Abstract
Environmental and endogenous genotoxic agents can result in a variety of alkylated and carboxymethylated DNA lesions, includingN3-ethylthymidine (N3-EtdT), O2-EtdT, O4-EtdT as well as N3-carboxymethylthymidine (N3-CMdT) and O4-CMdT. By using non-replicative double-stranded vectors harboring a site-specifically incorporated DNA lesion, we assessed the potential roles of alkyladenine DNA glycosylase (Aag), alkylation repair protein B homologue 2 (Alkbh2) or Alkbh3 in modulating the effects of N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT or O4-CMdT on DNA transcription in mammalian cells. We found thatthe depletion of Aagdid not significantly change the transcriptional inhibitory or mutagenic properties ofall five examined lesions, suggesting a negligible role of Aag in the repair of these DNA adducts in mammalian cells. In addition, our results revealed that N3-EtdT, but not other lesions, could be repaired by Alkbh2 andAlkbh3 in mammalian cells.Furthermore, we demonstraed the direct reversal of N3-EtdT by purified human Alkbh2 proteinin vitro.These findings provided important new insights into the repair of the carboxymethylated and alkylated thymidine lesions in mammalian cells.
Human cells are constantly exposed to various endogenous and exogenous agents that can induce damage to genomic DNA1. Unrepaired DNA lesions may inhibit DNA replication and transcription as well as induce mutations in these processes, which may eventually result in the development of cancer and other human diseases2,3.
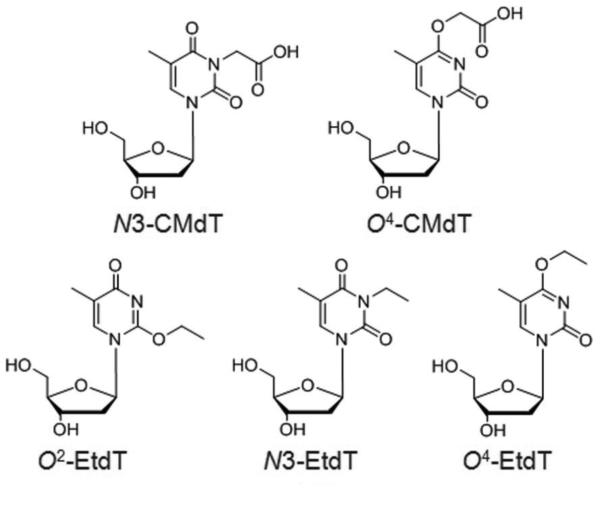
DNA alkylation damage is generally unavoidable due to theubiquitouspresence of environmental and endogenous alkylating agents4. TheN3, O2, and O4 of thymine are among the major alkylation sites in DNA and the resulting alkylated thymine lesions have been readily detected in various mammalian cells and tissues5-9. In particular, the regioisomeric N3-ethylthymidine (N3-EtdT), O2-EtdT, and O4-EtdT (Figure 1) lesions have been detected at significantly higher levels in leukocyte and saliva DNA of smokers than nonsmokers, and thus have been proposed as prospective biomarkers for ethylation damage and for cancer risk assessment8,9.
Figure 1.
Chemical structures of N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT, and O4-CMdT.
N-nitroso compounds (NOCs) represent another common type of DNA damaging agents that are widely present in processed meat, tobacco smoke, and other external and internal sources10-12. Metabolic activation of many NOCs can cause the formation of carboxymethylating agents (e.g., diazoacetate) which can modify DNA to give a battery ofcarboxymethylated DNA adducts13-15. It was recently reported that N3-carboxymethylthymidine (N3-CMdT) and O4-CMdT (Figure 1) are the major carboxymethylated thymidine lesions formed in isolated DNA exposed to diazoacetate, and may contribute to diazoacetate-induced signature mutations in TP53 gene found in gastrointestinal cancer14,16-18.
Several studies have examinedthe roles of translesion synthesis DNA polymerases and nucleotide excision repair (NER) proteins in modulating the effects of these DNA lesions on DNA replication or transcription in vitro and in cells17-23. For instance, it was found that DNA polymerase V is required for the error-prone bypass of all three regioisomeric EtdT lesions and the error-free bypass of N3-CMdT in Escherichia coli cells18,21. In addition, depletion of transcription-coupled NER proteins can exacerbate the inhibitory and mutagenic effects of N3-EtdT, N3-CMdT and O4-CMdT on DNA transcription in mammalian cells19,20.
