ABSTRACT
Ribosome biogenesis is an essential process initiated in the nucleolus. In eukaryotes, multiple ribosome biogenesis factors (RBFs) can be found in the nucleolus, the nucleus and in the cytoplasm. They act in processing, folding and modification of the pre-ribosomal (r)RNAs, incorporation of ribosomal proteins (RPs), export of pre-ribosomal particles to the cytoplasm, and quality control mechanisms. Ribosome biogenesis is best established for Saccharomyces cerevisiae. Plant ortholog assignment to yeast RBFs revealed the absence of about 30% of the yeast RBFs in plants. In turn, few plant specific proteins have been identified by biochemical experiments to act in plant ribosome biogenesis. Nevertheless, a complete inventory of plant RBFs has not been established yet. We analyzed the proteome of the nucleus and nucleolus of Arabidopsis thaliana and the post-translational modifications of these proteins. We identified 1602 proteins in the nucleolar and 2544 proteins in the nuclear fraction with an overlap of 1429 proteins. For a randomly selected set of proteins identified by the proteomic approach we confirmed the localization inferred from the proteomics data by the localization of GFP fusion proteins. We assigned the identified proteins to various complexes and functions and found about 519 plant proteins that have a potential to act as a RBFs, but which have not been experimentally characterized yet. Last, we compared the distribution of RBFs and RPs in the various fractions with the distribution established for yeast.
Keywords: Arabidopsis thaliana, nucleolus, nucleus, orthology, proteomics, rRNA processing, ribosome biogenesis
Abbreviations
- EJC
exon junction complex
- NMD
nonsense-mediated mRNA decay
- NoPDB
(NOPdb) nucleolar protein database
- RBF
ribosome biogenesis factor
- RP
ribosomal protein
Introduction
Ribosome biogenesis is a cellular process essential for viability of all pro- and eukaryotes. Ribosome maturation involves pre-rRNA processing, modification and folding as well as the incorporation of ribosomal proteins (RP).1-3 In eukaryotes, the 18S, 5.8S and 25/28S rRNAs are transcribed as a single pre-rRNA,4,5 whereas the 5S rRNA is transcribed separately.6 Pre-rRNA processing and folding requires a multitude of proteinaceous and RNA ribosome biogenesis factors (RBFs).2 Genetic and proteomic studies have identified around 255 proteins functioning in ribosome biogenesis in the yet best studied eukaryotic model organism Saccharomyces cerevisiae.2,5 Recent approaches have also targeted the inventory of RBFs in humans and plants.7-11 While the initial analysis by Wild and co-workers has focused on the confirmation of an RBF-like function of human proteins with similarity to yeast RBFs (153 factors),7 the subsequent study by Tafforeau and coworkers identified 286 nucleolar human RBFs, from which 74 do not have a yeast homolog.8 In parallel, orthologues of more than 200 yeast RBFs have been assigned in plants by bioinformatics means.9-11 In turn, about 30% of the yeast RBFs could not be assigned in plant genomes.10,11 Considering the 2 findings that (i) human RBFs without homology to yeast RBFs have been identified and that (ii) not all yeast RBFs are identified in plant genomes favors the existence of plant specific RBFs.
The major steps of ribosome biogenesis occur in the nucleus and the nucleolus,12,13 whereas only final steps in the biogenesis of both subunits occur in the cytoplasm.2,5 Although it has been established that ribosome biogenesis is not the only function of the plant nucleolus.14 It can be expected that most of the RBFs can be found in the nucleus and the nucleolus.2 While the proteome of the mammalian nucleolus has been explored in detail in organisms like human or mouse,e.g.15-18 not much is known about this compartment in plants. In the one available proteomic analysis of Arabidopsis thaliana nucleoli 217 proteins were identified, including ribosomal, exon junction complex, non-ribosomal and even non-nucleolar proteins.19,20 It could be demonstrated that some proteins redistribute under stress from the nucleus to the nucleolus (e.g., RSZp22; eIF4A-III; STRS1).21-23 In addition to ribosome biogenesis, the nucleolar compartment contributes to other processes like the nonsense-mediated mRNA decay (NMD) pathway in plants.14,20,24 However, the knowledge about the diversity of the processes in the plant nucleolus and about the plant nucleolar proteome per se is still sparse.
We analyzed the proteome of cytoplasm, nucleus and nucleolus of A. thaliana. By this we could confirm the previously detected proteins20 and extended the proteome of the A. thaliana nucleolus. We discuss the detection of RBFs, RPs, spliceosomal proteins and of proteins involved in NMD. Beside components of known complexes, we identified 319 A. thaliana proteins having characteristics that make a function in ribosome biogenesis possible, but the function of these proteins is not yet experimentally confirmed. We compare the distribution of the RBFs and RPs in the different sub-compartments with the distribution described for yeast. In general, we observe that plant RPs are found in compartments that suggest an earlier assembly than in yeast, whereas the distribution of plant RBFs in the various compartments by large parallels the regime established for yeast ribosome biogenesis. In addition, we confirmed RP phosphorylation in all 3 compartments and realized acetylation of RPs only in the nucleus and nucleolus, but not in the cytoplasm.
Results
Isolation of the nucleolus and proteomic analysis
We established a protocol for nucleolus isolation from cell culture as a prerequisite to analyze the proteome of the according fractions (Fig. 1A; Methods). We separated the cell lysate in cytoplasm and nucleus. Subsequently, the nucleus was further fractionated into nucleus and nucleolus. To judge the quality of the fractionation we used antibodies against several proteins whose localization is established (Fig. 1B). Using antibodies against Toc33 and Toc75, 2 components of the chloroplast translocon,25 we realized a presence of chloroplast proteins in the nuclear / nucleolar fraction below 10% when compared to the cytoplasmic fraction (Fig. 1B, C). The analysis of the distribution of the mitochondrial porin VDAC26 or cytosolic proteins (eIF1A, Lsg1)27 yielded a protein abundance of about 40-50% in the nucleus, while their abundance was reduced to 10% in the nucleolar fraction when compared to the cytoplasmic fraction (Fig. 1B, C). Proteins with dual localization in cytoplasm and nucleus showed a comparable abundance in the cytoplasm and nucleus, whereas they were largely depleted from the nucleolar fraction (∼10%; RPL15, RPL5, NOB1).28,29 In contrast, proteins expected to be present in the nucleolus were highly enriched in this fraction (ENP1, FIB).29,30
Figure 1.
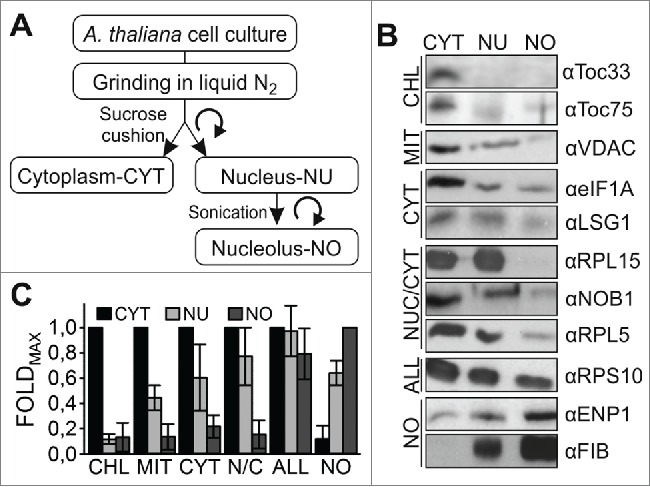
Strategy of cell fractionation. (A) Scheme of A. thaliana cell culture fractionation indicating the abbreviation of the fraction used subsequently. (B) The isolated cellular fractions were subjected to SDS-PAGE followed by western blotting with the indicated antibodies recognizing proteins from chloroplasts (CHL), mitochondria (MIT), cytosol (CYT), nucleus and cytosol (N/C), from all compartments (ALL) and from the nucleolus (NO). (C) The quantification of the western blots is shown including the standard deviation between samples and factors.
From our analysis we became confident that proteins found to be exclusively or highly enriched in the nucleolar fraction can be assigned as nucleolar proteins. Subsequently, we determined the proteome of the 3 fractions. All proteins were hydrolysed with trypsin and the peptides labeled during tryptic digestion using 16O and 18O containing water. The peptide pairs exhibiting a differential ratio were interrogated via nano-LC-MS/MS mass spectrometry (MS). MS allows a comparative and relative quantitation by peptide counting of changes in protein abundances between 2 compared samples. We compared the fractions of 16O-labeled cytoplasm vs. 18O-labeled nucleus, 16O-labeled nucleus vs. 18O-labeled cytoplasm, 16O-labeled cytoplasm vs. 18O-labeled nucleolus, 16O-labeled nucleolus vs. 18O-labeled cytoplasm, 16O-labeled nucleus vs. 18O-labeled nucleolus and 16O-labeled nucleolus vs. 18O-labeled nucleus for 3 independent replicates.31 Proteins were assigned to a certain fraction in case of detection in 2 of the 3 replicates (see material and methods).
