Abstract
Previous studies have demonstrated that E proteins induce activation-induced deaminase (AID) expression in activated B cells. Here, we examined the role of Id3 in germinal center (GC) cells. We found that Id3 expression is high in follicular B lineage cells but declines in GC cells. Immunized mice with Id3 expression depleted displayed a block in germinal center B cell maturation, showed reduced numbers of marginal zone B cells and class-switched cells, and were associated with decreased antibody titers and lower numbers of plasma cells. In vitro, Id3-depleted B cells displayed a defect in class switch recombination. Whereas AID levels were not altered in Id3-depleted activated B cells, the expression of a subset of genes encoding signaling components of antigen receptor-, cytokine receptor-, and chemokine receptor-mediated signaling was significantly impaired. We propose that during the GC reaction, Id3 levels decline to activate the expression of genes encoding signaling components that mediate B cell receptor- and or cytokine receptor-mediated signaling to promote the differentiation of GC B cells.
INTRODUCTION
Germinal centers (GCs) are secondary structures that develop in peripheral lymphoid organs following immunization or antigen stimulation (1). In GCs, B cells undergo class switch recombination (CSR), somatic hypermutation (SHM), and affinity-based selection (2). Upon B cell activation, CSR leads to the replacement of the immunoglobulin heavy chain (IgH) constant region (Cμ) with Cγ, Cα, or Cε, thus enabling secretion of antibodies (Ab) with different effector functions (3). In GC B cells, SHM introduces point mutations across the V region clusters, resulting in the generation of affinity Ab variants. Both CSR and SHM depend on enzyme activation-induced deaminase (AID) expression (4, 5). Based on differences in cellular density and constituents, GCs can be segregated into dark zones (DZ) and light zones (LZ). In the LZ, B cells span a network of follicular dendritic cells (FDC) to undergo selection upon interacting with antigen. In the DZ compartment, B cells undergo clonal expansion and affinity maturation, including CSR and SHM. Upon completion of the affinity maturation process, B cells exit the GC cycle to differentiate into Ab-secreting plasma cells (PCs) or memory B cells.
E proteins are members of the helix-loop-helix (HLH) family and include E12, E47, E2-2, and HeLa E box binding protein (HEB) (6, 7). E12 and E47 are encoded by the E2A gene and arise through alternative splicing (8). In early B cell progenitors, the E2A proteins act together with the transcriptional regulators FOXO1 and EBF1 to specify the B cell fate (9). The E2A gene products in later stages of B cell development orchestrate Ig(κ) locus assembly and receptor editing and maintain the GC compartment (10, 11). Finally, E proteins promote class switching by directly activating AID expression (12, 13).
Four proteins that antagonize the DNA binding activities of E proteins, Id1, Id2, Id3, and Id4, have been identified (14, 15). Id1 and Id4 expression in developing lymphocytes is low. Id2 and Id3 have been particularly well studied in the context of T cell development (6). Specifically, in developing T cell progenitors, they enforce the pre-T cell receptor (pre-TCR), γδ TCR, and αβ TCR checkpoints (16–20). Id3-deficient mice are closely associated with the development of CXCR5+ T follicular helper-like (TFH-like) cells and CXCR5+ effector regulatory T cells (Treg) (17, 20, 21). Finally, Id2 and Id3 act in the thymus to suppress the development and expansion of invariant NKT (iNKT) cells, Vγ1.1+ Vδ6.3+ innate γδ T cells, and innate variant TFH cells (20–26).
The intrinsic roles of Id proteins in B cell development have not been examined in great detail. Id1 and Id4 expression is barely detectable, and Id2 expression is low except for marginal zone B cells (27). Here, we demonstrate that Id3 expression is high in follicular B lineage cells but declines upon differentiating into GC B cells. Whereas Id3 expression appeared to be nonessential in early B cell progenitors, in immunized mice with Id3 expression depleted, antibody responses were severely impaired. We found that loss of Id3 expression in B cells of immunized mice perturbed marginal zone B cell development, GC formation, IgG1 CSR, and IgG1 secretion. Global transcriptome analyses revealed that AID expression was not impaired. However, Id3 expression was essential to regulate the expression of genes encoding proteins associated with antigen receptor-, cytokine receptor-, and chemokine receptor-mediated signaling. Finally, we found that Id3 expression was required to promote CSR. Based on these, as well as previous, observations, we propose that the E-Id protein axis controls the developmental progression of GC B cells.
MATERIALS AND METHODS
Mice.
C57BL/6, Id3fl/fl, Id3fl/−, Id3GFP, Cd19Cre, and Cγ1Cre mice were bred and housed under pathogen-free conditions at the University of California, San Diego, in accordance with the Institutional Animal Care and Use Guidelines. Id3-GFP, CD19-Cre, Cγ1-Cre, and Id3f/f mice were described previously (17, 28, 29, 30).
Immunizations.
Mice were intraperitoneally injected with 400 μg NP-KLH (4-hydroxy-3-nitrophenylacetyl [Biosearch] conjugated to keyhole limpet hemocyanin [Pierce]) mixed with monophosphoryl lipid A (MPL)-based adjuvant (Sigma) (31). For boost immunizations, 200 μg NP-KLH in phosphate-buffered saline (PBS) was administered more than 60 days after priming. Sheep red blood cells (SRBC) (Colorado Serum Co.; 31102) were injected into the peritoneum in a 100-μl suspension in PBS (10%).
Flow cytometry.