The E.coliN3-methyladenine DNA glycosylase II (AlkA) and its mammalian ortholog, alkyladenine DNA glycosylase (Aag), recognize and excise a chemically diverse array of alkylated DNA lesions24-27. In this vein, crude extractsof E.coli cells inducedfor the adaptive response remove O2-methyl-2′-deoxycytidine (O2-MedC) and O2-MedT from DNA and these activities are dependent on the presence of AlkA28.On the other hand, E.coli AlkB, a dioxygenase that requires non-heme Fe(II), molecular oxygen, and 2-oxoglutarate as cofactors,canrepaira variety of alkylated DNA lesions includingN3-methyl-2′-deoxycytidine (N3-MedC) and N3-MedT29-34. Among the nine mammalian AlkB homologs (i.e., Alkbh1-Alkbh8 and FTO), Alkbh2 and Alkbh3 were observed to be capable of reversing directly some simple N-alkylated DNA lesions29,35,36. Moreover, deficiency in these two genes confers elevated sensitivities of mammalian cells toward alkylating agents29,36-38.Therefore,it is important toinvestigatethe involvement ofAag, Alkbh2, and Alkbh3inthe repair ofN3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT and O4-CMdT lesions in mammalian cells.
RESULTS AND DISCUSSION
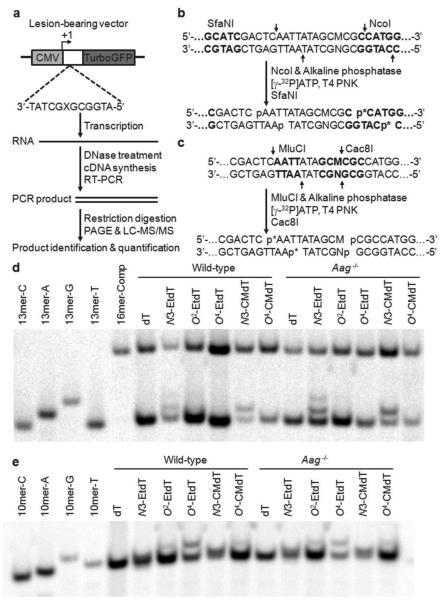
Using an established competitive transcription and adduct bypassassay(Figure 2a)39, we examined the potential roles of Aag, Alkbh2 and Alkbh3 in modulating the deleterious effects of N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT and O4-CMdT on transcription in mammalian cells. Briefly, non-replicating double-stranded plasmids containing a site-specifically incorporated DNA lesion or unmodified dT were constructed, and co-transfected with a non-lesion competitor into mammalian cells that are proficient or deficient in Aag, Alkbh2, or Alkbh3(Supplementary Figure S1). The RNA products were extracted from the cells, and treated with DNaseI to eliminate residual DNA contamination. The transcripts of interest were amplified by reverse transcription PCR (RT-PCR), and the resulting RT-PCR products were restriction digested to generate short oligodeoxyribonucleotides (ODN) fragments for polyacrylamide gel electrophoresis (PAGE)and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. With the use of two sets of restriction enzymes, i.e., NcoI/SfaNI and MlucI/Cac8I19,20, restriction fragments with single nucleotide differences at the lesion site were resolved by PAGE analysis (Figure 2b-e). The identities of the restriction fragments of interest were also confirmed by LC-MS/MS analysis (Supplementary Figures S2-S3).
Figure 2.
