cells spend within macrophages. Other differences we found between spore and yeast pathogenesis most likely reflect the additional time required for germination. Progression to secondary fungemia was clearly faster after yeast inoculation (Fig. 1G and 2F and G). Our observations that neutrophil function did not appear to impact secondary fungemia (Fig. 2F) or overwhelming fungemia (Fig. 3C) after spore inoculation could very well be overturned if sufficient time was allowed for secondary fungemia to be established. Beyond the issue of germination kinetics, it stands to reason that another possible factor in any pathogenic difference between spore and yeast inoculation would be the effect that germination within host phagocytes has on the regulation and expression of cryptococcal virulence factors. The fact that inoculated yeast still required some time inside phagocytes to become capable of brain invasion in this model underscores the fact that host interactions modulate pathogen responses to changing conditions throughout the time course of infection.
In conclusion, while in part expanding upon the findings about cryptococcal pathogenesis already made with this new model host (35, 36), we have also been able to reconcile some previously conflicting findings in other models by taking advantage of the unique access to both local and host-wide events that the zebrafish larva provides. Detailing the process chronologically and simultaneously in multiple tissues provides a chance to stitch together prior findings in specific tissues or cell types. This perspective has also enabled us to delineate the role of sustained, low-grade fungemia in overall pathogenesis. Future investigations with this model will focus on defining the host and cryptococcal factors required for this prolonged fungemia, with the hope that such factors may provide a specific target for timely diagnosis and intervention.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Huttenlocher and Hull labs for critical review of the manuscript and for animal care. Bruce Klein, Damian Krysan, and Lalita Ramakrishnan provided helpful discussion and criticism. Benjamin Knox provided insights from preliminary data not included in this publication. Heinz-Georg Belting and Markus Affolter kindly provided the Tg(flk:mCherry) line of zebrafish.
J.M.D. performed all experimental procedures except as noted and wrote and edited the manuscript. M.H. performed isolation of spores for inoculation. M.R.B. constructed the cryptococcal GFP expression strain pair. C.M.H. and A.H. provided material support and oversight of the experiments, with heavy editorial input on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00506-16.
REFERENCES
- 1.Faini D, Maokola W, Furrer H, Hatz C, Battegay M, Tanner M, Denning DW, Letang E. 2015. Burden of serious fungal infections in Tanzania. Mycoses 58(Suppl 5):S70–S79. [DOI] [PubMed] [Google Scholar]
- 2.Parkes-Ratanshi R, Achan B, Kwizera R, Kambugu A, Meya D, Denning DW. 2015. Cryptococcal disease and the burden of other fungal diseases in Uganda; where are the knowledge gaps and how can we fill them? Mycoses 58(Suppl 5):S85–S93. [DOI] [PubMed] [Google Scholar]
- 3.Desalermos A, Kourkoumpetis TK, Mylonakis E. 2012. Update on the epidemiology and management of cryptococcal meningitis. Expert Opin Pharmacother 13:783–789. doi: 10.1517/14656566.2012.658773. [DOI] [PubMed] [Google Scholar]
- 4.Perfect JR. 2015. Cryptococcosis (Cryptococcus neoformans and Cryptococcus gatti), p 2934–2948. In Bennett JE, Dolin R, Blaser M.J (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 8th ed Elsevier Saunders, New York, NY. [Google Scholar]
- 5.French N, Gray K, Watera C, Nakiyingi J, Lugada E, Moore M, Lalloo D, Whitworth JA, Gilks CF. 2002. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS 16:1031–1038. doi: 10.1097/00002030-200205030-00009. [DOI] [PubMed] [Google Scholar]
- 6.Meyer AC, Jacobson M. 2013. Asymptomatic cryptococcemia in resource-limited settings. Curr HIV/AIDS Rep 10:254–263. doi: 10.1007/s11904-013-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMullan BJ, Sorrell TC, Chen SC. 2013. Cryptococcus gattii infections: contemporary aspects of epidemiology, clinical manifestations and management of infection. Future Microbiol 8:1613–1631. doi: 10.2217/fmb.13.123. [DOI] [PubMed] [Google Scholar]
- 8.Coelho C, Bocca AL, Casadevall A. 2014. The intracellular life of Cryptococcus neoformans. Annu Rev Pathol 9:219–238. doi: 10.1146/annurev-pathol-012513-104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol 60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 10.Davis J, Zheng WY, Glatman-Freedman A, Ng JA, Pagcatipunan MR, Lessin H, Casadevall A, Goldman DL. 2007. Serologic evidence for regional differences in pediatric cryptococcal infection. Pediatr Infect Dis J 26:549–551. doi: 10.1097/INF.0b013e318047e073. [DOI] [PubMed] [Google Scholar]