Single-cell suspensions of bone marrow, lymph nodes, and spleens were prepared, and red blood cells were lysed, counted, and stained with the following antibodies: fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, allophycocyanin (APC)-, APC-Cy7-, Pacific Blue-, Alexa Fluor 700-, Alexa Fluor 780-, peridinin chlorophyll protein (PerCP)-Cy5.5-, PE-Cy7-, or biotin-labeled monoclonal antibodies purchased from BD Pharmingen or eBioscience, including B220 (RA3-6B2), CD19 (1D3), CD38 (90), IgD (11.26), GL7 (GL7), CD95 (Jo-2), CXCR4 (2B11), IgM (R6-60.2), CD86 (GL1), IgG1 (A85-1), CD21 (7G6), CD23 (B3B4), c-kit (ACK2), CD25 (PC61), CD138 (281-2), CD93 (AA4.1), Sca1 (E13-161.7), CD150 (TC15-12F12.2), Flt3 (A2F10), interleukin 7 receptor (IL-7R) (A7R34), Ly6D (49-H4), CD8 (53-6.7), Mac1 (M1/70), Gr1 (RB6-8C5), NK1.1 (PK136), Ter119 (TER119), TCRβ (H57), TCRγδ (GL3), CD3ε (2C11), CD4 (GK1.5), and CD8 (53-6.7). Biotinylated antibodies were labeled with streptavidin-conjugated Qdot-605 (Invitrogen). Clone 2.4 G2 anti-CD16-CD32 (eBioscience) was used to block Fc receptors. Dead cells were removed from sorting and analysis by propidium iodide (PI) staining (Sigma-Aldrich). Data were collected on an LSRII (BD Biosciences) and analyzed with FlowJo software (TreeStar). Sorting was performed on a FACSAria (BD).
ELISA, ELISpot assay, and in vivo BrdU labeling.
The numbers of antibody-secreting cells (ASCs) were determined as follows. Cells were cultured overnight at 37°C on 96-well MultiScreen-HA filter plates (Millipore) precoated with goat anti-mouse Ig capture antibodies (Southern Biotechnology Associates [SBA]). Spots were visualized with goat anti-mouse IgM or IgG1 antibodies conjugated to horseradish peroxidase (HRP), and color was developed with 3-amino-9-ethyl carbazole (Sigma-Aldrich). Serum immunoglobulins and NP-specific antibodies were measured by enzyme-linked immunosorbent assay (ELISA). NP-specific ASCs were detected by enzyme-linked immunospot (ELISpot) assay as described previously (32). For in vivo detection of cycling cells, mice were exposed to the thymidine analogue bromodeoxyuridine (BrdU) (0.8 mg/ml) in drinking water. DZ and LZ GC B cells were stained with Fas, GL7, CD86, and CXCR4; fixed; and stained with FITC-labeled anti-BrdU (Becton Dickinson) as described previously (33).
B cell isolation and cell culture.
B cells from spleens were isolated using a negative-selection protocol as described previously (Miltenyi Biotec). B cells were cultured in RPMI 1640 medium plus 5% fetal bovine serum (FBS), antibiotics, 2 mM l-glutamine, and β-mercaptoethanol (50 μM). The B cells were activated in complete medium at 1 × 106 cells/ml in the presence of lipopolysaccharide (LPS) (25 μg/ml; Sigma; L2654-1MG) and IL-4 (5 ng/ml; RD Systems; 404-ML-101/CF) as described previously (12). The cells were cultured at 37°C and 5% CO2, harvested at the times indicated, washed, and analyzed using flow cytometry.
RNA-seq analysis.
Transcriptome sequencing (RNA-seq) data were analyzed with the pipeline tool Omics Pipe using the RNA-seq count-based differential expression analysis pipeline (34, 35). Quality control of the raw fastq files was performed using the software tool FastQC (Babraham Bioinformatics). Sequencing reads were aligned to the mouse genome (mm10) using the STAR aligner (36). Read quantification was performed at the exon level using htseq-count with UCSC RefSeq annotation (35). The R BioConductor package DESeq2 was used to calculate size factors to normalize library sizes across replicates and to calculate means and variances based on a negative binomial distribution model to detect differentially expressed genes, based on an adjusted P value of <0.05 (37). Functional enrichment of the differentially expressed genes was performed using the ToppGene Suite and WebGestalt (38). An interaction network of the differentially expressed genes in the B cell receptor signaling KEGG pathway and the 20 most connected neighbors was created using interactions from GeneMANIA.
Statistical analysis.
P values were calculated with the two-tailed Student t test for two-group comparison as applicable using Microsoft Excel software. The statistical significance level was a P value of 0.05.
Accession number.
Sequencing data were deposited in the GEO database with accession number GSE84374.
RESULTS
E2A and Id3 expression in B lineage cells.