In vivo transcription assay. (a) A schematic diagram illustrating the procedures for assessing the impact of the DNA lesions on DNA transcription.‘X’ indicates N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT, O4-CMdT or dT, which was located on the transcribed strand of TurboGFP gene downstream of the CMV promoter. The +1 transcription start sites are indicated by arrowhead.Not shown is the lesion-free competitor vector, which has three more nucleotides near the lesion region than the undamaged control vector and is co-transcribed with the control or lesion-containing plasmids in mammalian cells. (b-c) Sample processing for NcoI/SfaNI-(b) or MlucI/Cac8I-(c) mediated restriction digestion/postlabeling assay (p* indicates 32P-labeled phosphate group). (d-e) Representativegel images showing the NcoI/SfaNI-(d) or MlucI/Cac8I-(e) produced restriction fragments of interest. The restriction fragment arising from the competitor vector, i.e., d(5′-CATGGCGATATGCTAT-3′), is designated as ‘16mer-Comp’; ‘13mer-C’, ‘13mer-A’, ‘13mer-G’, and ‘13mer-T’represent the standard synthetic ODNsd(5′-CATGGCGNGCTAT-3′), where ‘N’ is C, A, G, and T, respectively.‘10mer-C’, ‘10mer-A’, ‘10mer-G’, and ‘10mer-T’represent the standard synthetic ODNs d(5′-AATTATAGCM-3′), where ‘M’ is C, A, G, and T, respectively.
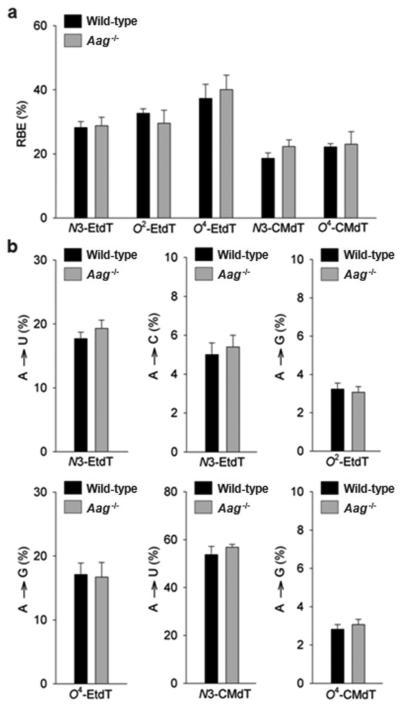
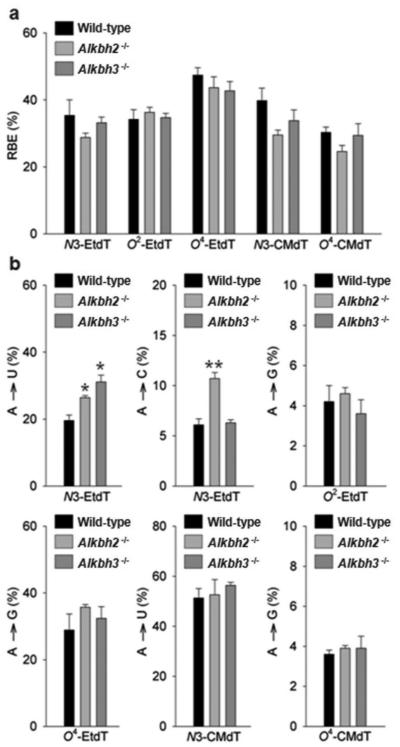
Our results showed that the fivelesions substantially inhibit DNA transcription and induce mutations, whereas depletion of Aag had a negligible effect on transcriptional alterations induced by these DNA lesions in mammalian cells (Figures 2b-e, and 3a-b, Supplementary Table S2 and S3). We also found that the absence of Alkbh2 or Alkbh3did not lead to a statistically significantchange (P > 0.05) in the transcriptional blockageor mutagenic properties ofO2-EtdT, O4-EtdT, N3-CMdT and O4-CMdT in mammalian cells (Figure 4a-b, Supplementary Figure S4, and Supplementary Table S4 and S5). In addition,the removal of Alkbh2 or Alkbh3fromthe wild-type background conferredno obvious impact on the bypass of N3-EtdT, but could lead to an altered mutational signature of N3-EtdT(Figure 4a-b, Supplementary Figure S4, and Supplementary Tables S4 and S5).We found that transcriptional bypass of N3-EtdT induced A✧Cand A✧U mutations at markedly higher frequencies (~11% and ~26%, respectively) in Alkbh2-deficient cells than in the repair-proficient cells (~6% and ~20%, respectively)(Figure 4a-b, Supplementary Figure S4, and Supplementary Table S5). We also observed that N3-EtdT induced a substantially higher degree of A✧U mutation (~31%) in Alkbh3-deficient cells than in the wild-type background (Figure 4a-b, Supplementary Figure S4, and Supplementary Table S5). These results suggest that,in mammalian cells, Alkbh2 and Alkbh3 may be involved in the removal of N3-EtdT, but not O2-EtdT, O4-EtdT, N3-CMdT or O4-CMdT.
