Abstract
Mycobacterium abscessus is an emerging pathogenic mycobacterium involved in pulmonary and mucocutaneous infections, presenting a serious threat for patients with cystic fibrosis (CF). The lack of an efficient treatment regimen and the emergence of multidrug resistance in clinical isolates require the development of new therapeutic strategies against this pathogen. Reverse genetics has revealed genes that are present in M. abscessus but absent from saprophytic mycobacteria and that are potentially involved in pathogenicity. Among them, MAB_3593 encodes MgtC, a known virulence factor involved in intramacrophage survival and adaptation to Mg2+ deprivation in several major bacterial pathogens. Here, we demonstrated a strong induction of M. abscessus MgtC at both the transcriptional and translational levels when bacteria reside inside macrophages or upon Mg2+ deprivation. Moreover, we showed that M. abscessus MgtC was recognized by sera from M. abscessus-infected CF patients. The intramacrophage growth (J774 or THP1 cells) of a M. abscessus knockout mgtC mutant was, however, not significantly impeded. Importantly, our results indicated that inhibition of MgtC in vivo through immunization with M. abscessus mgtC DNA, formulated with a tetrafunctional amphiphilic block copolymer, exerted a protective effect against an aerosolized M. abscessus challenge in CF (ΔF508 FVB) mice. The formulated DNA immunization was likely associated with the production of specific MgtC antibodies, which may stimulate a protective effect by counteracting MgtC activity during M. abscessus infection. These results emphasize the importance of M. abscessus MgtC in vivo and provide a basis for the development of novel therapeutic tools against pulmonary M. abscessus infections in CF patients.
INTRODUCTION
Mycobacterium abscessus, a rapidly growing mycobacterium (RGM), is the etiological agent of a wide spectrum of infections in humans. M. abscessus is responsible for severe and persistent pulmonary infections, disseminated cutaneous diseases, and posttrauma and postsurgery wound infections, mostly in immunocompetent patients and in cystic fibrosis (CF) patients (1–5). Infections by this pathogen lead to a fatality rate that is higher than those seen with other RGM species. Infection of CF patients with M. abscessus is a major health-related issue in most CF centers worldwide (1, 2, 6, 7) and is considered a contraindication to lung transplantation, leaving these patients with minimal therapeutic options.
M. abscessus provokes very challenging therapeutic issues because of its natural resistance to most available antibiotics (8–10) and is considered the most drug-resistant mycobacterial species. The lack of optimal therapeutic treatments and the emergence of multidrug resistance in M. abscessus stress the need for the discovery of new strategies to combat these infections.
Reverse genetics, based on the comparison between pathogenic and nonpathogenic species, allows the identification of genes missing in nonpathogenic species and encoding potential virulence factors. One major outcome of this comparison relies on (i) an understanding of the impact of the identified genes during infection and (ii) attempts at modulating the virulence phenotype by stimulating an efficient adaptive immune response. As an example, mining the M. abscessus genome sequence has revealed several “nonmycobacterial” virulence genes, presumably acquired by horizontal gene transfer (HGT) and absent from the saprophytic RGM Mycobacterium smegmatis (11). Interestingly, several of these genes are shared with other major CF pathogens, such as Pseudomonas aeruginosa or Burkholderia cenocepacia. This is the case with the phospholipase C (PLC)-encoding gene, which not only was important for virulence in an M. abscessus mouse model of infection (12) but also exhibited potent vaccine properties in CF (ΔF508 FVB) mice immunized with plc-encoding DNA through the production of high anti-PLC antibody titers (13). As such, certain virulence factors may be viewed as relevant antigen targets for the development of protective vaccines against human pathogens (14–16).
In addition to PLC, comparative genomics revealed another M. abscessus gene, MAB_3593, absent from M. smegmatis (11). MAB_3593 encodes a protein homologous to MgtC, a virulence factor shared by several intracellular pathogens, including Mycobacterium tuberculosis (17, 18). This factor contributes to intramacrophage survival of numerous pathogens as well as to bacterial adaptation in environments with limited Mg2+ concentrations (18). MgtC was found to be highly expressed in cellular infection models for several pathogens (19, 20), which emphasizes its importance during infection. MgtC expression in M. abscessus has not yet been evaluated in cellular models or in environments with poor Mg2+ concentrations, nor has its role during infection been addressed.
As a first step toward the elucidation of the involvement of MAB_3593 in the virulence of M. abscessus, we created a mutant strain in which the MAB_3593 gene encoding M. abscessus MgtC (referred to here as MgtCMAB) has been disrupted. We next analyzed the expression of MgtC in a low-Mg2+ environment and within infected cells, which prompted us to evaluate the protection against M. abscessus conferred to mice after immunization using a formulated mgtCMAB DNA preparation.
MATERIALS AND METHODS
Bacterial strains and growth culture conditions.
M. abscessus strains (derivatives of CIP 104536T, smooth variant) were grown at 37°C in Sauton's medium containing 0.025% tyloxapol (Sigma) or on LB plates, in the presence of zeocin (25 μg/ml), when required. Low-magnesium medium was obtained by replacing the magnesium sulfate in the Sauton's medium with a similar concentration of potassium sulfate. Escherichia coli (DH5α) was used for cloning and was grown in LB medium with zeocin (25 μg/ml) at 37°C. Salmonella enterica serovar Typhimurium strains, derived from wild-type strain 14028s, were grown in LB medium with ampicillin (100 μg/ml) at 37°C.
Cloning of mgtCMAB-like genes under the control of the Salmonella mgtC promoter and heterologous complementation of a Salmonella mgtC mutant strain.
The MAB_3593 and MAB_0146 genes from M. abscessus were amplified by PCR using CIP 104536T culture as the template and primers indicated in Table S1 in the supplemental material. Each PCR fragment was cloned at the SphI and EcoRI sites of plasmid pNM11, a pBR322 plasmid derivative that harbors the S. Typhimurium mgtC promoter (21). The resulting plasmids express the MgtC-like proteins from M. abscessus with transcriptional and translational regulation sites of the mgtC gene of S. Typhimurium. Mutation W227A was introduced in a plasmid carrying MAB_3593 using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions and the primers listed in Table S1. The S. Typhimurium NM14 strain (ΔmgtC mutant) (21) was transformed by electroporation with plasmids harboring the MAB_3593 or MAB_0146 gene. NM14 was also transformed with pNM11, which harbors the mgtC promoter only, used as a negative control, with pNM12, which expresses the S. Typhimurium mgtC gene as positive control (21), and with pNM13, which expresses M. tuberculosis MgtC (22). Growth in magnesium-deprived medium was performed in no-carbon essential (NCE) medium supplemented with 10 μM MgCl2, as described previously (23), and the optical density at 600 nm (OD600) was measured after 18 h of growth.