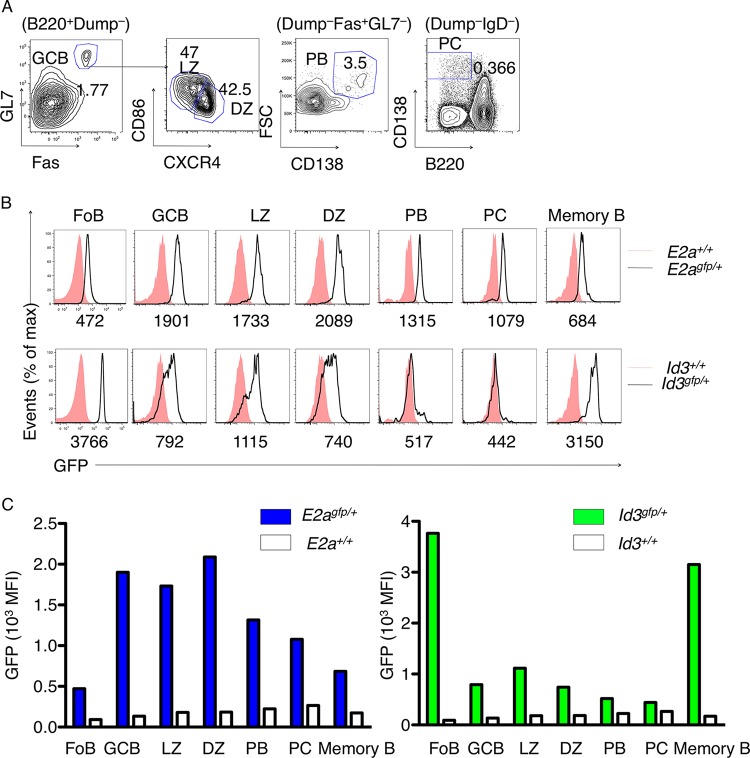
Previous studies have demonstrated that Id3-deficient mice display an impaired response to immunization (39). As a first approach to explore potential intrinsic roles for the E-Id protein axis in activated B lineage cells, we examined the expression of E2A and Id3 using E2a-green fluorescent protein (GFP) and Id3-GFP reporter mice (Fig. 1A). To this end, E2A-GFP and Id3-GFP reporter mice were immunized with SRBC. Two weeks postimmunization, GFP levels were examined in peripheral B cells derived from the spleen using flow cytometry. To identify the GC B cell population, peripheral B cells were examined for the expression of GL7 and Fas. The GC compartment can be segregated further into light- and dark-zone B cells using CD86 and CXCR4 as markers. CD86 levels are elevated in light-zone B cells, while CXCR4 expression is expressed at higher levels in dark-zone B cells. Using these markers in conjunction with flow cytometry, we found that follicular B cells expressed relatively low levels of E2A (Fig. 1B and C). E2A abundance was elevated in B cells upon differentiating into GC B cells, with slightly higher levels in the dark-zone than in the light-zone B cells (Fig. 1B and C). Id3 abundance was high in follicular B cells but declined in germinal center B cells, both the light and dark zones, and was barely detectable in plasma cells (Fig. 1B). The CD38+ IgG1+ “memory” compartment was associated with low levels of E2A expression but displayed high abundance of Id3 (Fig. 1B and C). Thus, during the transition from the naive B to the GC B cell stage, E2A levels steadily increase, whereas Id3 abundance declines.
FIG 1.
E2A and Id3 expression in peripheral B lineage cells. (A) Strategy for gating peripheral B cell compartments: GC (B220+ Dump− Fas+ GL7+), DZ (Fas+ GL7+ CXCR4hi CD86lo), LZ (Fas+ GL7+ CXCR4lo CD86hi), plasmablasts (PB) (Dump− Fas+ GL7− CD138+ FSChi), and PCs (Dump− IgD− B220− CD138+). (B) E2A and Id3 expression was monitored in the B cell compartments using E2A-GFP and Id3-GFP reporter mice. (C) Quantification of E2-GFP (left) and Id3-GFP (right) levels in peripheral B cell compartments. MFI, mean fluorescence intensity.
Role of Id3 in B cells of immunized mice.
To assess the role of Id3 in activated B lineage cells, CD19-Cre; Id3loxP/loxP mice were generated. The CD19-Cre mice carry the Cre recombinase gene in the first exon of the CD19 locus, permitting Cre expression under the control of regulatory elements associated with the CD19 locus (28). Bone marrow cells were isolated from control and CD19-Cre; Id3loxP/loxP mice and analyzed using flow cytometry. Expression of the cell surface markers B220 and CD19 was used to identify the B cell compartment. IgM and IgD expression was examined to segregate the immature from the mature B cell stages. The pro-B cell and pre-B cell compartments were characterized as IgM− IgD− c-kit+ CD25− and IgM− IgD− c-kit− CD25+, respectively. This analysis showed that the pro-B cell, immature-B cell, and mature-B cell compartments were not significantly altered in bone marrow cells with Id3 expression depleted (see Fig. S1 in the supplemental material).
To evaluate the role of Id3 in peripheral B cell development, splenocytes were examined for the expression of CD19, CD5, CD23, and CD93 (see Fig. S2 in the supplemental material). Marginal zone B cells (IgMhi CD23hi) were reduced in CD19-Cre; Id3loxP/loxP mice compared to control mice (see Fig. S2 in the supplemental material). The follicular B cell compartment (IgMhi IgDhi), however, was not affected upon depletion of Id3 expression (see Fig. S2 in the supplemental material).
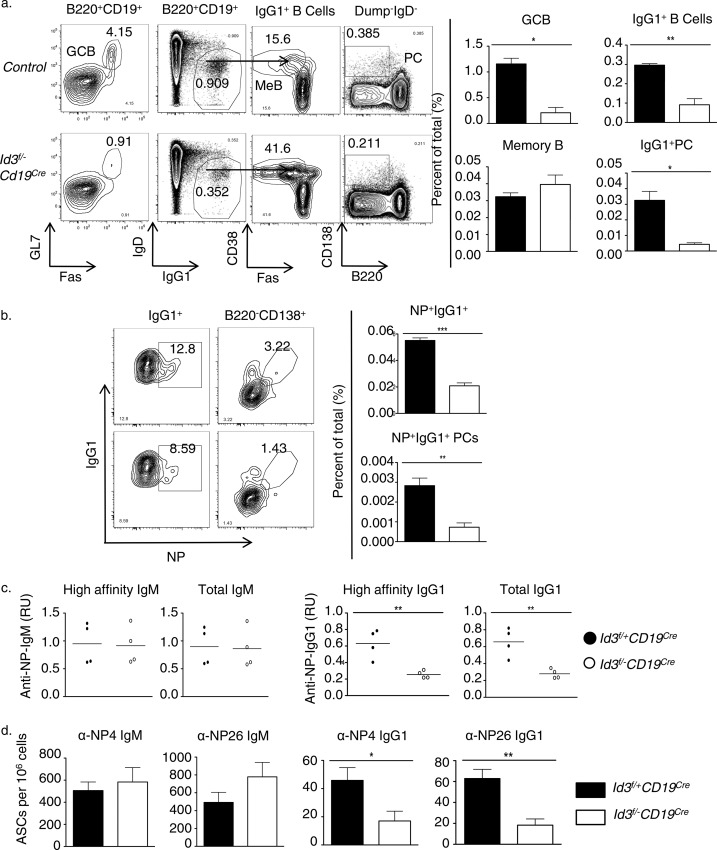
To evaluate the role of Id3 in immunized mice, control and CD19-Cre; Id3loxP/loxP mice were injected with NP-KLH. To examine for the presence of GC B cells, splenocytes were isolated from immunized control and CD19-Cre; Id3loxP/loxP mice, stained for the expression of GL7 and Fas, and analyzed using flow cytometry (Fig. 2). We found that 2 weeks postimmunization, CD19-Cre; Id3loxP/loxP mice showed significant impairment in GC formation as defined by the expression of GL7 and Fas (Fig. 2a). To determine the presence of class-switched cells, B220+ CD19+ cells were examined for the expression of IgG1. We found that the proportions of IgG1 switched cells and CD138hi B220lo plasma cells were also decreased in CD19-Cre; Id3loxP/loxP mice compared to control mice (Fig. 2a).