- 11.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, Casadevall A. 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107:E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol 37:3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liechty CA, Solberg P, Were W, Ekwaru JP, Ransom RL, Weidle PJ, Downing R, Coutinho A, Mermin J. 2007. Asymptomatic serum cryptococcal antigenemia and early mortality during antiretroviral therapy in rural Uganda. Trop Med Int Health 12:929–935. doi: 10.1111/j.1365-3156.2007.01874.x. [DOI] [PubMed] [Google Scholar]
- 14.Rajasingham R, Meya DB, Boulware DR. 2012. Integrating cryptococcal antigen screening and pre-emptive treatment into routine HIV care. J Acquir Immune Defic Syndr 59:e85–e91. doi: 10.1097/QAI.0b013e31824c837e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman D, Lee SC, Casadevall A. 1994. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect Immun 62:4755–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibuya K, Hirata A, Omuta J, Sugamata M, Katori S, Saito N, Murata N, Morita A, Takahashi K, Hasegawa C, Mitsuda A, Hatori T, Nonaka H. 2005. Granuloma and cryptococcosis. J Infect Chemother 11:115–122. doi: 10.1007/s10156-005-0387-X. [DOI] [PubMed] [Google Scholar]
- 17.Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. 2002. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J Infect Dis 186:522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- 18.Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM. 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun 77:3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopold Wager CM, Hole CR, Wozniak KL, Wormley FL Jr. 2016. Cryptococcus and phagocytes: complex interactions that influence disease outcome. Front Microbiol 7:105. doi: 10.3389/fmicb.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston SA, May RC. 2013. Cryptococcus interactions with macrophages: evasion and manipulation of the phagosome by a fungal pathogen. Cell Microbiol 15:403–411. doi: 10.1111/cmi.12067. [DOI] [PubMed] [Google Scholar]
- 21.Vu K, Eigenheer RA, Phinney BS, Gelli A. 2013. Cryptococcus neoformans promotes its transmigration into the central nervous system by inducing molecular and cellular changes in brain endothelial cells. Infect Immun 81:3139–3147. doi: 10.1128/IAI.00554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, Kim KS. 2012. Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell Microbiol 14:1544–1553. doi: 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. 2009. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun 77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lortholary O, Improvisi L, Nicolas M, Provost F, Dupont B, Dromer F. 1999. Fungemia during murine cryptococcosis sheds some light on pathophysiology. Med Mycol 37:169–174. [PubMed] [Google Scholar]
- 25.Griffiths EJ, Kretschmer M, Kronstad JW. 2012. Aimless mutants of Cryptococcus neoformans: failure to disseminate. Fungal Biol Rev 26:61–72. doi: 10.1016/j.fbr.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leopold Wager CM, Wormley FL Jr. 2014. Classical versus alternative macrophage activation: the Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol 7:1023–1035. doi: 10.1038/mi.2014.65. [DOI] [PubMed] [Google Scholar]
- 28.Mednick AJ, Feldmesser M, Rivera J, Casadevall A. 2003. Neutropenia alters lung cytokine production in mice and reduces their susceptibility to pulmonary cryptococcosis. Eur J Immunol 33:1744–1753. doi: 10.1002/eji.200323626. [DOI] [PubMed] [Google Scholar]
- 29.Wozniak KL, Kolls JK, Wormley FL Jr. 2012. Depletion of neutrophils in a protective model of pulmonary cryptococcosis results in increased IL-17A production by gammadelta T cells. BMC Immunol 13:65. doi: 10.1186/1471-2172-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coffey MJ, Phare SM, George S, Peters-Golden M, Kazanjian PH. 1998. Granulocyte colony-stimulating factor administration to HIV-infected subjects augments reduced leukotriene synthesis and anticryptococcal activity in neutrophils. J Clin Invest 102:663–670. doi: 10.1172/JCI2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vecchiarelli A, Monari C, Baldelli F, Pietrella D, Retini C, Tascini C, Francisci D, Bistoni F. 1995. Beneficial effect of recombinant human granulocyte colony-stimulating factor on fungicidal activity of polymorphonuclear leukocytes from patients with AIDS. J Infect Dis 171:1448–1454. doi: 10.1093/infdis/171.6.1448. [DOI] [PubMed] [Google Scholar]