Construction of the MAB_3593 mutant and the complemented strain.
The mgtC M. abscessus knockout (KO) mutant was obtained by allelic exchange in M. abscessus CIP-S with a strategy using phage recombinase as previously reported (24, 25). A linear DNA fragment (100 ng), carrying the zeocin cassette (from Streptoalloteichus hindustanus) as well as 1,000 bp of the region surrounding each side of the MAB_3593 gene (see primers in Table S1 in the supplemental material), was introduced into electrocompetent M. abscessus CIP-S bearing the pJV53 recombineering plasmid (26). Cells with a homologous recombination event were selected on LB agar plates supplemented with zeocin. Proper replacement of the mgtC gene by the zeocin cassette was checked by a PCR screen using forward and reverse primers outside the deleted region (see Table S1).
To construct a complemented strain, mgtC with its upstream region (500 bp) was first amplified with primers MgtC-reg-PvuII and MgtC-reg-HindIII (see Table S1 in the supplemental material) and cloned into the integrative pMVH361-hygromycin plasmid. This plasmid was then transformed in the mgtC M. abscessus KO mutant.
Macrophage infection assays.
J774 cells were maintained at 37°C in 5% CO2 using Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco). J774 cells were allowed to adhere in a 24-well plate at a density of 5 × 104 cells/well for 24 h at 37°C in 5% CO2.
For infection, M. abscessus cultures grown exponentially in Sauton's medium (OD600 between 0.8 and 1) were centrifuged and washed in phosphate-buffered saline (PBS) and bacterial clumps were disrupted by 6 successive passages through a 26-gauge syringe needle. The remaining aggregates were then eliminated with a short spin procedure (1 min at 1,100 rpm). Macrophages were infected at a multiplicity of infection (MOI) of approximately 2. The infection was allowed to proceed for 3 h before four PBS washes were performed to remove extracellular bacteria. Cells were then lysed with 0.1% Triton X-100–PBS, and the number of intracellular mycobacteria was counted by plating appropriate dilutions onto LB agar plates. To evaluate the multiplication rate, infected cells were incubated for 48 h in DMEM prior to cell lysis and bacterial enumeration. The ratio between the numbers of internalized bacteria at 24 h or 48 h and the numbers of internalized bacteria at 3 h after infection was calculated.
THP1 cells were maintained at 37°C in 5% CO2 using RPMI medium (Gibco) supplemented with 10% FBS and were differentiated into adherent macrophages by adding 60 nM phorbol myristate acetate (PMA) for 72 h. Infection was carried out as described above in a 24-well plate at a density of 2 × 105 cells/well.
RNA extraction, qRT-PCR, and analysis of protein expression by Western blotting.
RNA and cDNA were prepared from 5 ml of mid-logarithmic-phase bacterial cultures grown in Sauton's medium containing or not containing magnesium as previously described (27). Control experiments without reverse transcriptase were performed for each RNA sample to rule out DNA contamination. The 16S rRNA (rrs) gene was used as an internal control (28). The sequences of primers used for quantitative real-time PCR (qRT-PCR) are listed in Table S1 in the supplemental material. For isolation of bacterial RNA from M. abscessus-infected J774 macrophages, cells were seeded into a 100-cm2 tissue culture dish and infected at an MOI of 5. After a 2-h incubation period, cells were washed with phosphate-buffered saline (PBS) medium and then incubated for 1 h in DMEM–200 μg/ml amikacin. After 24 h, cells were harvested, washed with PBS, and lysed with PBS containing 0.1% Triton X-100. Bacteria were then pelleted by centrifugation at 13,000 rpm for 10 min at 15°C. Bacteria were finally resuspended by adding 500 μl PBS, and total RNA was isolated as described above.
To prepare bacterial lysates, M. abscessus cultures grown in Sauton's medium with or without magnesium were centrifuged at 4,000 × g and resuspended in Tris-buffered saline (TBS) (the resuspension volume was normalized according to OD600). M. abscessus lysates from macrophage cocultures were prepared following J774 infection by pelleting the cells by centrifugation at 900 × g for 5 min and resuspending the pellet in 5 ml cooled PBS–1% Triton X-100 for 5 min at room temperature. After the addition of 2.5 ml of PBS, the samples were centrifuged at 4,000 × g for 15 min at 4°C and resuspended in TBS and processed as described for the classical M. abscessus cultures. Samples were sonicated on ice three times for 30 s each time and centrifuged at 14,000 × g, and protein concentrations in the lysates were determined using the Bradford assay. The crude extracts were then separated using 12.5% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with mouse serum collected after DNA immunization (see below). The specificity of the mouse serum was checked using a crude lysate from M. smegmatis expressing the recombinant His-tagged MgtCMAB protein. A peroxidase-conjugated goat anti-mouse IgG (H + L, Southern Biotech) was then added (dilution, 1/5,000) prior to detection using the peroxidase substrate. Rat anti-KasA antibodies (29) were used as a loading control.
Serum samples from CF patients enrolled in the French study cohort “OMA” (4) were diluted 1:1,000 for Western blot analysis.
Immunization protocol in mice.
Plasmid pVAX1-MAB_3593 (3,654 bp) contains a kanamycin resistance gene and a MAB_3593 DNA insertion of 723 bp under the control of a previously reported cytomegalovirus (CMV) promoter (13). The mgtCMAB gene sequence was optimized for expression in eukaryotic cells by substitution of rare codons. The control plasmid pCMV β-galactosidase carries an ampicillin resistance gene and a DNA-E. coli β-galactosidase gene (3,141 bp) under the control of a CMV promoter (13). The immunization scheme shown in Fig. S1 in the supplemental material was followed because it was successfully used with other antigens as reported earlier (13). We used a synthetic delivery system composed of a tetrafunctional 704 block copolymer that has been shown to increase dramatically the vaccination efficiency over that seen with naked DNA in mouse model (30). The polymer was administered by intramuscular injection. For the aerosol challenge, mycobacterial inocula of M. abscessus CIP 104536T were prepared as described previously (31, 32). Five to seven 6-week-old homozygote ΔF508 FVB female mice (33) were aerosolized with M. abscessus on day 56, i.e., 14 days after the last immunization as previously described (13). Mice were sacrificed on days 1, 7, 14, and 21 after infection. Lungs, spleen, and liver were removed aseptically, and CFU counts were determined as described previously (34).