FIG 2.
Id3 is essential to promote the developmental progression of germinal center B cells and the generation of an effective antibody response in immunized mice. (a) Flow cytometric analysis of B cell subsets derived from spleens isolated from 8-week-old control (Id3fl/− Cd19WT and Id3fl+ Cd19Cre or Cd19WT), Cd19Cre Id3fl/−, or Cd19Cre immunized mice. The mice were immunized with NP-KLH plus adjuvant. (Left) The two left plots display Fas versus GL7 and IgG1 versus IgD expression, gated on the B220+ CD19+ compartment derived from spleens of immunized control and Cd19Cre Id3fl/− or Cd19Cre mice. The two right plots display Fas versus CD38 expression, gated on the IgG1+ IgD− compartment, and B220 versus CD138 expression, gated on the Dump+ IgD− compartment. The numbers in the plots indicate the percentages of Fas+ GL7+, IgG1+ IgD−, Fas− CD38+, and B220− CD138+ cells. (Right) Percentages of GC (Fas+ GL7+), IgG1+ B cells (IgG1+ IgD−), “memory-like” B cells (IgG1+ IgD− Fas− CD38+), and plasma cells (Dump+ IgD− B220− CD138+). (b) (Left) Flow cytometric analysis of NP-reactive IgG1-switched B cells (IgG1+ IgD− NP+) and NP-reactive PCs (Dump+ IgD− B220− CD138+ NP+) derived from spleens isolated from 8-week-old control and Cd19Cre Id3fl/− or Cd19Cre mice immunized with NP-KLH plus adjuvant. The numbers in the plots indicate the percentages of IgG1+ IgD− NP+ and Dump+ IgD− B220− CD138+ NP+ cells. (Right) Percentages of NP-reactive IgG1+ B cells and NP-reactive PCs. (c) Quantification of NP4-reactive IgM and IgG1 and NP26-reactive IgM and IgG1 in sera isolated from control and Id3-CD19 Cd19Cre Id3fl/− or Cd19Cre mice (n = 4 per group) 14 days after immunization with NP-KLH. (d) ELISpot analysis of NP4- and NP26-specific IgM and IgG1 ASCs in spleens isolated 14 days post-NP-KLH immunization. (a to d) The data are representative of at least four experiments (means and standard deviations [SD]; control, n = 4, and Id3f/− Cd19Cre, n = 4 independent biological replicates) (a, b, and d) or one experiment (means; control, n = 4, and Id3f/− Cd19Cre, n = 4 independent biological replicates) (c).
To examine the response to specific antigens, we compared the percentages of NP+ IgG1+ B cells and NP+ IgG1+ PCs in control versus CD19-Cre; Id3loxP/loxP mice. Consistent with the above-mentioned results, we found a smaller fraction of NP-specific IgG1+ B cells and PCs in the spleen of Id3-depleted mice compared to controls (Fig. 2b). Next, we measured NP-reactive-antibody levels in the sera isolated from immunized control and CD19-Cre; Id3loxP/loxP mice. Sera were tested for the ability to interact with 4 (NP4) or, alternatively, 26 (NP26) NP haptens to distinguish between high- and low-affinity NP antibodies (31). While serum titers of NP-specific IgM were unchanged, NP-reactive IgG1 levels were 2-fold lower in sera derived from CD19-Cre; Id3loxP/loxP mice than in sera from control mice (Fig. 2C). Consistent with the decreased titers of serum IgG1, the numbers of NP-specific IgG1+ ASCs in the spleens of immunized CD19-Cre; Id3loxP/loxP mice were reduced 2-fold compared to control mice (Fig. 2D). Collectively, these data indicated that in immunized mice, Id3 expression is essential to permit the developmental progression of marginal zone B cells, GC B cell development, and the development of class-switched cells.
Role of Id3 in cycling GC B cells.