- 32.Sun D, Zhang M, Liu G, Wu H, Li C, Zhou H, Zhang X, Shi M. 2016. Intravascular clearance of disseminating Cryptococcus neoformans in the brain can be improved by enhancing neutrophil recruitment in mice. Eur J Immunol doi: 10.1002/eji.201546239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Zhang M, Liu G, Wu H, Zhu X, Zhou H, Shi M. 2016. Real-time imaging of interactions of neutrophils with Cryptococcus neoformans demonstrates a crucial role of complement C5a-C5aR signaling. Infect Immun 84:216–229. doi: 10.1128/IAI.01197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, Sun D, Liu G, Wu H, Zhou H, Shi M. 2016. Real-time in vivo imaging reveals the ability of neutrophils to remove Cryptococcus neoformans directly from the brain vasculature. J Leukoc Biol 99:467–473. doi: 10.1189/jlb.4AB0715-281R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bojarczuk A, Miller KA, Hotham R, Lewis A, Ogryzko NV, Kamuyango AA, Frost H, Gibson RH, Stillman E, May RC, Renshaw SA, Johnston SA. 2016. Cryptococcus neoformans intracellular proliferation and capsule size determines early macrophage control of infection. Sci Rep 6:21489. doi: 10.1038/srep21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenor JL, Oehlers SH, Yang JL, Tobin DM, Perfect JR. 2015. Live imaging of host-parasite interactions in a zebrafish infection model reveals cryptococcal determinants of virulence and central nervous system invasion. mBio 6:e01425-15. doi: 10.1128/mBio.01425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JM, Ramakrishnan L. 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gratacap RL, Rawls JF, Wheeler RT. 2013. Mucosal candidiasis elicits NF-kappaB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Model Mech 6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knox BP, Deng Q, Rood M, Eickhoff JC, Keller NP, Huttenlocher A. 2014. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot Cell 13:1266–1277. doi: 10.1128/EC.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varela M, Romero A, Dios S, van der Vaart M, Figueras A, Meijer AH, Novoa B. 2014. Cellular visualization of macrophage pyroptosis and interleukin-1beta release in a viral hemorrhagic infection in zebrafish larvae. J Virol 88:12026–12040. doi: 10.1128/JVI.02056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Q, Yoo SK, Cavnar PJ, Green JM, Huttenlocher A. 2011. Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev Cell 21:735–745. doi: 10.1016/j.devcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvie EA, Green JM, Neely MN, Huttenlocher A. 2013. Innate immune response to Streptococcus iniae infection in zebrafish larvae. Infect Immun 81:110–121. doi: 10.1128/IAI.00642-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon-Chung KJ. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833. doi: 10.2307/3758800. [DOI] [PubMed] [Google Scholar]
- 44.Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol 101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 45.Alspaugh JA, Perfect JR, Heitman J. 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol 25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F. 2002. Getting started with yeast. Methods Enzymol 350:3–41. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
- 47.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. 2009. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell 8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chrisman CJ, Alvarez M, Casadevall A. 2010. Phagocytosis of Cryptococcus neoformans by, and nonlytic exocytosis from, Acanthamoeba castellanii. Appl Environ Microbiol 76:6056–6062. doi: 10.1128/AEM.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 50.Cosma CL, Swaim LE, Volkman H, Ramakrishnan L, Davis JM. 2006. Zebrafish and frog models of Mycobacterium marinum infection. Curr Protoc Microbiol Chapter 10:Unit 10B.12. [DOI] [PubMed] [Google Scholar]
- 51.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. 2005. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 52.Hachicho N, Reithel S, Miltner A, Heipieper HJ, Kuster E, Luckenbach T. 2015. Body mass parameters, lipid profiles and protein contents of zebrafish embryos and effects of 2,4-dinitrophenol exposure. PLoS One 10:e0134755. doi: 10.1371/journal.pone.0134755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westerfield M. 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- 54.Wang Y, Kaiser MS, Larson JD, Nasevicius A, Clark KJ, Wadman SA, Roberg-Perez SE, Ekker SC, Hackett PB, McGrail M, Essner JJ. 2010. Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development 137:3119–3128. doi: 10.1242/dev.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693–702. doi: 10.1016/S1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 56.Reference deleted.