Statistical analysis.
Student's t test and Fisher's exact test were used. A P value of <0.05 was considered significant.
RESULTS
M. abscessus encodes two MgtC-like proteins that belong to different phylogenetic clusters.
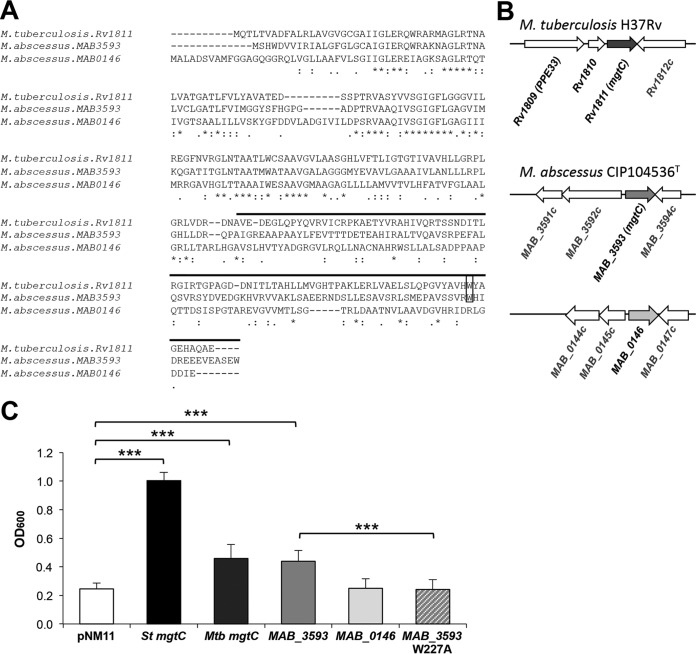
The presence of an MgtC-encoding gene (MAB_3593) in M. abscessus was recently reported (11). MAB_3593 exhibits 43% identity with and 59% similarity (at the protein level) to M. tuberculosis MgtC (Rv1811; here referred to as MgtCMTB) (Fig. 1A). However, the genomic organization around mgtC seen with MAB_3593 is distinct from that seen with Rv1811 (Fig. 1B), and MAB_3593 was probably selected by HGT acquisition in M. abscessus together with MAB_3592c (11) (Fig. 1B). MAB_3593 belongs to the same phylogenetic cluster as MgtCMTB, considered the cluster of MgtC proteins with an intracellular function (11, 35). This cluster includes MgtC members in other important CF pathogens, such as Pseudomonas aeruginosa, Burkholderia cenocepacia, Mycobacterium avium, and Stenotrophomonas maltophilia (see Fig. S2 in the supplemental material). Intriguingly, a second MgtC-like protein is encoded by the M. abscessus genome (MAB_0146) (http://bigsdb.web.pasteur.fr/mycoabscessus/mycoabscessus.html). MAB_0146 is more divergent than Rv1811, especially in the C-terminal domain (Fig. 1A), and belongs to a different phylogenetic clade that includes an MgtC-like protein from M. avium and one from another CF pathogen, the fungus Aspergillus fumigatus (36) (see Fig. S2).
FIG 1.
MgtC-like proteins encoded by M. abscessus genome. (A) Alignment of M. tuberculosis MgtC (MgtCMTB) and M. abscessus MgtC-like proteins (MAB_3593 and MAB_0146) was done with the T-COFFEE Multiple Sequence Alignment Server (tcoffee.crg.cat/). The upper line indicates the predicted soluble C-terminal part. Near the C terminus, a conserved tryptophan residue identified as important for MgtC function in Salmonella in Mg2+-deprived medium is indicated by a black rectangle. Conservation of residues is indicated below the sequences as follows: asterisk (*), residues that are identical in all sequences in the alignment; colon (:), conserved substitutions; period (.), semiconserved. (B) Genomic organization around mgtC-like genes. (C) Functional analysis of M. abscessus MgtC-like proteins by heterologous complementation of a Salmonella mgtC mutant strain compared to MgtCMTB. A Salmonella Typhimurium (St) mgtC mutant was transformed with plasmids harboring MAB_3593, MAB_0146, or mgtCMTB (Rv1811). A mutation was introduced in MAB_3593 to test the role of functional relevance of the W227 residue. The OD600 was measured after growth of complemented strains in medium with a low concentration of Mg2+ for 18 h. A negative control was provided with a strain harboring an empty vector (pNM11), whereas a positive control was provided with a strain expressing the Salmonella mgtC gene (mgtCST). Data represent the mean values plus standard errors of results from at least three independent experiments. Mtb, M. tuberculosis; ***, P < 0.001 (Student t test).
MAB_3593 (mgtCMAB) complements partially a Salmonella mgtC mutant.
Heterologous complementation studies were carried out using a Salmonella mgtC mutant with both M. abscessus mgtC-like genes, which provide a simple and rapid way to test for the functionality and activity of each copy (21, 22). In fact, the Salmonella mgtC mutant is defective for growth in low-magnesium (Mg2+) medium and we have previously shown that heterologous complementation with MgtCMTB significantly improves the growth of the Salmonella mgtC mutant in such a medium (22) (Fig. 1C). A partial complementation was also seen with MAB_3593 (Fig. 1C), to levels similar to those obtained with mgtCMTB, suggesting a conserved function with regard to adaptation to Mg2+ deprivation. In contrast, no in vitro complementation was noticed with MAB_0146 in low-Mg2+ medium. Interestingly, a tryptophan residue near the C-terminal end of MgtC that is important for MgtC function in low-Mg2+ medium (21) is conserved in MAB_3593 but not in MAB_0146 (Fig. 1A). Replacement of Trp227 residue by an Ala (W227A) in MAB_3593 suppressed the partial complementation (Fig. 1C), supporting the idea of the functional importance of this residue, as reported for other MgtC proteins. As MAB_3593 appears to be the sole M. abscessus MgtC-like protein that shares functional similarity with the Salmonella and M. tuberculosis MgtC proteins, it is referred to here as MgtCMAB.
An mgtCMAB mutant is defective for optimal growth in Mg2+-deprived medium.