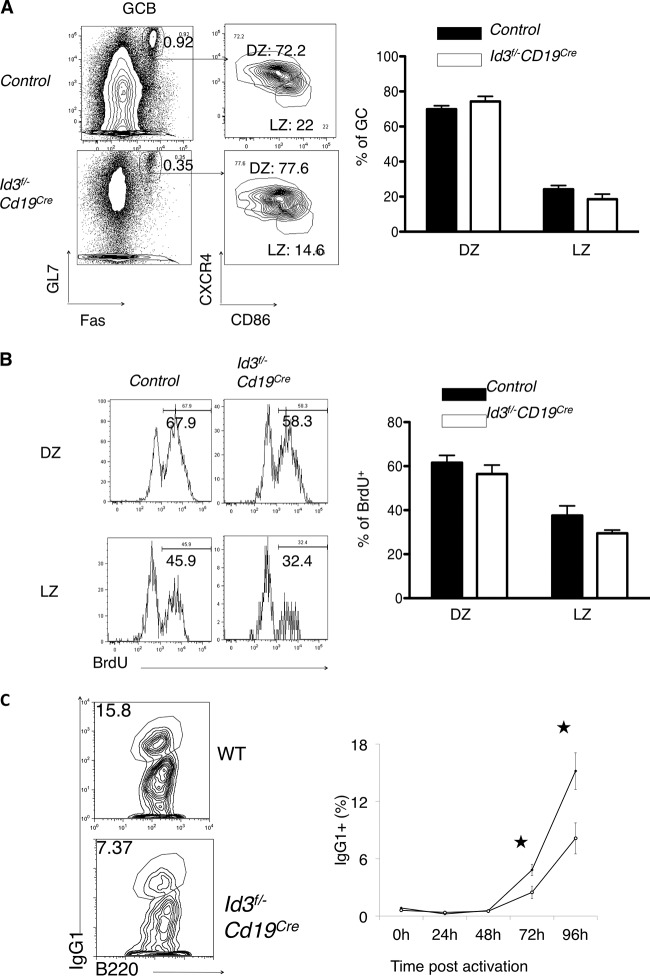
Since Id3 expression differs, albeit slightly, between the DZ and the LZ, GC formation was examined for the two developmental stages in immunized wild-type and Id3 mutant mice. As an immunogen, SRBC were chosen, since they elicit a rapid GC development response upon immunization. Control and CD19-Cre; Id3loxP/loxP mice were immunized with SRBC and analyzed by flow cytometry. To identify DZ versus LZ, B cells were examined for the expression of CXCR4 and CD86 (Fig. 3A). We found that although GC formation was impaired in CD19-Cre; Id3loxP/loxP mice immunized with SRBC, the ratio of DZ to LZ B cells was not affected (Fig. 3A). To determine whether Id3 expression affects the proliferation of GC cells, cycling cells were labeled in vivo using BrdU incorporation (Fig. 3B). We found that a slightly smaller fraction of DZ GC cells were cycling in CD19-Cre; Id3loxP/loxP mice than in control mice (Fig. 3B).
FIG 3.
Abnormal cell cycle progression and class switching in B cells derived from immunized Cd19Cre Id3fl/− and wild-type mice. (A) (Left) Flow cytometric analysis of subcompartmentalization into DZ and LZ of GC B cells isolated from spleens of both control and Cd19Cre Id3fl/− or Cd19Cre mice following immunization with sheep red blood cells. The numbers in the plots indicate the percentages of Fas+ GL7+, CXCR4hi CD86lo, and CXCR4lo CD86hi cells. (Right) Quantification of percentages of DZ and LZ cells in both control and Cd19Cre Id3fl/− and Id3fl/fl Cd19Cre mice. (B) (Left) BrdU incorporation of DZ (top) and LZ (bottom) cells, as gated in panel A. (Right) Percentages of BrdU+ cells within the DZ or LZ isolated from both control and Cd19Cre Id3fl/− or Cd19Cre mice immunized with SRBC. (C) IgG1 class switching is affected in B cells derived from Id3f/− CD19Cre mice. B cells were activated in vitro in the presence of LPS and IL-4 over a 4-day period. The percentages of IgG1+ expressors isolated from Id3f/− Cd19+/+ mice and Id3f/− CD19Cre mice during 24-h intervals are shown. Solid circles, wild-type B cells; open circles, B cells derived from Id3f/− CD19Cre mice. For all panels, data are representative of the results of two experiments (means and SD; control, n = 3, and Cd19Cre Id3fl/− or Id3fl/fl Cd19Cre, n = 3 independent biological replicates).
To examine in greater detail whether Id3 affects CSR and cell cycle progression ex vivo, B cells derived from both control and CD19-Cre; Id3loxP/loxP mice were cultured in the presence of LPS and IL-4. We found that IgG1 switching following activation by LPS+ IL-4 was perturbed in B cells isolated from CD19-Cre; Id3loxP/loxP mice compared to control mice (Fig. 3C). To examine whether the inability of Id3-depleted B cells to undergo CSR was caused by a defect in cell cycle progression, B cells isolated from wild-type and CD19-Cre; Id3loxP/loxP mice were labeled with carboxyfluorescein succinimidyl ester (CFSE) and cultured in the presence of LPS and IL-4 (see Fig. S3 in the supplemental material). We found that B cells depleted of Id3 expression were impaired in their ability to proliferate (see Fig. S3 in the supplemental material). Collectively, these data suggest that the proliferation deficit caused by Id3 deletion may contribute to loss of CSR.
Role of Id3 in secondary-antibody response.
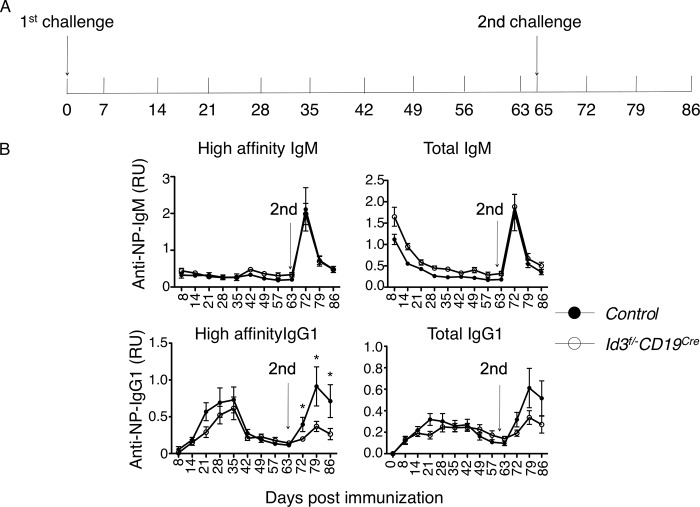
The data described above indicate that Id3 expression is required to generate an effective immune response. To investigate the role of Id3 in immunized mice in greater detail, we challenged both control and CD19-Cre; Id3loxP/loxP mice with NP-KLH plus adjuvant, followed by boosting with NP-KLH 2 months after the initial immunization (Fig. 4A). We found that CD19-Cre; Id3loxP/loxP mice displayed responses, checked both 7 days and 14 days post-secondary immunization, that were similar to those in control mice as they related to IgM levels, including both high-affinity (NP4-reactive IgM) and low-affinity (NP26-reactive IgM) antibodies (Fig. 4B). In contrast, the generation of NP4-reactive IgG1 antibodies was significantly perturbed in immunized and boosted CD19-Cre; Id3loxP/loxP mice compared to control mice (Fig. 4B).