Applying the described criteria we identified 2762 different proteins in the 3 fractions (Fig. 2A, B; Table S1). In total, we identified 2544 proteins in the nucleus and 1602 proteins in the nucleolar fraction (Fig. 2B). As expected from the marker analysis (FIB, Fig. 1B), the overlap between the nuclear and nucleolar fraction is large with 1429 proteins. In addition, the overlap of the nuclear and cytoplasmic proteome (1322 proteins) exceeds the overlap between the nucleolar and cytoplasmic proteome (780 proteins), especially as the majority of the latter proteins are present in all 3 fractions. Comparison of all proteins found in here with the 217 proteins previously identified in the nucleolar fraction (Fig. 2C, atNOPdb, left)20 yielded a re-discovery rate of 85% (Fig. 2C, right) with the largest discrepancy for the proteins subsequently assigned as nuclear proteins in the earlier dataset.
Figure 2.
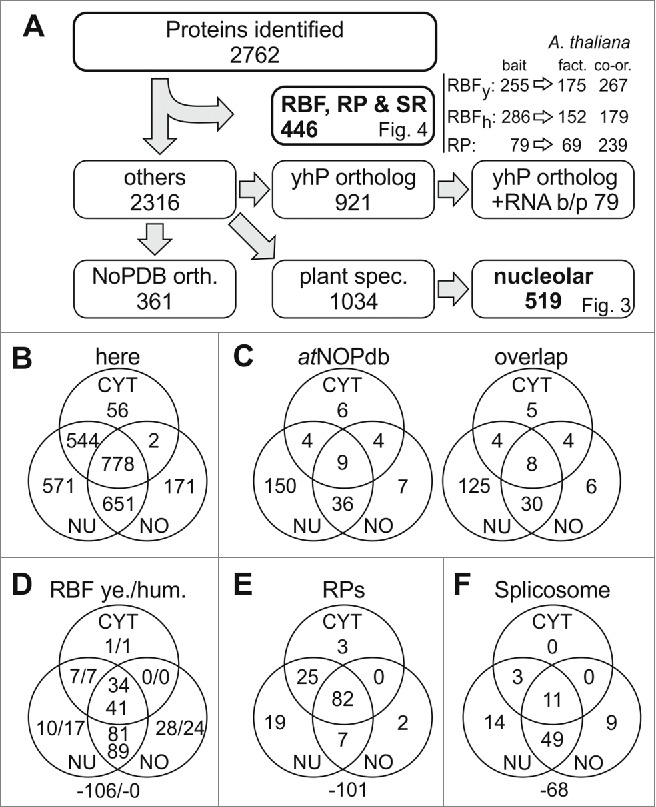
Classification of identified proteins. (A) Shown is the procedure of data processing. Identified proteins are listed in Table S1, identified co-orthologues to RBFs in yeast and humans in Table S2; to RPs in Table S4 and spliceosomal proteins in Table S5. For each pool the number of proteins is indicated and the according figure is referred to. The protein accessions of identified orthologues to proteins deposited in nucleolar database for human (NoPDB) are listed in Table S6, the sequences orthologous to other yeast and human proteins (yhP) are listed in Table S7,8. (B) Shown is the number of proteins discovered in the 3 different fractions. (C) Shown is the number of proteins in the 3 different compartments in the atNOPdb (left) and the number thereof found in our proteomic study (right). (D) Shown is the number of proteins identified in the 3 different fractions analyzed by proteomics with ortholog search to yeast RBFs (left) or human RBFs (right). The number outside the Venn-diagram gives the co-orthologues identified in the A. thaliana genome but not found here. (E, F) Shown is the proportion of found RPs (E) and splicing factors (F) in the various fractions. CYT, cytosol; NU, nucleus; NO, nucleolus.
Nuclear protein complexes of A. thaliana
We identified 161 proteins of the 267 orthologues and co-orthologues to yeast RBFs in our proteomic set (Fig. 2D, left numbers).10,11 Additionally, 4 orthologues to yeast RBFs were found only in one of the replicas in the according fraction and were excluded from the analysis. In total, the proteins represent 133 different yeast RBFs. As expected, all except of one RBF ortholog were found in the nucleus and nucleolus, but 42 are also present in the cytoplasm. Only the ortholog to RLI1 was found exclusively in the cytoplasmic fraction (Table S2), which is consistent with its function in the quality control function of this protein.32
Furthermore, we assigned (co-)orthologues to human RBFs.8 179 A. thaliana proteins are orthologous to 152 human RBFs (Fig. 2A; Table S3). All of the (co-)orthologues assigned to the human RBFs were detected in our proteome (Fig. 2D; right numbers). Again, only one protein (TUFM ortholog) was found exclusively in the cytoplasmic fraction. It is assigned as a translation elongation factor and thus, the functional assignment correlates with the detected localization. In combination, only 246 (co-)orthologues to yeast and human RBFs have been identified, because some of the plant proteins are orthologues of both, yeast and human RBFs.
We also investigated the distribution of the RPs in our proteomic data set. We identified 138 of the 239 RP paralogues assigned in A. thaliana (Fig. 2E).11,33 Worth mentioning, 3 additional RPs (RPL10A, RPS15C and RPL26B) were detected once in one of the fractions analyzed and were, therefore, omitted in our assignment. However, we detected at least one (co-)ortholog of all RPs except RPL29, L31, L39, L40, L41, S29 and S30. Interestingly, studies focusing on the ribosomal proteome had difficulties to detect L29, L39, L41, S29 and S30 as well.33 Two RPs have only been detected in the nucleolus according to our procedure, while 26 were only identified in the nucleus or nucleus/nucleolus fraction (Table S4). Manual inspection revealed that other orthologues to 18 of these proteins have been found in the cytoplasmic fraction, while RPL17, RPL28, RPL36 and RPS28 are only found in the nucleus/nucleolus fraction according to the quality criteria applied (Table S4).
It has been discussed that the nucleolus plays an important function in NMD of mRNA.14 We detected several proteins involved in this process in plants - UPF1 (At1g33980) and UPF3 (At5g47010)34 in the nucleolar and the nuclear fraction, and UPF2 (At2g39260)35 in the nucleolar fraction - while SMG7 (At5g19400) was only once detected in the nucleus and SMG7-like (At1g28260)36 was not found. Moreover, the exon junction complex (EJC) appears to be required for intron-based NMD.35,37 Four of the 6 components involved in this process were indeed observed in the nucleolar and the nuclear fractions (found: Mago, At1g02140; Y14, At1g51510; elF4A3, At3g19760; Barentsz2, At1g15280; not found: Barentsz1, At1g80000; PYM, At1g11400). Thus, based on the proteomic analysis, a function of the nucleolus in NMD appears to be likely.
A third complex for which the discovery of proteins was inspected is the spliceosome. Previously, 154 (co-)orthologues to 95 factors were experimentally or bioinformatically identified.38 We observed 86 of these proteins representing 67 different factors; the vast majority as expected in the nucleus (Fig. 2F; Table S5). Six additional proteins (representing co-orthologues to one additional factor) were detected once in one of the 3 fractions (Table S5).
In summary, 446 proteins found in either of the fractions and 351 proteins found in the nucleolar fraction have previously been assigned by various means as factor to be involved in ribosome biogenesis, splicing or non-mediated mRNA decay.
Acetylation and phosphorylation of identified proteins
Within our dataset we also have information about protein acetylation. To judge the specificity at first we analyzed the acetylation of histones, which are known to be exclusively acetylated in the nucleus. Indeed, we observed acetylation of 6 histones exclusively in the nucleus or nucleolus, but not in the cytoplasm (Table 1). In addition, for one Histone superfamily protein we observed an evidence for phosphorylation (Table 1).
Table 1.