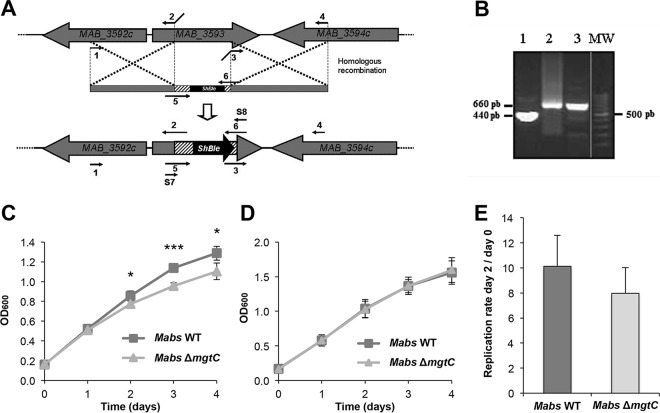
A M. abscessus mgtC KO mutant was generated by allelic exchange using the recombineering system (25) with insertion of a zeocin cassette in MAB_3593 (Fig. 2A). PCR screening was performed to check for the proper replacement of mgtCMAB by a zeocin cassette (Fig. 2B). We then investigated the phenotype of the ΔmgtCMAB mutant in low-Mg2+ medium and in macrophages. The ΔmgtCMAB mutant exhibited a growth defect compared to the wild-type strain in Mg2+-deprived medium (Fig. 2C) but not in medium supplemented with Mg2+ (Fig. 2D). Complementation of the mutation, with an integrative plasmid carrying the mgtC gene and its upstream promoter region, suppressed the growth defect in Mg2+-deprived medium (see Fig. S3 in the supplemental material). These results emphasize the contribution of MgtCMAB in the adaptive response of M. abscessus to low-Mg2+ concentrations. Similarly, the ΔmgtCMAB mutant exhibited a replication rate that was slightly lower than those seen with the parental strain in J774 murine macrophages (Fig. 2E) and THP1 human macrophages (see Fig. S4) after 2 days of infection. There was a similar finding at a shorter infection time (replication rates of 2.4 ± 0.44 for the wild-type strain and 2.1 ± 0.52 for mutant strain at 24 h postinfection). However, for both macrophage cell lines, the differences in the intracellular behaviors between the ΔmgtCMAB mutant and the control strain were not significant.
FIG 2.
Role of MgtCMAB in adaptation to magnesium (Mg) deprivation and intramacrophage environment. (A) Construction of the ΔmgtCMAB mutant by homologous recombination (HR). Primers were designed for amplification of nearly 1,000 bp of two M. abscessus regions encompassing mgtC and containing the initial and the final sequences of this gene and for cloning between them and the zeocin resistance gene (Streptoalloteichus hindustanus [ShBle]). The entire fragment was then electroporated into M. abscessus CIP-S containing the pJV53 plasmid for homologous recombination and integration into the M. abscessus chromosome. Numbers 1 to 6 correspond to primers AM-MgtC F, AM-MgtC R, AV-MgtC F, AV-MgtC R, Zeo-MgtC F, and Zeo-MgtC R, which are described in Table S1 in the supplemental material, respectively. Details of primers Screening 7 (S7) and Screening 8 (S8), used for mutant screening, are given also in Table S1. (B) Validation of the ΔmgtCMAB mutant by PCR using primers Screening 7 and Screening 8. PCR was performed on the M. abscessus CIP wild-type strain (lane 1); on the PCR product used for allelic exchange (lane 2); or on a zeocin-resistant clone selected after the recombination event (lane 3). Lane MW, DNA ladder (lane taken from a different area of the same gel); pb, base pairs. (C and D) Growth of the ΔmgtCMAB mutant in Mg2+-deprived liquid medium. The growth curves represent wild-type (WT) and ΔmgtCMAB strains grown in Sauton's medium without magnesium (C) or in regular Sauton's medium supplemented with magnesium (D). OD600 data over the growth period are indicated. The experiment was independently repeated three times (*, P < 0.05; ***, P < 0.001). Mabs, M. abscessus. (E) Replication of the mgtC mutant in the J774 macrophage cell line after 48 h. Results are expressed as means + standard deviations (SD) of the results from four independent experiments. The difference in replication rates between the two strains is not significant (P = 0.1, Student's t test).
mgtCMAB expression is induced in low-Mg2+ medium and within macrophages.
The observed defect of the ΔmgtCMAB mutant in Mg2+-deprived medium raises the issue of mgtC regulation as a consequence of the use of various Mg2+ concentrations. To address this issue, the expression of mgtCMAB was investigated in the wild-type M. abscessus strain at both the transcriptional and translational levels. Bacteria were grown in Sauton's medium supplemented with Mg2+ or not supplemented prior to mRNA extraction. Expression of the mgtCMAB transcripts was determined along with that of two other genes: MAB_3592c, located immediately upstream of mgtCMAB (Fig. 1B), and the mgtC-like MAB_0146 gene. Quantitative RT-PCR performed using the 16S rRNA gene as an internal control indicated that expression of mgtC was highly (about 50-fold) induced by Mg2+ deprivation. In sharp contrast, minor (2- to 3-fold) transcriptional induction was noticed for MAB_3592c or MAB_0146 in the absence of Mg2+ (Fig. 3A).
FIG 3.