FIG 4.
Id3 expression is required to generate an efficient secondary immune response. (A) Immunization strategy to investigate the role of Id3 in secondary immune responses. For the first challenge, both control and Id3-CD19 conditional knockout (cKO) mice were immunized with NP-KLH plus adjuvant. For the second challenge, mice were boosted with NP-KLH only. (B) Time course analysis of NP4- or NP26-reactive IgM and IgG1 levels in sera isolated from control and Cd19Cre Id3fl/−, Cd19Cre Id3fl/−, or Cd19Cre mice immunized with NP-KLH plus adjuvant at day 0 and boosted with NP-KLH at day 65 (arrows). The data are representative of the results of two independent experiments. Shown are means ± SD; n = 7 mice in the control group; n = 8 mice in the (Id3f/− Cd19Cre group). RU, ratio unit.
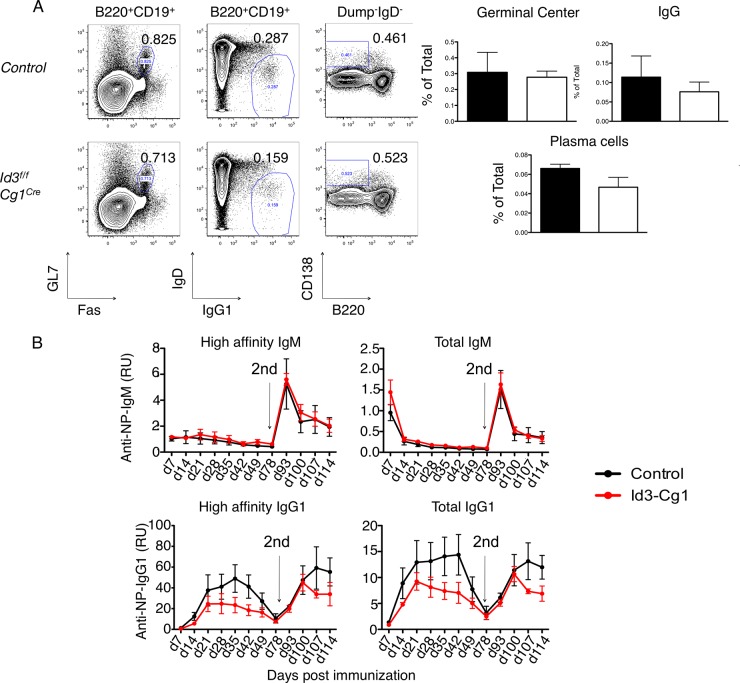
To determine whether Id3 also acts to orchestrate secondary immune responses, Cγ1-Cre; Id3fl/fl mice were generated (29). The Cγ1-Cre transgene allows depletion of Id3 beyond the GC checkpoint (29). Both control and Cγ1-Cre; Id3fl/fl mice were immunized with SRBC and examined 2 weeks postinjection. The floxed Id3 exons were efficiently excised (>90%) in Cγ1-Cre; Id3fl/fl mouse GC B cells (data not shown). We found that the fraction and the total number of GC B cells in immunized Cγ1-Cre; Id3fl/fl mice were comparable to those in immunized control mice (Fig. 5A). IgG1-switched B cell and plasma cell development also appeared to be unaffected in immunized Cγ1-Cre; Id3fl/fl mice (Fig. 5A). Following boosting, both the IgM and IgG1 recall responses in Cγ1-Cre; Id3fl/fl mice were comparable to those in control mice (Fig. 5B). Collectively, these results indicate that Id3 expression does not play an inhibitory role beyond the GC B cell stage against mounting an efficient immune response.
FIG 5.
Ablation of Id3 expression after germinal center entry does not impair germinal center formation, class switching, or the memory response. (A) (Left) Flow cytometric analysis of activated B cell subpopulations derived from spleens isolated from 8-week-old control or Cγ1Cre; Id3fl/fl mice immunized with NP-KLH plus adjuvant. The two left plots indicate Fas versus GL7 and IgG1 versus IgD expression, gated on the B220+ CD19+ compartment derived from spleens of immunized control or Cγ1Cre; Id3fl/fl mice. The right plots indicate B220 versus CD138 expression, gated on the Dump+ IgD− compartment. The numbers in the plots indicate the percentages of Fas+ GL7+, IgG1+ IgD−, and B220− CD138+ cells. (Right) Percentages of GC (Fas+ GL7+), IgG1+ B cells (IgG1+ IgD−), and plasma cells (Dump+ IgD− B220− CD138+) isolated from the spleens of immunized mice. (B) Time course analysis of NP4- or NP26-reactive IgM and IgG1 in sera isolated from control and Cγ1Cre; Id3fl/fl mice immunized with NP-KLH plus adjuvant at day 0 and boosted with NP-KLH at day 78 (arrows). The data are representative of the results of two independent experiments. Shown are means ± SD; n = 5 mice in the control group; n = 5 mice in the Id3f/f Cγ1Cre group.
Id3-depleted naive and activated B lineage cells are associated with distinct gene expression signatures.