Figure 3.
Relative bypass efficiencies (RBEs) (a) and mutagenic potentials (b) of N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT, and O4-CMdT during transcription in mammalian cellsthat are proficient (i.e., ‘Wild-type’) or deficient in Aag.The data represent the mean and standard error of the mean of results from three independent experiments.
Figure 4.
Bypass efficiencies (a) and mutagenic potentials (b) of N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT, and O4-CMdT during transcription in mammalian cellsthat are proficient or deficient in Alkbh2 or Alkbh3.The data represent the mean and standard error of the mean results from three independent experiments.‘*’, P< 0.05; ‘**’, P< 0.01. The P values were calculated by using unpaired two-tailed Student’s t-test.
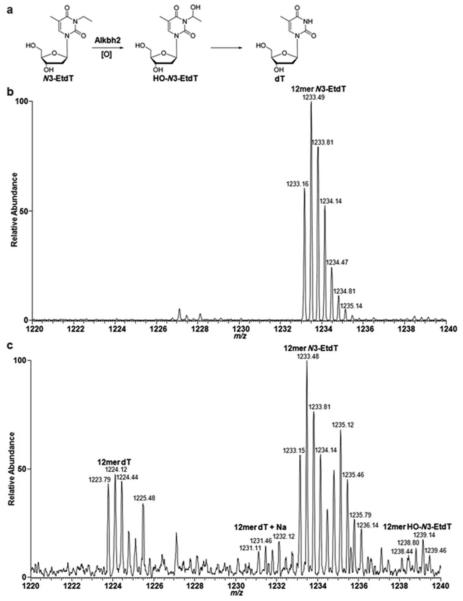
We also investigated the repair activity of Alkbh2orAlkbh3towardN3-EtdT and the other four lesions using an in vitroDNA repair assay. To this end, we incubated a 12-mer lesion-bearing oligodeoxynucleotide,or a duplex DNA with a single lesionlocated in the same sequence context, with purified human Alkbh2or Alkbh3proteins in HEPES buffer containing the requisite cofactors including2-oxoglutarate and ascorbate. We subsequently monitored the products of the in vitro repair reaction by using LC-MS and MS/MS analysis. In the reaction without the DNA repair protein, we observed the monoisotopic peak for the [M-3H]3− ion (m/z1233.15) of the 12-mer substrate containing the unaltered N3-EtdTlesion (Figure 5a, b). In the presence of the Alkbh2 protein, we found that the N3-EtdT, when located in the double-stranded but not single-stranded DNA, was partially converted to unmodified thymidine (the [M-3H]3− ion at m/z = 1223.79) and an intermediate product with the methylene functionality in the ethyl group being oxidized to its hydroxymethylene derivative (i.e., HO-N3-EtdT, the [M-3H]3− ion at m/z = 1238.44) (Figure 5a, cand Supplementary Figure S5a). In addition, we did not observe any repair activity of Alkbh2 protein towardO2-EtdT, O4-EtdT, N3-CMdT or O4-CMdT in vitro (Supplementary Figures S6and S7), which isin agreement with our cellular repair results.Unexpectedly, the presence of purified Alkbh3 protein did not result in detectablelevel of repair ofN3-EtdT-bearing DNA in the in vitro reaction under our experimental conditions(Supplementary Figure S5b).As a control, incubation of N3-MedC-bearing DNA with the Alkbh2 or Alkbh3 protein produced the undamaged product(Supplementary Figure S8), which is consistent with previous findings that N3-MedC is a substrate forthese repair proteins36,37.
Figure 5.