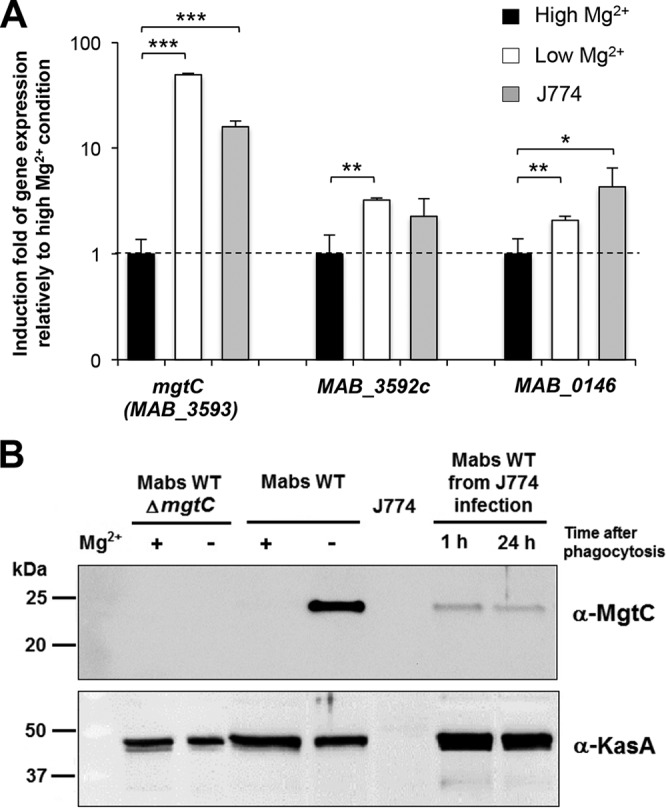
Expression of mgtCMAB in Sauton's medium and inside macrophages. (A) Quantification of mgtCMAB (MAB_3593), MAB_3592c, and MAB_0146 RNA expression by qRT-PCR using RNA isolated from the wild-type strain grown in medium with a high or low Mg2+ concentration or internalized for 24 h inside J774 macrophages. The 16S rRNA gene was used as an internal standard. Results are expressed as means + standard deviations of the data from three experiments performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student t test). (B) Expression of M. abscessus MgtC protein from wild-type M. abscessus grown under conditions of a high or low Mg2+ concentration or internalized for 1 or 24 h inside J774 macrophages. Lysate of M. abscessus mgtC mutant grown with or without Mg2+ was loaded as a negative control. Bacterial lysates equivalent to 4 μg (lysates from bacterial cultures) or 20 μg (lysates from infected macrophages) of total proteins were separated using 12.5% SDS-PAGE. For the detection of MgtCMAB, the membrane was immunostained using a hyperimmune serum specific for MgtCMAB (see Fig. S5 in the supplemental material), washed, and probed with peroxidase-conjugated goat anti-mouse (IgG; 1/5,000). As a control, the samples were immunostained with an immune serum specific for the KasA protein of M. tuberculosis.
Western blotting was subsequently performed to analyze the expression of MgtCMAB at the protein level in cultures grown in Sauton's medium with or without Mg2+. Crude bacterial lysates were probed using anti-MgtCMAB antibodies and hyperimmune sera obtained after DNA immunization of mice (see below). Before testing M. abscessus lysates, the specificity of the serum was first verified on a lysate from a M. smegmatis strain overexpressing a recombinant His-tagged MgtCMAB protein (see Fig. S5 in the supplemental material). As shown in Fig. 3B, MgtCMAB was not detected in lysates from bacilli grown in medium containing Mg2+. However, expression of MgtCMAB was readily detected when M. abscessus was grown in medium without Mg2+, further supporting the hypothesis of strong induction at the protein level mediated by Mg2+ depletion. The KasA protein used as a control was similarly detected in the two media. As anticipated, no immunoreactive band was detected in the lysate of the ΔmgtCMAB mutant grown under inducing and noninducing conditions.
We next monitored gene expression in bacteria internalized in J774 macrophages. After 24 h of macrophage infection, the mgtCMAB-specific transcripts were found to be strongly induced (by about 15-fold) compared to the expression level in plankton-growing cultures (Fig. 3A). Comparatively, MAB_0146 was mildly induced (about 4-fold) in macrophages (Fig. 3A). Consistent with this gene expression pattern, the MgtC protein was detected in bacterial lysate recovered from macrophages (Fig. 3B).
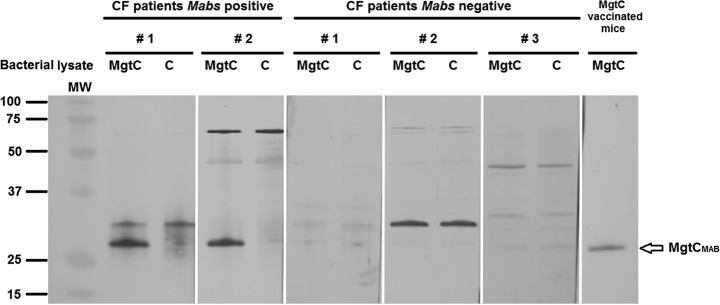
To further confirm the in vivo MgtCMAB expression results, we evaluated the antibody responses of a M. abscessus-infected CF patient during infection. Sera from infected CF patients recognized an MgtCMAB recombinant protein expressed in M. smegmatis (Fig. 4). The specificity of the hybridization signal was shown using, as negative control, a M. smegmatis lysate that did not express MgtCMAB (Fig. 4, lanes C). Moreover, sera from M. abscessus-negative patients did not recognize MgtCMAB (Fig. 4).
FIG 4.
Presence of specific MgtC antibodies in the serum of CF patients infected with M. abscessus. Sera (diluted 1:1,000) from two CF patients infected with M. abscessus and three CF patients negative for infection with M. abscessus were tested using Western blotting against a total lysate of M. smegmatis overexpressing the recombinant MgtCMAB protein tagged with a C-terminal polyhistidine (lanes MgtC) or control M. smegmatis lysate (lanes C). Serum from mice immunized with mgtCMAB DNA against the M. smegmatis lysate overexpressing the recombinant MgtCMAB protein is included as a positive control (right lane). The blots are issued from a single membrane and hybridized with different sera. MW, molecular weight.
Taken together, these results indicate that MgtCMAB is not expressed under standard in vitro growth conditions but that its expression is highly induced under conditions of Mg2+ deprivation, within infected macrophages, and during infection in CF patients as shown by the presence of MgtCMAB antibodies. We therefore reasoned that neutralizing the activity of MgtC through the production of specific antibodies may protect against M. abscessus infection in mice and would allow us to arrest the MgtC contribution in vivo.
Formulated mgtCMAB DNA immunization protects CF mice against M. abscessus infection.