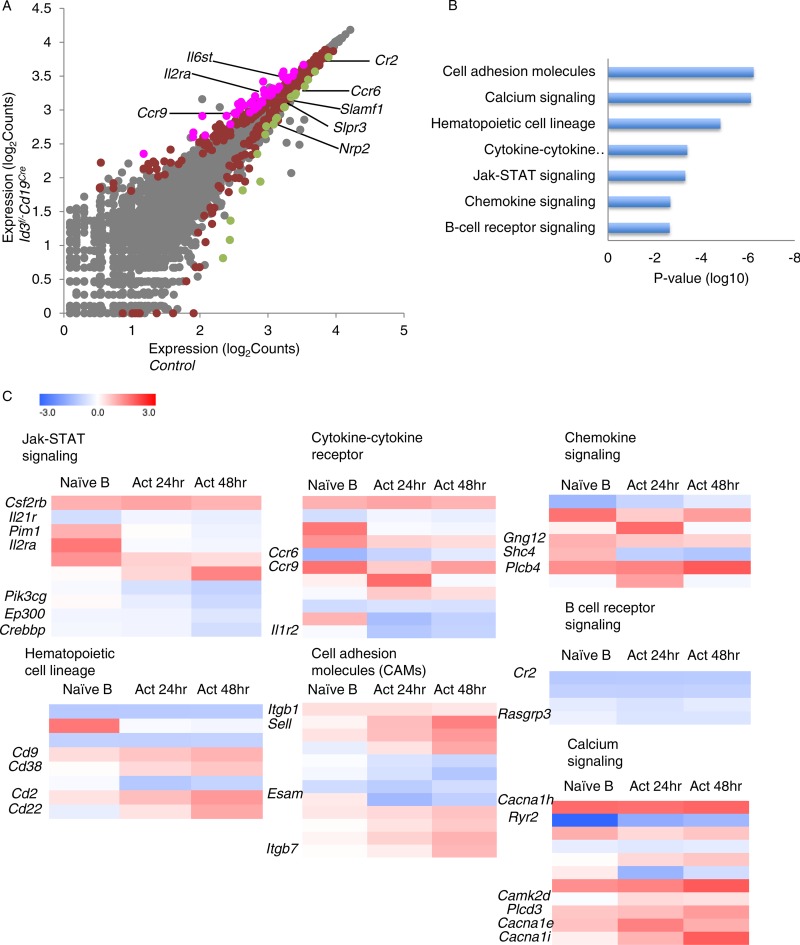
To determine how Id3 orchestrates GC B cell fate, we examined the transcription signatures from Id3-depleted activated B lineage cells. To accomplish this objective, B cells derived from control and CD19-Cre; Id3loxP/loxP mice were activated in vitro in the presence of LPS and IL-4 for 24 or 48 h. RNA was isolated from naïve as well as activated control and CD19-Cre; Id3loxP/loxP mice and analyzed by RNA-seq. One hundred twenty-five genes were differentially expressed (adjusted P < 0.05; 38 downregulated; 87 upregulated) in Id3-depleted versus wild-type naive B cells. Prominent target genes included IL21R, Cr2, Ccr6, Slamf1, Slpr3, Nrp2, Il6st, Il2ra, and Ccr9 (Fig. 6A; see Table S1 in the supplemental material). Surprisingly, however, the RNA-seq analysis revealed that AID levels were not elevated in activated B cells depleted of Id3 expression. Hierarchical clustering revealed a subset of differentially expressed transcripts that were associated with genes encoding cell adhesion molecules, calcium signaling, hematopoietic cell lineage, cytokine-cytokine receptor, JAK-STAT signaling, chemokine signaling, and B cell receptor signaling (Fig. 6B and C). RNA-seq analysis of activated B cells isolated 24 h and 48 h poststimulation showed that 36 and 191 genes, respectively, including the chemokine receptor Ccr6, were differentially expressed in the absence of Id3 (see Tables S2 and S3 in the supplemental material). Collectively, these data indicate that in activated B cells, Id3 does not control AID expression but rather acts to regulate the expression of a subset of genes encoding signaling components that mediate antigen receptor-, cytokine receptor-, and chemokine receptor-mediated signaling.
FIG 6.
Transcription signatures of activated B lineage cells depleted of Id3 expression reveal a spectrum of Id3 target genes encoding factors involved in chemokine receptor-, cytokine receptor-, and antigen receptor-mediated signaling. (A) RNA-seq analysis of sorted naive B cells isolated from control versus CD19-Cre; Id3f/f mice (log2 abundance values for RefSeq genes). The points represent genes differentially expressed in CD19-Cre; Id3f/f mice (red; P < 0.05), genes upregulated >2-fold in CD19-Cre; Id3loxP/loxP mice (magenta; P < 0.05), genes downregulated >2-fold in CD19-Cre; Id3loxP/loxP mice (green; P < 0.05), and genes not significantly changed (gray) compared to control cells. Selected genes are labeled. (B) Selected modules in terms of gene ontology and their associated P values affected by depletion of Id3 expression. (C) Heat maps for differentially expressed genes in each category. RNA transcripts derived from control or Id3-depleted naive B cells 24 h and 48 h after in vitro activation were quantified, and the log2 values of the fold changes between control and Id3-deficient samples are shown.
DISCUSSION
Previous studies have demonstrated critical roles for Id3 at multiple checkpoints in lymphocyte development (6, 7, 39, 40). Here, we extended these studies and found that depletion of Id3 expression impaired the ability of activated B lineage cells to undergo cellular expansion, CSR, and GC development. Previous studies indicated that E proteins control CSR by directly activating AID expression (12, 40, 41). However, surprisingly, we found that the absence of Id3 did not regulate CSR by modulating AID levels. How, then, are E and Id proteins linked to modulate CSR? We propose the following. In naive B cells, Id3 levels are high, whereas E protein abundance is low. Upon antigen receptor- and cytokine-mediated signaling, Id3 levels steadily decline. Depletion of Id3 does not lead to premature activation of AID expression but rather appears to impair the expression of genes encoding factors that mediate antigen receptor- and/or cytokine-mediated signaling. Lower levels of signaling components in Id3-depleted cells then lead to perturbed GC development. Furthermore, we suggest that declining levels of Id3 are not sufficient to activate E2A-mediated induction of AID expression. Rather, we propose that in activated B cells, E2A protein abundance steadily increases to reach maximum levels at the GC B cell stage, when it becomes insensitive to the dosage of Id3, acting in concert with other transcriptional regulators to induce AID expression. Thus, we propose that Id3 acts in cycling B cells to permit activation of genes associated with antigen receptor- and cytokine receptor-mediated signaling, whereas E proteins act in concert with other transcriptional regulators to induce the expression of AID.