Repair of N3-EtdT by Alkbh2in vitro.(a) A schematic diagram illustrating the repair of N3-EtdT by Alkbh2. (b,c) LC-MS for monitoring the products from the in vitro incubation reactions of a duplex DNA, that is, d(5′-ATGGCG(N3-EtdT)GCTAT-3′)/d(5′-TTATAGCACGCCATGG-3′) in the absence (b) or presence (c) of purified human Alkbh2 protein. Shown are the high-resolution “ultra zoom-scan” MS results for monitoring the [M-3H]3− ions of the initial 12mer N3-EtdT-bearing ODN (i.e., 12mer N3-EtdT) as well as its repaired product d(5′-ATGGCGTGCTAT-3′) (i.e., 12mer dT) and an intermediate product (i.e., 12merHO-N3-EtdT).
A previousstudy suggested thatthe bypass of a strongly blocking lesion (e.g.N3-MedT) by AlkB can confer distinct effects on miscoding properties of the DNA lesion during DNA replication in E.coli cells30. In this vein, the absence of AlkB did not considerably change the inhibitory effect of N3-MedT on DNA replication, but couldlead to a markedincrease in T→A transversion in E.coli cells30.In line with this previous finding, our results demonstrated that the mammalian AlkB homologues (i.e., Alkbh2andAlkbh3) could manifest a significant change in the effect of N3-EtdT on the fidelity, but not the efficiency, of DNA transcription in mammalian cells. In this respect, we found that depletion of Alkbh2leads to elevated frequencies of A→U and A→C mutations, whereas depletion of Alkbh3can only increase A→Umutation opposite the N3-EtdT lesion.Further studies are needed to elucidate the distinct impacts of Alkbh2 and Alkbh3 on the mutagenic properties of N3-EtdT during transcription in mammalian cells in the future.
We alsofound that Alkbh2 is capable of repairing the N3-EtdT lesion within double-stranded DNAin vitro by a direct reversal mechanism.In keeping with our findings, previous in vitro studies showed that both theE.coliAlkB and the human Alkbh2 demethylate N3-MedT in DNA, but that Alkbh3 is much less efficient at repairing N3-MedT than the other two proteins40.In addition, it was reported that N3-MedC is much more efficiently removed by Alkbh2 from double-stranded DNA than from single-stranded DNA 36. We found that Alkbh2 is less efficient at removing N3-EtdT from double-stranded DNA than N3-MedC from single-stranded DNA under our in vitro repair conditions, indicating that N3-EtdT may not be as good a substrate as N3-MedC for Alkbh2. On the other hand, we did not observe the direct reversal of N3-EtdT by purified human Alkbh3 protein under the conditions tested.suggesting that other DNA repair cofactor(s)may be required for Alkbh3-mediated removal of N3-EtdT in mammalian cells. In this vein, a previous study showed that ASCC3 helicase can interact with Alkbh3 to facilitate DNA alkylation repair 41.
Different from N3-EtdT, we found that O2-EtdT, O4-EtdT and O4-CMdT lesions are not substrates for Alkbh2orAlkbh3in vitroor in mammalian cells. Our results further supported the notion that AlkB and its mammalian homologs repair only those alkylated DNA lesions that are attached via the nitrogen atoms, but not oxygen atoms of the nucleobases30,31,36,37,40,42. On the other hand, we found that neither Alkbh2 nor Alkbh3 is involved in the repair of N3-CMdT in vitroor in mammalian cells. The distinct repair activities of the mammalian AlkB homologues forN3-EtdT and N3-CMdT may be attributed to the unique chemical properties of these two lesions. These results suggest that the addition of a hydrophilic and bulky carboxyl group, but not a hydrophobic and relatively small ethyl group, to the N3 position of thymine may inhibit the binding of the lesion to the active site of mammalian AlkB homologue (e.g.Alkbh2 or Alkbh3).
In conclusion,we systematically investigated, for the first time, the potential roles of Aag, Alkbh2 and Alkbh3 in the repair of regioisomeric ethylated and carboxymethylated thymidine lesions. Our results revealed thatAag displayed a negligible role in the transcriptional alterations induced by N3-EtdT,O2-EtdT, O4-EtdT, N3-CMdT and O4-CMdT, suggesting that Aag is not involved in repairing these lesions in mammalian cells. We also found that,among these five DNA adducts, only N3-EtdT constitutes a substrate for the mammalian AlkB homologues (i.e., Alkbh2 or Alkbh3). Together, these findings provide important new insights into the repair of DNA lesions induced by alkylating and carboxymethylating agents in mammalian cells.