Histone acetylation. Shown are all Histones identified. Given is the tentative name, the ATG number, the identification (x) in cytoplasm (CYT), nucleus (NUC) or nucleolus (NOC) as well as the number of replicas in which acetylation was observed. In case that acetylation (A) or phosphorylation (P) is observed, the last column gives the number and the modified amino acid in the 3 letter code. Numbers in bold show detection in 16O and 18O samples.
| Localization |
Modification |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein function | ATG | CYT | NUC | NOC | CYT | NUC | NOC | Modified AA | |
| Histone superfamily protein | AT1G07660 | X | X | 4 | 1 | A | Lys-32/92 | ||
| Histone B2 | AT5G22880 | X | X | X | 3 | A | Lys-28/67/93 | ||
| Histone deacetylase 2B | AT5G03740 | X | X | 1 | 2 | A | Lys-123/287 | ||
| Histone deacetylase HDT4 | AT2G27840 | X | |||||||
| Histone deacetylase HD2A | AT3G44750 | X | X | 2 | 1 | A | Lys-12 | ||
| Histone superfamily protein | AT1G01370 | X | X | ||||||
| Histone H2A 6 | AT5G59870 | X | X | ||||||
| Histone H2A 7 | AT5G27670 | X | X | 2 | 1 | A | Lys-129 | ||
| Histone acetyltransferase | AT5G56740 | X | X | ||||||
| Histone superfamily protein | AT3G09480 | X | X | ||||||
| Histone superfamily protein | AT1G21970 | X | |||||||
| Histone deacetylase 2B | AT5G22650 | X | X | X | 1 | A | Lys-15 | ||
| Histone superfamily protein | AT1G09200 | X | 1 | P | Thr-108 | ||||
Next, we inspected the acetylation of other proteins. To ensure the specificity we only considered proteins for which a specific acetylated peptide was observed in at least 2 out of the 3 replicates. Based on this criterion we observed 28 acetylated proteins (Table 2). For only 3 proteins acetylation was observed in the cytoplasmic fraction.
Table 2.
Protein acetylation in the different fractions. Shown are all proteins for which one acetylated peptide was observed in a certain cellular compartment at least in 4 of 6 replicas. Given is the tentative function, the ATG number, the identification (x) in cytoplasm (CYT), nucleus (NUC) or nucleolus (NOC) as well as the number of replicas in which acetylation was observed. In case that acetylation is observed, the last column gives the number and the modified amino acid in the 3 letter code. A-Orthologue to protein in hNoPDB, B-Orthologue to human RBF, C-Orthologue to yeast RBF. Numbers in bold show detection in 16O and 18O samples.
| Localization |
Acetylation |
|||||||
|---|---|---|---|---|---|---|---|---|
| Protein function | ATG | CYT | NUC | NOC | CYT | NUC | NOC | Modified AA |
| HSP70 | At3g09440 | x | x | x | 4 | 4 | Lys-74/91/113/163/191/252/284/334/429/430/440 | |
| HSP70 | At5g02500 | x | x | x | 5 | 4 | Lys-56/74/91/163/191/252/334/363/429/430 | |
| BIP-Hsp70 | At5g42020 | x | x | x | 4 | Lys-87/160/172/191/219 | ||
| BIP-HSP70 | At5g28540 | x | x | x | 4 | Lys-87/160/172/191/219 | ||
| mtHSC70-1 | At4g37910 | x | x | x | 4 | Lys-120/174/187/431 | ||
| HSP90 | At5g56030 | x | x | x | 4 | Lys-45/417/449/456/653 | ||
| Histone | At1g07660 | x | x | 4 | Lys-32/92 | |||
| Elongation factorb | At1g07920 | x | x | x | 6 | 5 | Lys-172/179/261/318/392/417 | |
| ATP carrier1 | At3g08580 | x | x | x | 4 | Lys-95/105/116/136/323/374 | ||
| RNA helicase | At3g22330 | x | x | x | 4 | Lys-165/445 | ||
| Aldolasea | At3g52930 | x | x | x | 4 | Lys-24/38/87/103/106/196/285/316/338/357 | ||
| EIF3ea | At3g57290 | x | x | x | 5 | Lys-5 | ||
| Nucleosidediphosphatekinase | At4g09320 | x | x | x | 4 | Lys-106 | ||
| NOP1c | At4g25630 | x | x | x | 4 | Lys-115/125/153/287 | ||
| Transducinb | At5g11240 | x | x | 5 | Lys-7 | |||
| Cobalamin | At5g17920 | x | x | x | 4 | Lys-28/47/166/173/189/259/286/372/400/426 | ||
| RPS2 | At2g41840 | x | x | x | 4 | Lys-206/232 | ||
| RPS4 | At2g17360 | x | x | x | 4 | Lys-80/134 | ||
| RPS5 | At1g56070 | x | x | x | 4 | Lys-628/633/751/830 | ||
| RPP0 | At3g09200 | x | x | x | 4 | Lys-148/151 | ||
| RPP2A | At2g27710 | x | x | x | 5 | Lys-2/44/49 | ||
| RPP2A | At2g27720 | x | x | x | 5 | Lys-24/49 | ||
| RPL4 | At3g09630 | x | x | x | 4 | Lys-148/187/312/365 | ||
| RPL4 | At5g02870 | x | x | x | 4 | Lys-149/188/266/313 | ||
| RPL5b | At5g39740 | x | x | x | 4 | Lys-58 | ||
| RPL9 | At1g33120 | x | x | x | 4 | Lys-128/144 | ||
| RPL11 | At2g42740 | x | x | x | 4 | Lys-37/51 | ||
| RPL15 | At4g16720 | x | x | x | 4 | Lys-83/93 | ||
We observed 6 acetylated chaperones of the Hsp70 and Hsp90 family. It is known that acetylation of Hsp70 and Hsp90 is a mechanism of chaperone deactivation and in humans is required for autophagosome creation.39-41 In addition, we found 3 acetylated proteins that are orthologues to human or yeast RBFs. AT1G07920 is assigned as ortholog to the human translation elongation factor EF-1 α and AT5G11240 as ortholog to the human WDR43, a protein that was classified as RBF.8 Furthermore, AT4G25630 is an ortholog to the yeast RBF Nop1.10,11 Moreover, 12 RPs were found to be acetylated, but this acetylation was exclusively found in the in the nuclear fraction.
Furthermore, we inspected the occurrence of phosphopeptides. However, it needs to be mentioned that neither the purification procedure nor the experimental set up was optimized to protect proteins against dephosphorylation, e.g. by the addition of sodium orthovanadate. It is well known that phosphoproteins are unstable during tissue extraction and protein isolation.42 Therefore, we considered every protein for which at least one phosphopeptide was observed (Table 3). Moreover, we might miss a phosphorylation signal in some cases or fractions, because the phosphoproteins have been degraded during protein isolation. Nevertheless, we detected 30 phosphoproteins. In contrast to acetylation, which was mostly observed for proteins in the nuclear fraction, phosphoproteins were equally abundant in all fractions. Six of the phophoproteins are RPs that have been identified as phosphoproteins before.43 In contrast, the previously reported phosphorylation of RPS6 and RPL1343 has not been detected in here, most likely due to the before mentioned reasons. Moreover, phosphorylation of RPs was exclusively identified in the nuclear fraction.
Table 3.
Protein phosphorylation in the different fractions. Shown are all proteins for one at least one phosphorylated peptide was observed in any of the replicas. Given is the tentative function, the ATG number, the identification (x) in cytoplasm (CYT), nucleus (NUC) or nucleolus (NOC) as well as the number of replicas in which phosphorylation was observed. In case that phosphorylation is observed, the last column gives the number and the modified amino acid in the 3 letter code. A-Orthologue to protein in hNoPDB, B-Orthologue to human RBF, C-Orthologue to yeast RBF, D-Protein without ortholog in yeast or human, esplicing factor. Numbers in bold show detection in 16O and 18O samples.