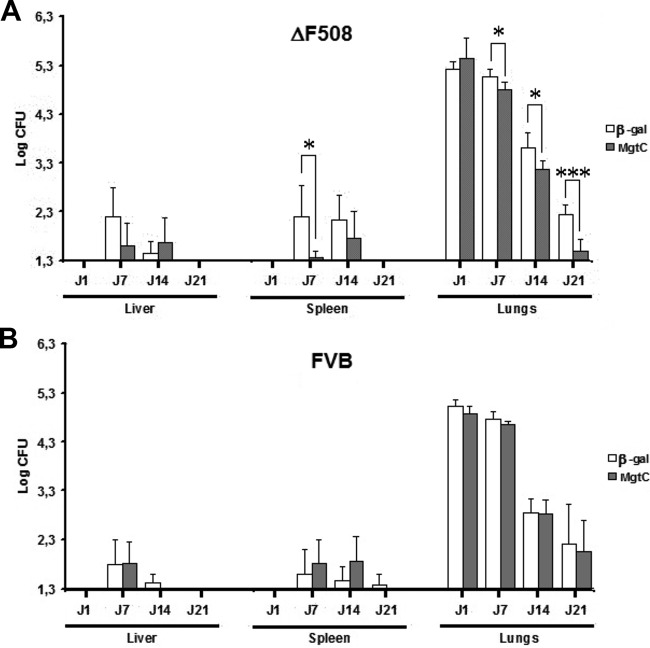
To further evaluate the impact of MgtC expression in vivo by an alternative approach to the ex vivo macrophage infection experiment, an MgtC immunotargeting approach was considered. Indeed, the pronounced induction of MgtCMAB expression in infected host cells is an important element in considering MgtCMAB as a potent immune target. Our recent success using a DNA-based immunization approach (13) prompted us to develop a plasmid DNA with a codon-optimized mgtCMAB gene transcribed under the control of a strong CMV promoter in formulation with a tetrafunctional amphiphilic block copolymer. This preparation was used to immunize CF mice bearing the cystic fibrosis transmembrane conductance regulator (CFTR) ΔF508 mutation, currently the most frequently encountered CFTR mutation in CF patients, as well as their wild-type FVB littermates. In addition to formulated mgtCMAB DNA, the corresponding β-galactosidase gene DNA was included as a control. Following immunization, mice were challenged using the aerosol route with 4 × 108 CFU of M. abscessus, as illustrated in Fig. S1 in the supplemental material (13). As shown in Fig. 5A, infection with M. abscessus was cleared more rapidly from the lungs of ΔF508 (CF) mice immunized with formulated mgtCMAB than from those of mice that had received the β-galactosidase-encoding DNA at 7 (P = 0.0178), 14 (P = 0.0145), and 21 (P = 0.0001) days postinfection. CFU counts also decreased significantly faster in the spleen (day 7; P = 0.0159) of immunized ΔF508 mice than in those of the control mice. Furthermore, only two (33%) of six mgtC-immunized ΔF508 mice were still culture positive in the lungs at 21 days postchallenge (Table 1) whereas seven (100%) of seven mice receiving the β-galactosidase control plasmid remained culture positive (P = 0.02). Only one (20%) of five mgtC-immunized ΔF508 mice remained culture positive in the spleen 7 days postchallenge, whereas five (83%) of six mice receiving the β-galactosidase control plasmid were culture positive at that time point. It is, however, noteworthy that the formulated mgtC DNA failed to protect the infected parental FVB littermates, thus highlighting a potent effect of the CFTR ΔF508 mutation in unmasking the protective effect against an aerosol challenge in CF mice (Fig. 5B and Table 1).
FIG 5.
Bacterial persistence of M. abscessus CIP in liver, spleen, and lungs of tetrafunctional amphiphilic block copolymer formulating MgtCMAB DNA-vaccinated or β-galactosidase DNA-vaccinated ΔF508 FVB mice (A) or the wild-type FVB littermates (B) after an aerosol infection challenge. Lungs, spleen, and liver of mice infected with an aerosolized solution containing 4 × 108 bacterial cells/ml of CIP S were collected and homogenized by dislocation. The homogenates of lungs, spleen, and liver were serially diluted and plated for CFU counts. Results are expressed as the log units of CFU for formulated mgtCMAB DNA-vaccinated mice (gray bars) or formulated β-galactosidase (β-gal) DNA-vaccinated mice (control group, white bars) at days 1, 7, 14, and 21 postinfection (*, P < 0.05; ***, P < 0.001).
TABLE 1.
Number of mice with residual liver, spleen, and lung infection on day 7 (liver and spleen) or 21 (lungs) post-aerosol challenge compared to the total number of immunized mice
| Organ(s) and day post-aerosol challengea | Mouse strain | No. of infected mice/total no. of mice tested |
P value | |
|---|---|---|---|---|
| β-Gal DNAb | mgtCMAB DNA | |||
| Liver | ||||
| 7 | ΔF508 | 5/6 | 2/5 | 0.24 |
| 7 | WT FVB | 4/6 | 5/6 | 1 |
| Spleen | ||||
| 7 | ΔF508 | 5/6 | 1/5 | 0.08 |
| 7 | WT FVB | 2/6 | 4/6 | 0.56 |
| Lungs | ||||
| 21 | ΔF508 | 7/7 | 2/6 | 0.02 |
| 21 | WT FVB | 6/8 | 7/8 | 1 |
Days with significant results based on the CFU analysis (Fig. 5) are taken into account.
β-Gal, β-galactosidase.
DISCUSSION
Genome sequence comparison between pathogenic and nonpathogenic microorganisms has revealed the potential of key virulence determinants as vaccine candidates. MAB_3593, uncovered from whole-genome sequencing of M. abscessus (11), has emerged as an attractive gene candidate as it encodes MgtC, a well-known virulence factor that is present in various bacteria and shared by several CF opportunistic pathogens but absent from M. smegmatis, a nonpathogenic RGM.
MgtC has been shown to promote bacterial growth in magnesium-depleted medium in all bacteria tested so far, including M. tuberculosis and M. marinum (17, 20, 37). In addition, expression of mgtC was shown to be highly induced by low Mg2+ concentrations in M. marinum (37), as well as in the nonrelated bacterial species S. Typhimurium (38) and P. aeruginosa (20). Despite its unique genomic location and organization (differing, for example, from that in M. marinum [37]), a similar result was observed in M. abscessus, with a 50-fold induction rate for the mgtCMAB transcripts found upon Mg2+ deprivation. That Mg2+-dependent expression of MgtC is conserved in phylogenetically distantly related organisms is very likely linked to the conserved function of MgtC in the adaptation to Mg2+ fluctuations, which is supported by the contribution of MgtCMAB to optimal growth in Mg2+-deprived broth medium. A second MgtC-like protein is encoded by the M. abscessus genome, which belongs to a phylogenic cluster distinct from MAB_3593, is poorly regulated by Mg2+ (2-fold induction), and fails to promote growth in low Mg2+ concentrations in a heterologous experimental system. The presence of two MgtC-like proteins has also been reported earlier in P. aeruginosa, where MgtC (PA4535) is regulated by Mg2+ deprivation and promotes growth in low-Mg2+ medium whereas the second MgtC-like protein (PA2558) does not (20, 21). However, the precise function(s) of these MgtC-like components remains unknown and whether they can compensate for a potential MgtC defect in macrophages remains to be established.