Recent studies have indicated that a deficiency in Id3 expression is closely associated with the development of Burkitt lymphoma (42, 43). Previously, we inspected the transcriptomes of Burkitt lymphomas depleted of the expression of E2A for overlap in Id3 target genes in activated murine B lineage cells. Interestingly, expression of Ccr6 (C-C motif chemokine receptor 6 gene), an Id3 target gene, was modulated by depletion of E2A expression in human Burkitt lymphoma cells (34). Thus, it appears that the regulation of Ccr6 by the E2A/Id3 axis is conserved between human and murine germinal center B cells. More recent studies have indicated that depletion of both Id2 and Id3 expression in T lineage cells leads to the rapid development of T cell lymphoma (20). Is Id3 expression also essential to suppress the development of lymphoma in mice? We have not observed lymphomagenesis in mice depleted of Id3 expression in B lineage cells, but it is conceivable that Id2 and Id3 act redundantly to suppress the development of murine B cell lymphoma.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Murre laboratory for thoughtful discussion. We thank Yuan Zhuang for the kind gift of Id3f/f and Robert Rickert for CD19-Cre mice.
This study was supported by funding to the CCBB from a CTRI grant (UL1TR001442) and by grants from the NIH to C.M. (AI00880, AI09599, and AI102853).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00150-16.
REFERENCES
- 1.Victora GD, Nussenzweig MC. 2012. Germinal centers. Annu Rev Immunol 30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.Chandra V, Bortnick A, Murre C. 2015. AID targeting: old mysteries and new challenges. Trends Immunol 36:527–535. doi: 10.1016/j.it.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teng G, Papavasiliou FN. 2007. Immunoglobulin somatic hypermutation. Annu Rev Genet 41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Zan H, Pon EJ, Mai T, Casali P. 2012. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Rothenberg EV. 2014. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol 32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bain G, Murre C. 1998. The role of E-proteins in B- and T-lymphocyte development. Semin Immunol 10:143–153. doi: 10.1006/smim.1998.0116. [DOI] [PubMed] [Google Scholar]
- 8.Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer, daughterless, MyoD and myc proteins. Cell 56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 9.Mansson R, Welinder E, Ashber J, Lin YC, Benner C, Glass CK, Sigvardsson M, Murre C. 2012. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci U S A 109:21028–21033. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck K, Peak M, Ota T, Nemazee Murre DC. 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med 206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Sayegh CE, Quong MW, Agata Y, Murre C. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol 4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 13.Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. 2010. B cell specific and stimulation-responsive enhancers depresses Aicda by overcoming the effects of silencers. Nat Immunol 11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 14.Benezra R, Davis RL, Lockshon D, Turner DL, Weintaub H. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 15.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix protein Id2. Nature 397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 16.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. 2007. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki M, Rivera RR, Miyazaki K, Lin C, Agata Y, Murre C. 2011. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naïve fate of T cells. Nat Immunol 12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zúñiga-Pflücker JC, Wiest DL. 2009. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity 31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Wu D, Jiang N, Zhuang Y. 2013. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol 191:5052–5064. doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki M, Miyazaki K, Chen SY, Chandra V, Agata Y, Chang A, Kawamoto H, Murre C. 2015. The E-Id protein axis modulates the activities of the PI3K-AKT-mTORC1-Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion and lymphomagenesis. Genes Dev 29:409–425. doi: 10.1101/gad.255331.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, Dong C. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda-Hayakawa I, Mahlios J, Zhuang Y. 2009. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol 182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verykokakis M, Krishnamoorth V, Iavarone A, Lasorella A, Sigvardsson M. 2013. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol 191:5973–5983. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. 2012. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity 36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alonzo ES, Sant'Angelo DB. 2011. Development of PLZF-expressing innate T cells. Curr Opin Immunol 23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Cruz LM, Stradner MH, Yang CY, Goldrath AW. 2014. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J Immunol 192:2227–2236. doi: 10.4049/jimmunol.1302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murre C. 2005. Helix-loop-helix proteins and lymphocyte development. Nat Immunol 6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 28.Rickert RC, Roes J, Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, Rodig S, Kutok J, Tarakhovsky A, Schmidt-Supprian M, Rajewsky K. 2010. Constitutive canonical NF-kB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell 18:580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z, Li H, Han M, Xu T, Wu X, Zhuang Y. 2011. Modeling Sjogren's syndrome with id3 conditional knockout mice. Immunol Lett 135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cumano A, Rajewsky K. 1985. Structure of primary anti-(4-hydroxy-3-nitrophenyl)acetyl (NP) antibodies in normal and idiotypically suppressed C57BL/6 mice. Eur J Immunol 15:512–520. doi: 10.1002/eji.1830150517. [DOI] [PubMed] [Google Scholar]
- 32.Czerkinsky C, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. 1983. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods 65:109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 33.Carayon P, Bord A. 1992. Identification of DNA-replicating lymphocyte subsets using a new method to label the bromo-deoxyuridine incorporated into the DNA. J Immunol Methods 147:225–230. doi: 10.1016/S0022-1759(12)80012-3. [DOI] [PubMed] [Google Scholar]
- 34.Fisch K, Meissner T, Gioia L, Ducom J-C, Carland T, Loguercio S, Su A. 2015. Omics Pipe: a community-based framework for reproducible multi-omics data analysis. Bioinformatics 31:1724–1728. doi: 10.1093/bioinformatics/btv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 36.Dobin A, Davis C, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550–558. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. 1999. Impaired immune responses and B cell proliferation in mice lacking the Id3 gene. Mol Cell Biol 19:5969–5980. doi: 10.1128/MCB.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quong M, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C. 2004. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J Exp Med 199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quong MW, Harris DP, Swain SL, Murre C. 1999. E2A activity is induced during B cell activation to promote immunoglobulin class switch recombination. EMBO J 18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Staudt LM. 2012. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Dave SS. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.