- 57.Shiau CE, Kaufman Z, Meireles AM, Talbot WS. 2015. Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLoS One 10:e0117513. doi: 10.1371/journal.pone.0117513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Y, Williams DA. 2002. RAC2 GTPase deficiency and myeloid cell dysfunction in human and mouse. J Pediatr Hematol Oncol 24:791–794. doi: 10.1097/00043426-200212000-00027. [DOI] [PubMed] [Google Scholar]
- 59.Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr Biol 16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 60.Nicola AM, Robertson EJ, Albuquerque P, Derengowski Lda S, Casadevall A. 2011. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. mBio 2:e00167-11. doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ibrahim AS, Filler SG, Alcouloumre MS, Kozel TR, Edwards JE Jr, Ghannoum MA. 1995. Adherence to and damage of endothelial cells by Cryptococcus neoformans in vitro: role of the capsule. Infect Immun 63:4368–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roseff SA, Levitz SM. 1993. Effect of endothelial cells on phagocyte-mediated anticryptococcal activity. Infect Immun 61:3818–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaoka H, Sakaguchi N, Sano K, Ito M. 1996. Intravascular granuloma induced by intravenous inoculation of Cryptococcus neoformans. Mycopathologia 133:149–158. doi: 10.1007/BF02373022. [DOI] [PubMed] [Google Scholar]
- 64.Chen SH, Stins MF, Huang SH, Chen YH, Kwon-Chung KJ, Chang Y, Kim KS, Suzuki K, Jong AY. 2003. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J Med Microbiol 52:961–970. doi: 10.1099/jmm.0.05230-0. [DOI] [PubMed] [Google Scholar]
- 65.Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J Clin Invest 120:1683–1693. doi: 10.1172/JCI41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coenjaerts FE, Hoepelman AI, Scharringa J, Aarts M, Ellerbroek PM, Bevaart L, Van Strijp JA, Janbon G. 2006. The Skn7 response regulator of Cryptococcus neoformans is involved in oxidative stress signalling and augments intracellular survival in endothelium. FEMS Yeast Res 6:652–661. doi: 10.1111/j.1567-1364.2006.00065.x. [DOI] [PubMed] [Google Scholar]
- 67.Feldmesser M, Tucker S, Casadevall A. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol 9:273–278. doi: 10.1016/S0966-842X(01)02035-2. [DOI] [PubMed] [Google Scholar]
- 68.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. 2014. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen-Chi M, Laplace-Builhe B, Travnickova J, Luz-Crawford P, Tejedor G, Phan QT, Duroux-Richard I, Levraud JP, Kissa K, Lutfalla G, Jorgensen C, Djouad F. 2015. Identification of polarized macrophage subsets in zebrafish eLife 4:e07288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Accetta D, Syverson G, Bonacci B, Reddy S, Bengtson C, Surfus J, Harbeck R, Huttenlocher A, Grossman W, Routes J, Verbsky J. 2011. Human phagocyte defect caused by a Rac2 mutation detected by means of neonatal screening for T-cell lymphopenia. J Allergy Clin Immunol 127:535–538. doi: 10.1016/j.jaci.2010.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.