MgtCMAB is not expressed in standard planktonic cultures, but, importantly, both the transcription of mgtCMAB and the production of MgtCMAB are significantly induced within infected macrophages. This publication presents the first report of the detection of an MgtC protein in infected macrophages. An induction by the intramacrophage environment has also been shown at the transcriptional level for S. Typhimurium mgtC (19) and P. aeruginosa mgtC (20). It can be inferred that, despite different genomic organizations, a conserved regulatory mechanism facilitates mgtC induction within host cells. The host signal that drives mgtC expression is unlikely to be associated with Mg2+ deficiency and remains to be clearly identified. Phagosome acidification has been proposed to contribute to the optimal expression of P. aeruginosa mgtC (20), and a combination of acidification and antimicrobial peptides has been reported to activate the Salmonella sensor that regulates mgtC within macrophages (39).
Although MgtC contributes to intramacrophage survival in several bacterial pathogens (18), the present findings suggest that MgtCMAB does not significantly contribute to M. abscessus intramacrophage survival under our experimental conditions, even though a slight replication defect was observed for the ΔmgtCMAB mutant. A similar behavior has been described for a M. marinum mgtC mutant and was correlated, potentially, to fast and important escape of the bacteria to the cytoplasm (37). The lack of a strong phenotype of the ΔmgtCMAB mutant in macrophages may also be linked to a similar phenomenon, supported by the view that M. abscessus is not a strict intracellular pathogen per se (A. L. Roux, A. Viljoen, A. Bah, R. Simeone, A. Bernut, T. Deramaudt, M. Rottman, J. L. Gaillard, L. Majlessi, R. Brosch, I. Vergne, C. de Chastellier, L. Kremer, and J. L. Herrmann, submitted for publication). In addition, we cannot exclude the possibility that the loss of MgtCMAB function inside macrophages is at least partially compensated by the MgtC-like protein encoded by the MAB_0146 gene, whose expression is also induced intracellularly, although at a lower level. Moreover, our results do not preclude the possibility of a contribution of MgtC to M. abscessus virulence in other infection models, and experiments should be conducted in animal models to further investigate the role of MgtCMAB during infection.
MgtC expression was observed in vivo, indirectly demonstrated by antibody production in M. abscessus-vaccinated mice and in CF patients infected with M. abscessus (Fig. 4). Similar results were observed with M. abscessus PLC expression in vivo (13). Reactive T cells and production of antibodies against PLC were observed after PLC DNA vaccination, both responses being required for effective protection of PLC-vaccinated CF mice against an M. abscessus aerosol challenge (13). Although we did not look at the T cell response, we hypothesized that the production of antibodies against MgtC induced by the DNA vaccination using a similar synthetic delivery system comprising a tetrafunctional block copolymer (13) might be an important component of the protection conferred to animals with CF that were vaccinated against an aerosol challenge. This protective efficacy was judged for its ability to reduce the bacterial burden in the lungs of infected mice and the more rapid lung clearance in the context of a CF mutation, validating the potential of our vaccine-based approach. Unexpectedly, this protection occurred in the context of a CFTR defect (ΔF508 mutation) but not in the parental mice and thus supports the idea of the importance of MgtC in M. abscessus pathogenicity in CF mice.
Vaccination represents a useful approach in the fight against multidrug-resistant bacteria, of which M. abscessus remains a notorious example. The search for vaccine targets has largely benefited from reverse vaccinology, primarily based on in silico analyses of annotated genomes to select the most appropriate proteins as vaccine antigens (14). However, information in addition to that obtained from in silico genome analyses, such as the selection of proteins upregulated during infection, can be used to further prioritize antigens for assessment of their vaccine properties. Our findings using the mgtCMAB DNA formulation confirm this hypothesis. The development of a relevant murine model for M. abscessus vaccination is still a challenge. Despite the natural clearance occurring around day 21 to day 30 of infection, we were able to see a more rapid decrease in bacterial burden in mgtCMAB-vaccinated mice, compared to beta-galactosidase-vaccinated mice. In addition, the protective effect triggered by the formulated mgtCMAB DNA vaccine appears even greater than that triggered by our previously described plc-based vaccine candidate, considering the percentage of culture-positive lungs at 21 days postinfection (13).
In conclusion, among all RGM species, M. abscessus is the only one that is a recognized respiratory pathogen in CF patients possessing several virulence factors acquired by HGT, such as MgtC (this work) or PLC (13). Since MgtC is also expressed in P. aeruginosa and B. cenocepacia, two major opportunistic pathogens in CF patients, we speculate that with regard to immune cross-reactivity, the mgtCMAB vaccine might also be beneficial in protection against both mycobacterial and Gram-negative infections in CF patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Vaincre La Mucoviscidose (French Cystic Fibrosis Foundation; IC0902 and RF2012 0600689 to V.L.M.). C.B. is supported by an MRT fellowship from the French Ministry of Research and the Fondation for Medical Research (FRM FDT20140930905). V.L.M. and A.B. are supported by the program DIMYVIR (ANR-13-BSV3-0007-01). B.P. is an employee of In-Cell-Art, which commercializes tetra functional block copolymers for vaccination.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00359-16.