METHODS
Materials
All chemicals, enzymes, [γ-32P]ATP, and unmodified ODNs unless otherwise specifiedwere purchased fromSigma-Aldrich, New England BioLabs, Perkin-Elmer and Integrated DNA Technologies, respectively. Purified human Alkbh2 and Alkbh3 proteins as well as the mouse embryonic fibroblast (MEF) cells that are proficient or deficient in Alkbh2 or Alkbh3were prepared as described previously 36,38. The Aag+/+ and Aag−/−MEF cells were kindlyprovided by Prof. Leona Samson43,44. Cells were grown in Dulbecco's Modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin(Invitrogen),and 100 μg/mL streptomycin (ATCC) in a 37°C incubator with 5% CO2.
Genotyping
DNA was isolated from MEF cells using a high-salt method 45, and the targeted Alkbh2,Alkbh3, or Aagdeletions were confirmed by PCR analysis as described previously 25,38. The primers are listed in Supplementary Table S1, and the PCR amplification with Phusion high-fidelity DNA polymerase (New England Biolabs) started at98°C for 2 min; then, 35 cycles at 98°C for 10 s, 58°C for 30 s, and 72°C for 5 s, and a final 5-min extension at 72°C. The PCR products were seperated on 2% (w/v) agarose gelsin the presence ofSYBR Safe DNA Gel Stain by electrophoresis, and then visualized underUVA light.
In vivo transcription assay
Non-replicating pTGFP-Hha10 plasmids containing a single site-specific lesion (i.e., N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT or O4-CMdT)as well as the undamaged control and competitor plasmid substrates were prepared as described previously19,20,46. The N3- or O2-EtdT-bearing plasmids were premixed with the competitor vector at a molar ratio of 12:1 (lesion/competitor), and the other lesion-bearing or lesion-free control plasmids were mixed individually with the competitor vector at a molar ratio of 3:1. The mixed DNA templates were then transfected into the Aag−/−, Alkbh2−/− or Alkbh3−/− cells as well as the corresponding wild-type cells following the previously published procedures19,20,46.
RNA extraction and RT-PCR
The RNAproducts arising from in vivo transcription were extracted using Total RNA Kit I (Omega) and were further treated witha DNA-free kit (Ambion) to eliminate DNA contamination following the manufacturer’s instructions. The transcripts of interest were reverse transcribed and the resulting cDNA products were PCR amplified as described elsewhere19,20,46.
PAGE analysis
For NcoI/SfaNI-mediated restriction digestion/postlabeling assay, a portion of the RT-PCR products was treated with 5 U NcoI and 1 U shrimp alkaline phosphatase in 10 μLof 1 × NEB buffer 3.1 at 37°C for 1 h and then at 70°C for 20 min. The dephosphorylated restriction fragments were radiolabeled with 5 U T4 polynucleotide kinase and ATP (50 pmol cold, premixed with 1.66 pmol [γ-32P]ATP) and were further digested with 2 U SfaNI in 20 μLof 1 × NEB buffer 3.1 at 37°C for 2 h. The resulting DNA mixtures were resolved by 30% native PAGE (acrylamide:bis-acrylamide=19:1) and quantified by phosphorimager analysis19,20,46. The MluCI/Cac8I-mediated restriction digestion/postlabeling assay was conducted in a similar fashion and the detailed experimental procedures were described recently19,20,46. The frequency of base misincorporation and the relative bypass efficiency (RBE) of DNA lesions were determined as described previously19,20,46.
LC-MS/MS analysis
LC-MS/MS identification of transcription products was performed as previously described19,20,46. Briefly, RT-PCR products were treated with 50 U MluCI, 25 U Cac8Iand 20 U shrimp alkaline phosphatase in 200 μLof 1 × CutSmart buffer at 37°C for 4 h. After phenol/chloroform extraction and ethanol precipitation, the DNA pellet was desalted with 70% ethanol and subjected to LC-MS/MS analysis following previously described procedures 19,20,46. The LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific) was set up for monitoring the fragmentation of the [M-3H]3− ions of the complementary 10-mer ODNs, i.e., d(AATTATAGCM), where ‘M’ designates A, T, C, or G.