| Localization |
Phosphorylation |
|||||||
|---|---|---|---|---|---|---|---|---|
| Protein function | ATG | CYT | NUC | NOC | CYT | NUC | NOC | Modified AA |
| ADL6d | At1g10290 | x | x | x | 1 | 2 | 4 | Ser-533/639/837; Thr-532 |
| Histone | At1g09200 | x | 1 | Thr-108 | ||||
| PDR7d | At1g15210 | x | x | 1 | Thr-893 | |||
| atNTT2d | At1g15500 | x | x | x | 1 | 4 | Ser-588/589 | |
| PGM3 | At1g23190 | x | x | x | 1 | Thr-56 | ||
| Thiamin bindinga | At1g24180 | x | x | x | 4 | Thr-369/Tyr-372 | ||
| MOS2 | At1g33520 | x | x | x | 1 | 1 | Tyr-51 | |
| pre-mRNA processing 40Ae | At1g44910 | x | x | x | 2 | Ser-387 | ||
| ADL3d | At1g59610 | x | x | x | 1 | 4 | 3 | Ser-533/844; Thr-532 |
| ABC transporterd | At1g59870 | x | x | x | 1 | Thr-920 | ||
| CCAR1b | At2g03150 | x | 3 | Ser-906 | ||||
| DNA binding proteind | At2g33620 | x | x | 2 | 2 | Ser-313 | ||
| Clathrina | At3g08530 | x | x | x | 1 | 1 | Thr-67; Ser-77 | |
| HD2Ad | At3g44750 | x | x | 1 | 1 | Ser-103 | ||
| Aldolasea | At3g52930 | x | x | x | 1 | Tyr-169 | ||
| Ubiquitin | At4g23040 | x | x | 1 | Ser-362 | |||
| FPR4c | At4g25340 | x | x | x | 2 | 1 | Ser-209 | |
| Chromatin silencingd | At4g31880 | x | x | x | 1 | 2 | 3 | Thr-289 |
| Tubulina | At4g38680 | x | x | 1 | Ser-14 | |||
| atNADP | At5g11670 | x | x | x | 1 | 1 | Thr-139 | |
| TUA3a | At5g19770 | x | x | x | 1 | Ser-379 | ||
| ADL1d | At5g42080 | x | x | 1 | Ser-168 | |||
| DNA Polb | At5g64420 | x | x | x | 1 | Ser-44 | ||
| 14-3-3 kappa | At5g65430 | x | x | x | 1 | 1 | Ser-70/71 | |
| RPP0 | at3g09200 | x | x | x | 1 | Ser-305 | ||
| RPP1 | at1g01100 | x | x | x | 1 | 1 | Ser-102 | |
| RPP1 | at4g00810 | x | x | x | 1 | 1 | Ser-103 | |
| RPP1 | at5g47700 | x | x | x | 2 | Ser-103 | ||
| RPP2A | at2g27710 | x | x | x | 1 | Ser-105 | ||
| RPP2A | at2g27720 | x | x | x | 1 | Ser-105 | ||
Among the identified phosphoproteins in the nucleus and in the cytoplasm are the ATP:ADP antiporter (atNTT2, AT1G15500) and the atNADP-malic enzyme 2 (atNADP; AT5G11670), which are involved in photosynthesis and calvin cycle.44 While our results confirmed the earlier determined phosphorylation site of the atNTT2 (Ser-588/589), for atNADP we identified a phosphorylation site (Thr-139) distinct from the previously described site of phosphorylation at the N-terminus.44 Interestingly, for atNTT2 we found acetylated peptides as well, but only in the nuclear fraction.
Classification of proteins identified in A. thaliana
The human database NoPDB contains a larger number of proteins compared to the A. thaliana nucleolar database.18,19 Thus, we analyzed the orthologous groups of the remaining 2316 identified proteins to the human nucleolar proteins. We detected 361 (co-)orthologues in NoPDB from the 2316 A. thaliana proteins (Fig. 2A; Table S6), 63% of them are found in the nucleolus.
We found 921 proteins (˜55% proteins found in the nucleolus) that are orthologues to yeast or human proteins not present in NoPDB (Fig. 2A; Table S7, S8). This might reflect the distinct structural and functional properties of the plant nucleolus.12,13 79 proteins thereof have a domain characteristic for RNA binding or processing (Table S8), but only for 35% of these the function is experimentally confirmed (Table S8).
“Plant specific” proteins identified in the nucleolus of A. thaliana
For 1034 of the 2316 detected A. thaliana proteins no orthologues to human or yeast proteins could be identified (Fig. 2A). Seventy of these “plant specific” proteins were detected in the nucleolar fraction only (Fig. 3A; Table S9). Twenty-five of these 70 proteins are proposed to be involved in transcription, chromatin remodelling, and histone activity or at least in DNA or RNA binding supported in part by experimental evidence (Fig. 3B). In addition, for 22 of the nucleolar “plant specific” proteins a function could not be proposed. Seventeen of the “plant specific” nucleolar proteins are assigned as metabolic enzymes, signal transduction modules or proteases and 6 are assigned as proteins of the cytoskeleton, membranes or other organelles, which is in agreement with the low contamination rate observed by immunodetection (Fig. 1).
Figure 3.
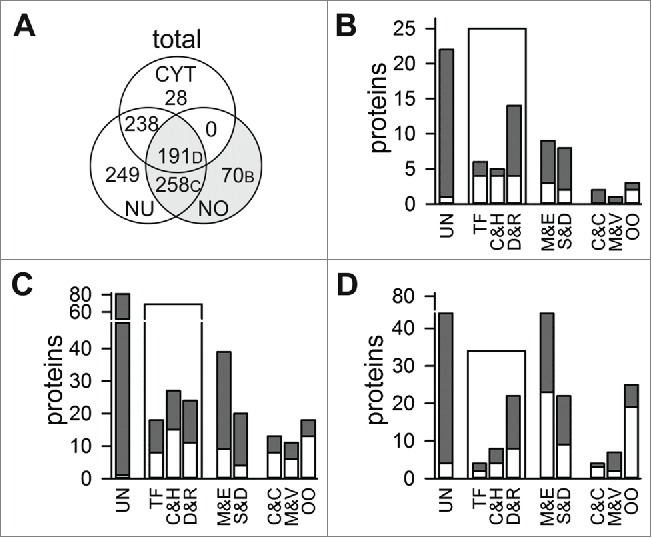
Plant specific nucleolar proteins. (A) Shown is the distribution of proteins without orthologues in yeast and human in total numbers. (B-D) Classification of proteins found exclusively in the nucleolar fraction (B; Table S9), in nucleolar and nuclear fraction (C; Table S10) and in the 3 fractions analyzed (D; Table S11). Total number is given in gray, protein function confirmed by publication is shown in white (UN, unknown; M&E, metabolism and enzymatic function; TF, transcription factor and regulation activity; C&H, chromatin remodeling, histone and histone modification; D&R, DNA/RNA binding or processing; S&D, signal transduction and protein degradation; C&C, definition of intracellular structures and proteins of the cytoskeleton; M&V, membrane proteins and proteins involved in vesicular transport; OO, proteins of other organelles).
In total, 258 “plant specific” proteins are found in both, nucleus and nucleolus (Fig. 3A). 88 of these proteins are of unknown function, while 69 of them are globally categorized as RNA/DNA binding or modifying (Fig. 3C; Table S10). 59 “plant specific” proteins of these 2 compartments are classified as enzymes or signal transduction or protein degradation modules, but for the majority this assignment is not yet supported by experimental evidence (Fig. 3C; gray bar vs. white bar). In addition, 41 of the “plant specific” proteins identified are assigned as proteins of the cytoskeleton or are involved in definition of cellular structures (C&C), as membrane or vesicular transport proteins (M&V), or as proteins of other organelles. However, 7 of 13 C&C proteins are related to microtubule function and 4 in nuclear structure definition, while among the membrane proteins 3 nucleoporins are found. Worth mentioning, most of the proteins of other organelles are membrane proteins of chloroplasts.
Analyzing the 191 “plant specific” proteins found in all 3 fractions (Fig. 3D; Table S11) shows that the majority of the proteins is assigned as enzymes or unknown (M&E: 56; unknown: 44), whereas 35 are DNA and RNA binding and processing proteins. As expected, in the pool of “plant specific” proteins identified in all fractions we observed a drastically larger portion of proteins from other organelles (OO, ˜11%) in comparison to the other 2 analyzed proteome sub-pools.
In total, we identified 148 “plant specific” proteins of unknown function in the nucleolus, irrespective whether the protein was found in another fraction as well (Fig. 3B-D). Moreover, 68 “plant specific” proteins in the nucleolus are proposed to function in RNA/DNA binding or processing, 35 in signal transduction and 68 “plant specific” proteins in the nucleolus are classified as enzymes. None of these proteins has yet been characterized.