REFERENCES
- 1.Griffith DE. 2003. Emergence of nontuberculous mycobacteria as pathogens in cystic fibrosis. Am J Respir Crit Care Med 167:810–812. doi: 10.1164/rccm.2301001. [DOI] [PubMed] [Google Scholar]
- 2.Olivier KN, Weber DJ, Wallace RJ, Faiz AR, Lee J-H, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 3.Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, Welinder-Olsson C. 2007. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol 45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roux A-L, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet MA, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard JL, Herrmann JL; JL for the OMA Group. 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol 47:4124–4128. doi: 10.1128/JCM.01257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux AL, Catherinot E, Soismier N, Heym B, Bellis G, Lemonnier L, Chiron R, Fauroux B, Le Bourgeois M, Munck A, Pin I, Sermet I, Gutierrez C, Véziris N, Jarlier V, Cambau E, Herrmann JL, Guillemot D, Gaillard JL; OMA group. 2015. Comparing Mycobacterium massiliense and Mycobacterium abscessus lung infections in cystic fibrosis patients. J Cyst Fibros 14:63–69. doi: 10.1016/j.jcf.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Esther CR Jr, Henry MM, Molina PL, Leigh MW. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Pediatr Pulmonol 40:39–44. doi: 10.1002/ppul.20222. [DOI] [PubMed] [Google Scholar]
- 7.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown-Elliott BA, Wallace RJ Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Elliott BA, Nash KA, Wallace RJ Jr. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann J-L, Daffé M, Brosch R, Risler J-L, Gaillard J-L. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakala N′Goma JC, Le Moigne V, Soismier N, Laencina L, Le Chevalier F, Roux AL, Poncin I, Serveau-Avesque C, Rottman M, Gaillard JL, Etienne G, Brosch R, Herrmann JL, Canaan S, Girard-Misguich F. 2015. Mycobacterium abscessus phospholipase C expression is induced during coculture within amoebae and enhances M. abscessus virulence in mice. Infect Immun 83:780–791. doi: 10.1128/IAI.02032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Moigne V, Rottman M, Goulard C, Barteau B, Poncin I, Soismier N, Canaan S, Pitard B, Gaillard JL, Herrmann JL. 2015. Bacterial phospholipases C as vaccine candidate antigens against cystic fibrosis respiratory pathogens: the Mycobacterium abscessus model. Vaccine 33:2118–2124. doi: 10.1016/j.vaccine.2015.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Rappuoli R. 2000. Reverse vaccinology. Curr Opin Microbiol 3:445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 15.Hoft DF, Brusic V, Sakala IG. 2011. Optimizing vaccine development. Cell Microbiol 13:934–942. doi: 10.1111/j.1462-5822.2011.01609.x. [DOI] [PubMed] [Google Scholar]
- 16.Le Moigne V, Gaillard JL, Herrmann JL. 2016. Vaccine strategies against cystic fibrosis pathogens. Hum Vaccin Immunother 12:751–756. doi: 10.1080/21645515.2015.1102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchmeier N, Blanc-Potard A, Ehrt S, Piddington D, Riley L, Groisman EA. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol Microbiol 35:1375–1382. doi: 10.1046/j.1365-2958.2000.01797.x. [DOI] [PubMed] [Google Scholar]
- 18.Alix E, Blanc-Potard AB. 2007. MgtC: a key player in intramacrophage survival. Trends Microbiol 15:252–256. doi: 10.1016/j.tim.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 20.Belon C, Soscia C, Bernut A, Laubier A, Bleves S, Blanc-Potard AB. 2015. A macrophage subversion factor is shared by intracellular and extracellular pathogens. PLoS Pathog 11:e1004969. doi: 10.1371/journal.ppat.1004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rang C, Alix E, Felix C, Heitz A, Tasse L, Blanc-Potard AB. 2007. Dual role of the MgtC virulence factor in host and non-host environments. Mol Microbiol 63:605–622. doi: 10.1111/j.1365-2958.2006.05542.x. [DOI] [PubMed] [Google Scholar]
- 22.Alix E, Godreuil S, Blanc-Potard AB. 2006. Identification of a Haarlem genotype-specific single nucleotide polymorphism in the mgtC virulence gene of Mycobacterium tuberculosis. J Clin Microbiol 44:2093–2098. doi: 10.1128/JCM.00278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanc-Potard AB, Groisman EA. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J 16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medjahed H, Reyrat J-M. 2009. Construction of Mycobacterium abscessus defined glycopeptidolipid mutants: comparison of genetic tools. Appl Environ Microbiol 75:1331–1338. doi: 10.1128/AEM.01914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 26.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat Rev Microbiol 6:851–857. doi: 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gannoun-Zaki L, Alibaud L, Carrère-Kremer S, Kremer L, Blanc-Potard AB. 2013. Overexpression of the KdpF membrane peptide in Mycobacterium bovis BCG results in reduced intramacrophage growth and altered cording morphology. PLoS One 8:e60379. doi: 10.1371/journal.pone.0060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, Sapriel G, Roux AL, Conlon K, Honoré N, Dillies MA, Ma L, Bouchier C, Coppée JY, Gaillard JL, Gordon SV, Loftus B, Brosch R, Herrmann JL. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol 90:612–629. doi: 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 29.Kremer L, Dover LG, Morbidoni HR, Vilchèze C, Maughan WN, Baulard A, Tu SC, Honoré N, Deretic V, Sacchettini JC, Locht C, Jacobs WR Jr, Besra GS. 2003. Inhibition of InhA activity, but not KasA activity, induces formation of a KasA-containing complex in mycobacteria. J Biol Chem 278:20547–20554. doi: 10.1074/jbc.M302435200. [DOI] [PubMed] [Google Scholar]
- 30.McIlroy D, Barteau B, Cany J, Richard P, Gourden C, Conchon S, Pitard B. 2009. DNA/amphiphilic block copolymer nanospheres promote low-dose DNA vaccination. Mol Ther 17:1473–1481. doi: 10.1038/mt.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile J-F, Daffé M, Perronne C, Soudais C, Gaillard J-L, Rottman M. 2007. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun 75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottman M, Catherinot E, Hochedez P, Emile J-F, Casanova J-L, Gaillard J-L, Soudais C. 2007. Importance of T cells, gamma interferon, and tumor necrosis factor in immune control of the rapid grower Mycobacterium abscessus in C57BL/6 mice. Infect Immun 75:5898–5907. doi: 10.1128/IAI.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Doorninck JH, French PJ, Verbeek E, Peters RH, Morreau H, Bijman J, Scholte BJ. 1995. A mouse model for the cystic fibrosis delta F508 mutation. EMBO J 14:4403–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catherinot E, Roux AL, Macheras E, Hubert D, Matmar M, Dannhoffer L, Chinet T, Morand P, Poyart C, Heym B, Rottman M, Gaillard JL, Herrmann JL. 2009. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J Clin Microbiol 47:271–274. doi: 10.1128/JCM.01478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanc-Potard AB, Lafay B. 2003. MgtC as a horizontally-acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J Mol Evol 57:479–486. doi: 10.1007/s00239-003-2496-4. [DOI] [PubMed] [Google Scholar]
- 36.Gastebois A, Blanc Potard AB, Gribaldo S, Beau R, Latgé JP, Mouyna I. 2011. Phylogenetic and functional analysis of Aspergillus fumigatus MGTC, a fungal protein homologous to a bacterial virulence factor. Appl Environ Microbiol 77:4700–4703. doi: 10.1128/AEM.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belon C, Gannoun-Zaki L, Lutfalla G, Kremer L, Blanc-Potard AB. 2014. Mycobacterium marinum MgtC plays a role in phagocytosis but is dispensable for intracellular multiplication. PLoS One 9:e116052. doi: 10.1371/journal.pone.0116052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 39.Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI. 2008. The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol 69:503–519. doi: 10.1111/j.1365-2958.2008.06303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.