In vitro DNA repair assay
For N3-EtdT, O2-EtdT, O4-EtdT, N3-CMdT or O4-CMdT, a 12-mer ODN d(5′-ATGGCGXGCTAT-3′) (‘X’ indicates a lesion) or a duplex DNA, i.e., d(5′-ATGGCGXGCTAT-3′)/d(5′-TTATAGCACGCCATGG-3′) was used as DNA substrate for in vitro repair studies. In addition, a 17-mer N3-CMdC-bearing ODN d(5′-GCGCAAAXCTAGAGCTC-3′) was employed as a control DNA substrate for Alkbh2- and Alkbh3-mediated repair reactions in vitro36. The DNA substrate (10 pmol) was incubated with purified Alkbh2 or Alkbh3 protein (50 pmol) at 37°C for 2h in a reaction mixture containing 50 mM HEPES-KOH, 75 μM Fe(NH4)2(SO4)2, 1 mM α-ketoglutarate, 2 mM ascorbate, 50 μg/ml bovine serum albumin. MgCl2(10 mM) was added to optimize the Alkbh2-mediated repair reaction as described elsewhere37. The proteins in the reaction mixture were removed by phenol/chloroform extraction, and the DNA products were ethanol precipitated in the presence of 2.5 mg carrier DNA (e.g. pTGFP-Hha10 plasmid DNA). After desalting with 70% ethanol, the DNA pellet was dissolved in water and subjected to LC-MS/MS analysis using the similar procedures as described above.
Supplementary Material
Acknowledgments
Acknowledgments
We thank Prof. LeonaSamson for kindly providing the wild-type and Aag−/−mouse embryonic fibroblast cells. This work was supported by the National Institutes of Health to Y.W.(DK082779 andES025121 and T.R.O.(CA176622 and CA107399)and X.D.was supported in part by an NRSA T32 training grant (ES018827).Research reported in this publication was also supported in part by the National Cancer Institute of the National Institutes of Health under award number CA33572 (City of Hope).The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Additional information
Supplementary information is available in the online version of the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- (1).Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- (2).Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Anna L, Kovacs K, Gyorffy E, Schoket B, Nair J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis. 2011;26:523–527. doi: 10.1093/mutage/ger011. [DOI] [PubMed] [Google Scholar]
- (6).Den Engelse L, De Graaf A, De Brij RJ, Menkveld GJ. O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis. 1987;8:751–757. doi: 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- (7).Swenberg JA, Dyroff MC, Bedell MA, Popp JA, Huh N, Kirstein U, Rajewsky MF. O4-ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulates in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1692–1695. doi: 10.1073/pnas.81.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen HJ, Wang YC, Lin WP. Analysis of ethylated thymidine adducts in human leukocyte DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal. Chem. 2012;84:2521–2527. doi: 10.1021/ac203405y. [DOI] [PubMed] [Google Scholar]
- (9).Chen HJ, Lee CR. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol. Lett. 2014;224:101–107. doi: 10.1016/j.toxlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- (10).Tricker AR. N-nitroso compounds and man: sources of exposure, endogenous formation and occurrence in body fluids. Eur. J. Cancer Prev. 1997;6:226–268. [PubMed] [Google Scholar]
- (11).Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- (12).Joosen AM, Kuhnle GG, Aspinall SM, Barrow TM, Lecommandeur E, Azqueta A, Collins AR, Bingham SA. Effect of processed and red meat on endogenous nitrosation and DNA damage. Carcinogenesis. 2009;30:1402–1407. doi: 10.1093/carcin/bgp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cupid BC, Zeng Z, Singh R, Shuker DE. Detection of O6-carboxymethyl-2'-deoxyguanosine in DNA following reaction of nitric oxide with glycine and in human blood DNA using a quantitative immunoslot blot assay. Chem. Res. Toxicol. 2004;17:294–300. doi: 10.1021/tx0340706. [DOI] [PubMed] [Google Scholar]