Cellular distribution of “plant specific” nucleolar proteins
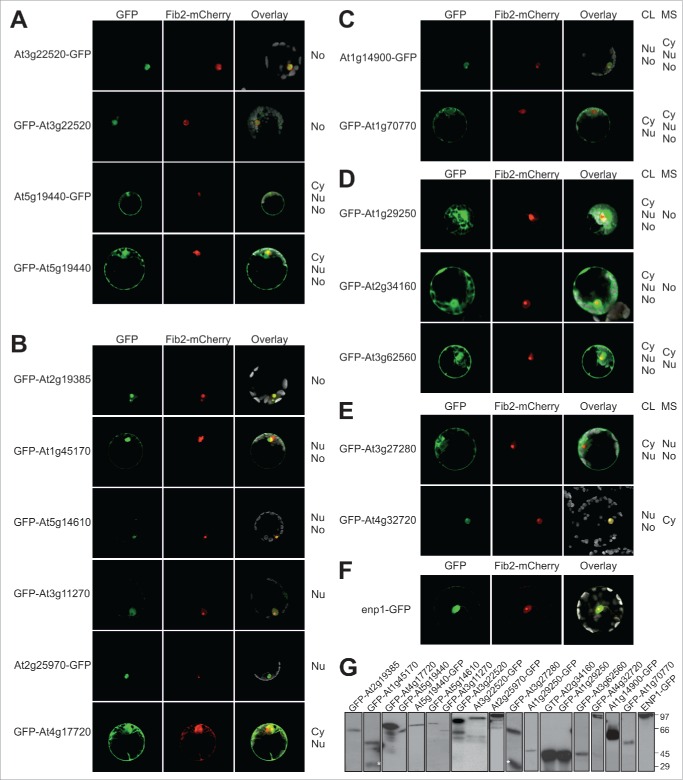
By mass spectrometry we identified “plant specific” proteins. We analyzed the subcellular localization of a randomly selected set of these proteins to confirm the identified localization and to justify the prediction of putative RBFs based on their nucleolar identification. We cloned the CDS of selected proteins in a reporter for expression with N-terminal or C-terminal GFP fusion and analysis in A. thaliana protoplasts (Fig. 4A-E). We confirmed that Fibrillarin2 fused to mCherry can be used to monitor nucleolar localization (Fig. 4F). In addition, based on the AT3G22520 detected in the nucleolus by proteomics and AT5G19440 found in all 3 fractions we demonstrate that N- or C-terminal GFP fusion yielded a comparable result and that the protein distribution as judged from the GFP fluorescence is consistent with the proteomic analysis (Fig. 4A). In total, we analyzed 15 proteins (Fig. 4A-E). For additional 6 proteins we observed the same distribution by fluorescence microscopy as by mass spectrometry (Fig. 4B), namely for AT2G19385 a nucleolar localization (panel 1), for AT1G45170 and AT5G14610 a nuclear/nucleolar localization (panel 2 and 3), for AT3G11270 and AT2G25970 a nuclear localization (panel 4 and 5) and for AT4G17720 in the cytosol and nucleus (panel 6). Note, while expression of GFP-AT1G45170 we observed a small portion of GFP only (Fig. 4G), which most likely accounts for the weak cytosolic signal observed.
Figure 4.
Intracellular distribution of identified proteins in A. thaliana protoplasts. A. thaliana protoplasts were co-transformed with Fibrillarin-mCherry (A-F) and plasmids coding GFP fusions of indicated genes (A-E) or ENP1-GFP coding plasmid (F). The GFP fluorescence (first panel) and mCherry fluorescence (second panel) were merged with the chlorophyll autofluorescence (third panel). Representative images of at least 3 independent transformations are shown. In (A) examples for N- and C-terminal GFP fusions are shown, in (A, B) examples with identical distribution observed by MS-analysis and in vivo analysis are presented, in (C) examples with reduced cellular distribution, in (D) examples with more complex cellular distribution and in (E) examples with a different cellular distribution observed by GFP fusion when compared to MS analysis is shown. (G) A. thaliana protoplasts from A to E were harvested, lysed and proteins subjected to Western Blot analysis with antibodies against GFP. The star marks free GFP, which is particularly dominant for AT3G27280.
For two proteins detected in all fractions by proteomic means, we realized a less complex distribution (Fig. 4C). Expression of AT1G14900-GFP yields GFP fluorescence in the nucleus and nucleolus only, while expression of GFP-AT1G70770 yields GFP fluorescence in the cytosol and in the nucleus but not in the nucleolus (Fig. 4D). For three proteins we observed GFP signal in all fractions, while the proteins were not found in all fractions by proteomic means (AT1G29250, nucleolus; AT2G34160, nucleolus; AT3G62560, cytosol and nucleus). Finally, for 2 proteins we found different localizations, while AT3G27280 was found in the nucleus and nucleolus by proteomic means, we realized a cytosolic and nuclear GFP signal (Fig. 4E). However, we noticed a significant portion of GFP in this case, which might account for the cytosolic GFP signal (Fig. 4G). For AT4G32720 we realized a nuclear / nucleolar distribution by GFP fluorescence analysis, while the protein was only detected in the cytosolic fraction (Fig. 4F).
Thus, for more than 50% of all analyzed proteins we observed the exact same result by proteome analysis of the fractions and by analysis of the protein distribution in cells by expression of GFP fusion proteins. For about 30% we realized an overlap of the 2 different approaches, because for about 10% the existence in all fractions was not confirmed, while for 20% we found the protein in all fractions by analysis of the distribution of the GFP fusion protein. Only for about 10% we observed a distinct result between GFP fluorescence analysis and proteome determination after fractionation. Considering that GFP fluorescence analysis can also be error prone, we conclude that by large the localization determined by MS analysis is confirmed.
Discussion
We identified 1602 proteins in the nucleolar fraction (Fig. 2). 56 of them have been previously deposited in the atNOPdb19 data base (Fig. 2), while 32 proteins have been found in the analysis of nuclear proteome before and after cold stress45 and 20 proteins have been found in a study of the nuclear matrix46 (Table S13). Based on the immunostaining (Fig. 1), the analysis of the distribution of some identified proteins (Fig. 4) and the detection of organellar and membrane proteins (Fig. 3B-D), the 822 proteins identified in the nucleolar as well as nuclear/nucleolar fraction are very likely nucleolar proteins, while the localization of the 780 proteins found in all 3 fractions remains to be confirmed by further experiments. Inspecting the identified proteins, 446 proteins in total and 351 proteins of the nucleolar fraction were assigned as RBFs, RPs, spliceosomal proteins or proteins involved in NMD based on existing reports10,11,33-38 or based on orthology to human RBFs8 (Fig. 2; Table S3). We identified 75% of the assigned spliceosomal proteins, which could either mean that our protein detection rate in the nucleus is about 75% or that not all factors are required for a functional spliceosome in cell cultures. We provide confirmatory evidence for the earlier proposal that the nucleolus acts in NMD14,20 and found 246 co-orthologues to either yeast or human RBFs. Inspecting the other identified proteins, additional RBFs might be present among the about 600 proteins of not yet assigned function.
Inspecting ribosome assembly in A. thaliana one can consider the orthologues to yeast and human RBFs8,10,11 (Fig. 2; Table S3), because existing experimental evidence suggests that many of them are functionally related.e.g.,27-29 However, in the not yet experimentally analyzed proteins identified here RBFs might be present as well, because the investigation of mammalian RBFs revealed that some have an ortholog in yeast but with distinct function or do not have a yeast ortholog at all.8 Consequently, at least the not yet characterized RNA binding proteins, plant specific nucleolar proteins of unknown or proteins with signaling function (kinases etc.), as well as proteins of unknown function orthologous to human proteins of the NoPDB15,18 could be considered as putative RBFs for future experiments (> 380; Table S6, S8-S11).
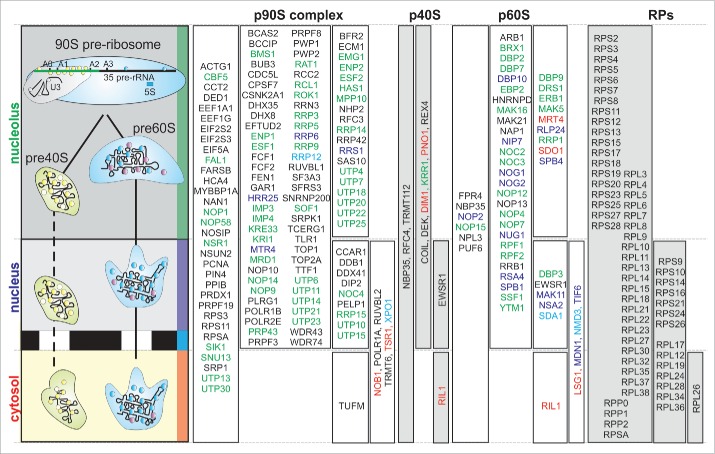
Based on our discovery of proteins in the 3 fractions we can now propose a first model of the timing of ribosome assembly in plants considering the (co-)orthologues to yeast and human RBFs. Almost all (except of 5 RBFs) proteins identified in A. thaliana and classified as (co)orthologues to RBFs could be assigned to different pre-ribosomal complexes based on literature evidence via the ortholog in human or yeast (Fig. 5).8,11 The assigned localization of plant RBFs is by large in good agreement with the observed distribution of the yeast RBFs.2,3,47,48 Interestingly, some RBFs proposed to act in the nucleolus in yeast are only found in the nucleus/cytosol (Fig. 5, green), while the RBFs Dim1, Pno1, Mrt4 and Sdo1 are expected to be in the cytoplasm based on the yeast system, are either found in the nucleolar or in the nucleolar and nuclear fraction (Fig. 5, red). Summerizing, our results suggest that most of the plant RBFs are assembeled with the preribosomal complexes in the nucleolus, even those acting in the cytoplasm. However, the distribution of the RBFs suggests that the overall assembly mechanism is conserved between mammals,8 fungi2 and plants49 (Fig. 5).
Figure 5.
Distribution of RBFs and RPs in A. thaliana. Shown is a scheme of ribosome biogenesis (left) to indicate the compartments. The compartment in which the according orthologues to yeast or human RBF or RP were found (Table S1) was used to extract the localization. Results for co-orthologues were unified and the name of the yeast (first choice) or human ortholog is depicted. The assignment of RBFs from yeast and human to different complexes was extracted from literature (for human: early = p90S, small = p40S and large = p60S).8,11 RBFs are color coded according the localization of the function in yeast (red letter: cytosol, blue: export complex, violet: nuclear, green: nucleolar; black, no assignment).
Similarly, it is widely accepted that many RPs associate very early with the rRNAs during ribosome assembly.e.g.2,50 In yeast, only RPS10, S26, L10, L24, L29, L40, L42, P0, P1 and P2 are discussed to assemble as late as in the cytosol.2,48 Consistently, most of the RPs identified here are found in the nucleolus as well (Fig. 5). L29 and L40 are not identified in here and RPL42 is not assigned in A. thaliana.11,33 In turn, we identified S10, S26 and L24 in the nucleus and L10, P0, P1 and P2 even in the nucleolus (Fig. 5). RPL10, P0 and P2 have been detected in the nucleolus in the previous study.20 Remarkably, P0 and P2 are found to be acetylated in the nuclear fraction (Table 2), and evidence is provided that they are phosphorylated in the nucleolus and the nucleus (Table 3), but both modificatons are not detected in the cytoplasmic fraction. At stage it is discussed that N-terminal acetylation of cytosolic ribosomal proteins observed in eukaryotes controls the translational activity of the ribosome,44,51 but nothing is known about the function of RP acetylation in the nucleus.
Although distinction of the maturation path exist between A. thaliana and S. cerevisiae, many essential processing steps are overlapping.49 Three processing events mark the initiation of the maturation cascade, namely processing in the 5′ ETS of the pre-rRNA, as well as cleavage in ITS1 (A2 or A3) and at B0 in the 3′-ETS to terminate transcription (Fig. 6). The enzymes involved in the 5′-ETS processing are not yet fully discovered, but it is experimentally documented that atXRN2 is involved in this process in A. thaliana.52 Remarkably, we did not identify this enzyme in our proteomic analysis. In yeast, Rnt1 was identified as central processing enzyme for B0,53,54 however, an ortholog to Rnt1 was not identified in the proteome of A. thaliana.10,11 In turn, the A. thaliana ortholog of Rcl1, which is likely involved in A2 cleavage in yeast,53,54 was found in the nuclear and nucleolar fraction. This suggests that A2 cleavage might occur in the nucleolus of plants although A3 cleavage is discussed as the main route to separate for small and large ribosomal subunit maturation in ITS1.49 Subsequently, 18S and 5.8S-25S maturation take independent routes. For 40S maturation only the action of Nob1 is required for D-cleavage in the cytosol, which is confirmed for A. thaliana.29 In line, Nob1 was identified in both, the nucleoplasm and the cytosol.
Figure 6.
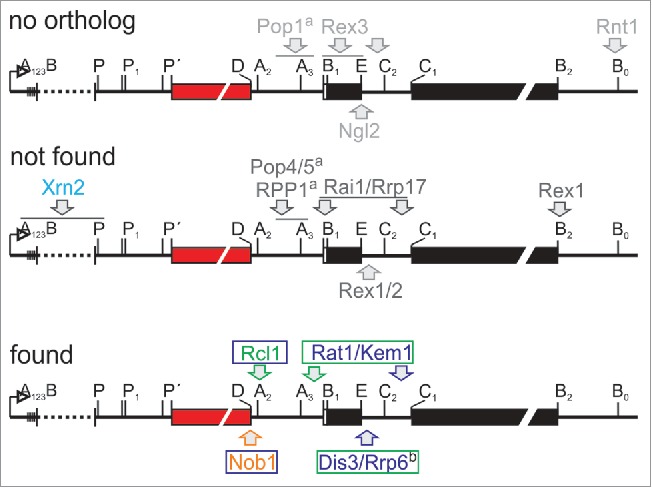
Orthologues to rRNA processing enzymes identified in A. thaliana. Shown is the primary transcript of the rRNA is shown.49 The enzymes assigned to act in rRNA processing in yeast were extracted from literature and assigned to the side of action.53,54 The enzymes for which no ortholog was identified in A. thaliana are shown on top, the enzymes to which orthologues are assigned which have not been found in our study are shown in the middle and the enzymes for which the orthologues have been identified in here are shown on the bottom. The color code on the bottom is: green/blue/orange arrow - cleavage in the nucleolus/nucleus/cytosol; the color of the letters and frames indicate the fraction in which the ortholog was identified according to Fig. 4. Xrn2 is highlighted in cyan as it is involved in 5′ETS maturation in plant but was not found in here. a…MRP RNAse component, b…Exosomal component.
For the maturation of the rRNAs of the large ribosomal subunit several steps are required. For processing of the 3′ ETS the function of Rex1/RNH70 is essential in yeast.53,54 While an ortholog exists in the proteome of A. thaliana, the protein was not identified in here. Processing at the 5′ region of 5.8S requires the action of the RNAse MRP, as well as of Rai1, Rat1, Rrp17 and Xrn1/Kem1 (Fig. 6).53,54 Orthology searches established the existence of orthologues to Pop4, Pop5 and RPP1 of the RNase MRP,10,11 but the proteins were not found in our proteomic analysis. In turn, orthologues of Rai1, Rat1, Rrp17 and Xrn1/Kem1 exist,10,11 but only Rat1 and Xrn1/Kem1 were found in the nucleolar and nuclear fraction. This is consistent with the proposed nucleolar occurrence of this activity.10,11 While the enzyme for cleavage at C2 is not yet identified, it is described that the exosome Rex1-3 and finally, in the cytoplasm, Ngl2 are involved in 3′ maturation of the 5.8S in yeast.53,54 Orthologues to the key enzymes of the exosome, namely Rrp44/Dis3, and Rrp610,11 have been observed in the nucleolar and nuclear fraction, although the action is discussed to take place in the nucleoplasm in yeast.53,54 In turn, while orthologues to Rex1 and Rex2 have been described,10,11 the 2 proteins have not been discovered in our proteomic study. Moreover, an ortholog to Ngl2 is not established.10,11 Maturation of the 5′ region of the 25S rRNA requires the action of Rai1, in addition to the above mentioned Rat1, Rrp17 and Xrn1/Kem1 in the nucleoplasm.53,54 Thus, while 5′ maturation of the rRNA is yet to be discovered in general, the plant enzymes catalyzing the 3′ETS processing, the A3 cleavage and the final processing of the 3′ region of the 5.8S remain to be identified. Whether the enzymes involved in these processes are among the proteins identified in here in the nucleolar fraction remains to be experimentally approached as such function cannot be inferred by bioinformatics strategies. For example, RNT1 is characterizes by a so called Ribonuclease III domain, which is present in 8 proteins in A. thaliana (Dicer like (DCL1-4): AT1G01040, AT3G03300, AT3G43920, AT5G20320; RNAs III like (RTL1-3): AT4G15417, AT3G20420, AT5G45150; unknown function: AT4G37510) not found in our study. For the 4 DCLs a dicer like function is established.55 While RTL2 is globally expressed, RTL1 is exclusively expressed in roots and RTL3 was not found to be expressed under standard growth conditions.56 For RTL2 (AT3G20420) an initial study suggested a function in 3′ ETS processing,55 which was subsequently questioned and a function in double-stranded RNA cleavage was discovered, which might hold true for RTL1 as well.57 Thus, the function of RTL1/2, which are only similar but not orthologous to RNT1, remains to be clarified. Finally, the protein of unknown function (AT4G37510) is only expressed in the mature rosette, but not in pollen or embryonic tissues,58 which questions in function in the essential process of ribosome biogenesis. Therefore, the plant enzymes involved in 3′ETS maturation of the pre-rRNA need to be experimentally discovered.