- (14).Wang J, Wang Y. Chemical synthesis of oligodeoxyribonucleotides containing N3- and O4-carboxymethylthymidine and their formation in DNA. Nucleic Acids Res. 2009;37:336–345. doi: 10.1093/nar/gkn946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wang J, Wang Y. Synthesis and characterization of oligodeoxyribonucleotides containing a site-specifically incorporated N6-carboxymethyl-2'-deoxyadenosine or N4-carboxymethyl-2'-deoxycytidine. Nucleic Acids Res. 2010;38:6774–6784. doi: 10.1093/nar/gkq458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gottschalg E, Scott GB, Burns PA, Shuker DE. diazoacetate-induced p53 mutations in vitro in relation to formation of O6-carboxymethyl- and O6-methyl-2'-deoxyguanosine DNA adducts: relevance for gastrointestinal cancer. Carcinogenesis. 2007;28:356–362. doi: 10.1093/carcin/bgl150. [DOI] [PubMed] [Google Scholar]
- (17).Swanson AL, Wang J, Wang Y. In vitro replication studies of carboxymethylated DNA lesions with Saccharomyces cerevisiae polymerase eta. Biochemistry. 2011;50:7666–7673. doi: 10.1021/bi2007417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yuan B, Wang J, Cao H, Sun R, Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).You C, Wang J, Dai X, Wang Y. Transcriptional inhibition and mutagenesis induced by N-nitroso compound-derived carboxymethylated thymidine adducts in DNA. Nucleic Acids Res. 2015;43:1012–1018. doi: 10.1093/nar/gku1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).You C, Wang P, Dai X, Wang Y. Transcriptional bypass of regioisomeric ethylated thymidine lesions by T7 RNA polymerase and human RNA polymerase II. Nucleic Acids Res. 2014;42:13706–13713. doi: 10.1093/nar/gku1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhai Q, Wang P, Wang Y. Cytotoxic and mutagenic properties of regioisomeric O2-, N3- and O4-ethylthymidines in bacterial cells. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhai Q, Wang P, Cai Q, Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).You C, Wang Y. Mass spectrometry-based quantitative strategies for assessing the biological consequences and repair of DNA adducts. Acc. Chem. Res. 2016;49:205–213. doi: 10.1021/acs.accounts.5b00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J. Biol. Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- (27).Lee CY, Delaney JC, Kartalou M, Lingaraju GM, Maor-Shoshani A, Essigmann JM, Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (Aag) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- (30).Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- (32).Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl. Acad. Sci. U. S. A. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Li D, Delaney JC, Page CM, Yang X, Chen AS, Wong C, Drennan CL, Essigmann JM. Exocyclic carbons adjacent to the N6 of adenine are targets for oxidation by the Escherichia coli adaptive response protein AlkB. J. Am. Chem. Soc. 2012;134:8896–8901. doi: 10.1021/ja3010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li D, Fedeles BI, Shrivastav N, Delaney JC, Yang X, Wong C, Drennan CL, Essigmann JM. Removal of N-alkyl modifications from N2-alkylguanine and N4-alkylcytosine in DNA by the adaptive response protein AlkB. Chem. Res. Toxicol. 2013;26:1182–1187. doi: 10.1021/tx400096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 2009;42:519–529. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- (37).Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, Falnes PO, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nay SL, Lee DH, Bates SE, O'Connor TR. Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts. DNA Repair. 2012;11:502–510. doi: 10.1016/j.dnarep.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).You C, Dai X, Yuan B, Wang J, Brooks PJ, Niedernhofer LJ, Wang Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012;8:817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li D, Delaney JC, Page CM, Chen AS, Wong C, Drennan CL, Essigmann JM. Repair of DNA Alkylation Damage by the Escherichia coli Adaptive Response Protein AlkB as Studied by ESI-TOF Mass Spectrometry. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/369434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- (44).Roth RB, Samson LD. 3-Methyladenine DNA glycosylase-deficient Aag null mice display unexpected bone marrow alkylation resistance. Cancer Res. 2002;62:656–660. [PubMed] [Google Scholar]
- (45).Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).You C, Wang Y. Quantitative measurement of transcriptional inhibition and mutagenesis induced by site-specifically incorporated DNA lesions in vitro and in vivo. Nat. Protoc. 10:1389–1406. doi: 10.1038/nprot.2015.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.