In summary, the plant RBF/RP assembly path according to the sub compartments of the cell parallels by large the one in yeast and mammals, but some distinctions are observed, particularly with respect to RP assembly. Thus, while we estbalish an initial inventory and assembly mode, plant specific RBFs, the function of post-translational modifications and reason for the distinct RP assembly needs to be investigated in future to understand the ribosome biogenesis pathway in plants.
Materials and methods
Cell culture growth and fractionation
Arabidopsis thaliana cell culture was grown in 1x MS salts, 3% (w/v) sucrose, 1 mg/L 2,4 dichlorophenoxyacetic acid, 4 mg/L nicotinic acid, 4 mg/L pyridoxine hydrochloride, 40 mg/L thiamine hydrochloride, 400 mg/L myo-inositol at 25°C and 150 rpm in the dark and passaged weekly. The cell fractionation method was accomplished as established59 with the following modifications: 40 mL of cell culture was ground in liquid nitrogen, resuspended in HNB buffer (5% Sucrose, 5% Glycerol, 25 mM HEPES (pH 7,5), 25 mM NaCl, 5 mM MgCl2, 1 mM EDTA (pH 8,0), 2 mM CaCl2 and 10 µl/ml Plant Protease Inhibitor Cocktail (PIC, Sigma)) and incubated on ice for 15 minutes. One percent Nonidet-P40 (final concentration) was added to the homogenate and vortexed vigorously. A cushion of 10% sucrose in HNB was laid under the homogenate and centrifuged at 2150 × g (4°C, 10 min). The supernatant contained the cytoplasmic fraction. The pellet (nucleus) was resuspended in NEB500 buffer (5% Glycerol, 25 mM HEPES (pH 7,5), 500 mM NaCl, 1 mM EDTA (pH 8,0), 0,1% ß-mercaptoethanol, 0,2% NP-40, 10 µl/ml PIC). After incubation for 15 min at 4°C and centrifugation at 2150 × g the pellet was resuspended in NEB500 buffer and sonified (Bandelin, Sonopuls, HD70; 30 sec with a cycle of 5 sec on and 5 sec off with an amplitude of 20%). After incubation on ice for 15 min the lysate was centrifuged at 20.800 × g and the supernatant containing the nuclear extract was removed.
Antibody generation and western blot analysis
The primary antibodies Fib (Fibrillarin monoclonal antibody 38F3, Thermo Scientific) and VDAC1 (voltage-dependent anion-selective channel protein 1 polyclonal antibody, Agrisera) were diluted 1:400 and 1:1000, respectively, in 1% w/v non-fat milk powder in 1x PBST. The ateIF4A antibody (Thermo Scientific) was diluted 1:4000. Antibodies against atENP1, atNOB1, atLSG1, atRPL5, atRPL15, atRPS10, atTOC33 and atTOC75-V were described previously.27-29,60,61
The different fractions were directly resuspended in cracking solubilisation buffer (5% (w/v) SDS, 8 M urea, 40 mM Tris–HCl pH 6.8, 0,1 mM EDTA, 0,4 mM mL−1 bromophenol blue, 147 mM β-mercaptoethanol, 1 mM PMSF). After incubation at 60°C for 10 min the protein amount was quantified by amido black protein quantification62 and equal amounts were loaded onto SDS-PAGE, which was used for western blot.63,64 Immunodetection was performed by standard protocols.65
Mass spectrometric analysis
Each sample (2 mg/ml) has been tryptically digested by using either normal water (16O) or 18O-enriched water (Sigma, 329878, 97%) according to well established protocols.31 All samples were digested overnight (37°C) with trypsin (Promega) in a ratio of 1:25. Digestion was stopped with 1% acetic acid and the peptide solution was desalted with C-18 ZipTip (Millipore) following the manufacturer's instructions. Pairwise combination of digested samples has been performed by the following scheme: fractions of 16O-labeled cytoplasm vs. 18O-labeled nucleus, 16O-labeled nucleus vs. 18O-labeled cytoplasm, 16O-labeled cytoplasm vs. 18O-labeled nucleolus, 16O-labeled nucleolus vs. 18O-labeled cytoplasm, 16O-labeled nucleus vs. 18O-labeled nucleolus and 16O-labeled nucleolus vs. 18O-labeled nucleus.
LC-MS/MS measurements have been performed as already described.66 In brief, reverse phase separation of tryptic peptides before mass spectrometric analyses was performed on a Proxeon Easy nano-LC system with a binary buffer system consisting of 0.1% acetic acid, 2% acetonitrile in water (buffer A) and 0.1% acetic acid in 100% acetonitrile (buffer B). Peptide separation was achieved using a linear gradient of buffer B from 5 up to 25% within 35 min. MS data were generated using the Orbitrap Velos MS equipped with a nano-electrospray ion source (PicoTip Emitter, New Objective). After a first survey scan (r = 60,000) MS/MS data were recorded for the 20 highest mass peaks in the linear ion trap at a collision-induced energy of 30%. The exclusion time to reject masses was set to 60 s and the minimal ion signal for MS/MS was 2000.
Proteins were initially identified by searching against an A. thaliana specific database (n = 35386) generated from TAIR1067 using the SEQUEST algorithm v3.5 (Sorcerer v4.04, Sage-N Research Inc.). Search parameters were 10 ppm Parent mass tolerance and 0.6 Da for fragment ion mass tolerance. Carbamidomethylation modification of cysteins (+57 amu), methionine oxidation (+16 amu), C-terminal 18O-modification (+2 amu), phosphorylation (+80 amu) and acetylation (+42 amu) were specified as variable modifications. Peptides were annotated on a false positive rate of 0.59% calculated by the Peptide Prophet algorithm. For protein assembling and grouping Trans-Proteomic Pipeline (v4.4.0, Protein Prophet) was used, whereas only proteins with at least 2 significant peptides were considered for identification and Sequest identifications required at least XCorr scores of greater than 2.5 for double and 3.0 for triple charged ion. Additionally, Scaffold (version Scaffold_4.2.0, Proteome Software Inc.) was used to group pairwise comparison and validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm with Scaffold delta-mass correction.68 Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.69 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Pairwise fractionation measurements were used meaning nucleus-nucleolus, nucleus-cytoplasm and nucleolus-cytoplasm fractions were labeled differently (O16 / O18). For each pair of fractions 6 independent replicates were analyzed, in which the labeling was switched for 3 independent cell culture replicates to exclude a bias by the label.
Identification of acetylation and phosphorylation modification
Proteins were identified as described above with search parameters: 10 ppm parent mass tolerance and 1.0 Da for fragment ion mass tolerance. Peptides were annotated on a false positive rate of 0.57% for acetylation modification and a false positive rate of 0.52% for phosphorylation modification calculated by the Peptide Prophet algorithm. Peptide identifications were accepted if the peptide thresholds were greater than 95.0%. Protein identifications were accepted if the protein thresholds were greater than 95.0% and contained at least 2 identified peptides. The replicas, where a modification either acetylation or phosphorylation was observed, was count and written in the tables see result section. To assure an accurate peptide and phosphorylation site assignment, identifications proofed by using a combination of SEQUEST X-Corr score calculation and fragmentation pattern validation (Scaffold, Proteome Software Inc.). Phosphorylated peptides containing incomplete b- or y-ion fragmentation patterns were manually inspected and validated using Excalibur QUAL BROWSER software (Thermo Scientific). Finally, identified and confirmed phosphorylated peptides were cross-checked by using PhosPhAt 4.0 and SwissProt databases for already described phosphorylation sites (see Table S15; S16).
Peptide identification
Mass spectrometric results were analyzed by ‘Scaffold’ and peptides identified were used for protein identification.70 Information for A. thaliana proteins were extracted from TAIR10.67 ‘Scaffold’ was used to define the fraction (nucleus, nucleolus, cytoplasm) in which each protein was detected based on the pairwise fractionation measurements. We used the following filter criteria: (i) Only proteins with a p-value (protein identification probability) > 95% in each replicate were considered. (ii) Each pairwise comparison including 6 replicates we filtered for fractions showing a signal in at least 3 replicates. (iii) Signals of pairwise fractions were compared to exclude false positives due to missing pairwise fraction data. This overlapping experimental approach ensures a second verification step excluding indistinct signals for one or more fractions.
Assignment of homologs
After assignment of all detected proteins via MS/MS to the different Arabidopsis thaliana fractions, the existence of all proteins identified in the Arabidopsis (AtNoPDB)19 and human nucleolar database (NoPDB)18 or in previous studies45,46 was analyzed. Next, Arabidopsis RBF (co-)orthologues to yeast RBFs7-11 identified based on HaMStR71 and OrthoMCL72; Arabidopsis ribosomal proteins co-)orthologues identified by the same approach or literature search;11,33 Arabidopsis spliceosomal proteins38 or Arabidopsis proteins involved in NMD14 identified in our study were extracted. For the remaining Arabidopsis proteins, orthology to yeast and human proteome was assigned by global analysis using OrthoMCL72 and InParanoid73 as previously established.74 In brief: protein sequences of Arabidopsis, yeast and human proteome were downloaded and used for pairwise ortholog search via InParanoid (Arabidopsis – yeast; Arabidopsis – human) and detection of clusters of orthologues via OrthoMCL. Both approaches were used to build merged clusters of (co-)orthologues from yeast, human and Arabidopsis. Cluster of orthologous sequences containing Arabidopsis protein identifier detected via MS/MS were extracted, Arabidopsis proteins detected via MS/MS showing no (co-)ortholog for human or yeast in one of both approaches were selected as plant specific and used for domain architecture analysis.
Functional domain analysis
For domain architecture analysis the Pfam database (Version 26.0)75 and the PfamScan76 and HMMER77 were used. Protein family scan from Pfam was performed to predict functional domains of proteins. The whole Arabidopsis proteome fasta file was scanned for functional domains. In addition, 336 different Pfam domains involved in RNA binding or processing75,78 were selected to identify proteins with a RNA binding domain as putative RNA binding factors or RBFs. The description of the 336 different Pfam domains is deposited in the Pfam database (http://pfam.sanger.ac.uk/).
Analysis of protein distribution in protoplasts
The sequence of each chosen gene was amplified from A. thaliana cDNA using specific oligonucleotides (Table S14) and cloned in the pRTdS vector system29 to generate N-terminal GFP fusions for expression analysis in protoplasts. A. thaliana protoplast isolation, transformation and visualization by Confocal Laser Scanning Microscopy (CLSM) were carried out like described.61
Supplementary Material
Biographies
Denise Palm: Institute for Molecular Biosciences, Goethe University Frankfurt, Max von Laue Str. 9, 60438 Frankfurt, Germany; TEL: 0049-69-79829294; E-MAIL: palm@em.uni-frankfurt.de
Dr. Stefan Simm: Institute for Molecular Biosciences and Cluster of Excellence Macromolecular Complexes. Goethe University Frankfurt, Max von Laue Str. 9, 60438 Frankfurt, Germany; TEL: 0049-69-79829289; E-MAIL: Simm@bio.uni-frankfurt.de
Katrin Darm: Department of Otorhinolaryngology, Head and Neck Surgery, University Medicine Greifswald, Ferdinand-Sauerbruch-Straße DZ7 J.05.06, 17475 Greifswald, Germany; TEL: 0049-3834-866206; E-MAIL: kdarm@uni-greifswald.de
Dr. Benjamin L. Weis: Institute for Molecular Biosciences, Goethe University Frankfurt, Max von Laue Str. 9, 60438 Frankfurt, Germany; current address: Hochschule Biberach, Institut of Applied Biotechnology, Karlstraße 11, 88400 Biberach; TEL: 0049-7351-582390; E-MAIL: BenjaminWeis@gmx.de
Maike Ruprecht: Institute for Molecular Biosciences, Goethe University Frankfurt, Max von Laue Str. 9, 60438 Frankfurt, Germany; TEL: 0049-69-79829293; E-MAIL: m.ruprecht@bio.uni-frankfurt.de
Prof. Dr. Enrico Schleiff: Institute for Molecular Biosciences, Cluster of Excellence Macromolecular Complexes and Buchman Institute for Molecular Life Sciences, Goethe University Frankfurt, Max von Laue Str. 9, 60438 Frankfurt, Germany; TEL: 0049-69-79829287; E-MAIL: schleiff@bio.uni-frankfurt.de
Dr. Christian Scharf: Department of Otorhinolaryngology, Head and Neck Surgery and Interfaculty Institute of Genetics and Functional Genomics, University Medicine Greifswald, Ferdinand-Sauerbruch-Straße DZ7 J.05.06, 17475 Greifswald, Germany; TEL: 0049-3834-866218; E-MAIL: scharf@uni-greifswald.de
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
ES & CS designed research; D.P. & B.L.W. performed fractionation; D.P., K.D. & C.S. performed mass spectrometric analysis, S.S. & C.S. performed bioinformatic analysis; D.P., C.S. & E.S. analyzed data; C.S. & E.S wrote manuscript; all were involved in final editing and all approved the manuscript.
Acknowledgments
The A. thaliana cell culture was a kind gift from A. Batschauer (Marburg). This work was supported by the Deutsche Forschungsgemeinschaft in the frame of the SFB902/B9 and through the Cluster of Excellence: Macromolecular complexes (ES).
References
- 1.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; http://dx.doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 2.Woolford JL, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013; 195:643-81; PMID:24190922; http://dx.doi.org/ 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turowski TW. Tollervey D Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA 2015; 6:129-39; PMID:25176256; http://dx.doi.org/ 10.1002/wrna.1263 [DOI] [PubMed] [Google Scholar]
- 4.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA 2015; 6:191-209; PMID:25327757; http://dx.doi.org/ 10.1002/wrna.1267 [DOI] [PubMed] [Google Scholar]
- 5.Henras AK, Plisson-Chastang C, O'Donohue M-F, Chakraborty A, Gleizes P-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 2015; 6:225-42; PMID:25346433; http://dx.doi.org/ 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciganda M, Williams N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2012; 2:523-33; PMID:NOT_FOUND; http://dx.doi.org/ 10.1002/wrna.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 2010; 8:e1000522; PMID:21048991; http://dx.doi.org/ 10.1371/journal.pbio.1000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tafforeau L, Zorbas C, Langhendries J-L, Mullineux S-T, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DLJ. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 2013; 51:539-51; PMID:23973377; http://dx.doi.org/ 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 9.Xu R, Zhang S, Huang J, Zheng C. Genome-wide comparative in silico analysis of the RNA helicase gene family in Zea mays and Glycine max: a comparison with Arabidopsis and Oryza sativa. PLoS One 2013; 8:e78982; PMID:24265739; http://dx.doi.org/ 10.1371/journal.pone.0078982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, Bohnsack MT, Schleiff E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res 2014; 42:1509-23; PMID:24234440; http://dx.doi.org/ 10.1093/nar/gkt1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simm S, Fragkostefanakis S, Paul P, Keller M, Einloft J, Scharf KD, Schleiff E. Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum. Bioinform Biol Insights 2015; 9:1-17; PMID:25698879; http://dx.doi.org/10.4137/BBI.S20751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JWS, Shaw PJ. Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 1998; 10:649-57; PMID:9596627; http://dx.doi.org/ 10.1105/tpc.10.5.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stępiński D. Functional ultrastructure of the plant nucleolus. Protoplasma 2014; 251:1285-306; PMID:24756369; http://dx.doi.org/ 10.1007/s00709-014-0648-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw P, Brown J. Nucleoli: composition, function, and dynamics. Plant Physiol 2012; 158:44-51; PMID:22082506; http://dx.doi.org/ 10.1104/pp.111.188052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad Y, Boisvert F-M, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res 2009; 37:181-4; PMID:NOT_FOUND; http://dx.doi.org/ 10.1093/nar/gkn804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature 2005; 433:77-83; PMID:15635413; http://dx.doi.org/ 10.1038/nature03207 [DOI] [PubMed] [Google Scholar]
- 17.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 2007; 17:749-60; PMID:17446074; http://dx.doi.org/ 10.1016/j.cub.2007.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. NOPdb: Nucleolar Proteome Database. Nucleic Acids Res 2006; 34:218-20; PMID:NOT_FOUND; http://dx.doi.org/ 10.1093/nar/gkj004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown JW, Shaw PJ, Shaw P, Marshall DF. Arabidopsis nucleolar protein database (AtNoPDB). Nucleic Acids Res 2005; 33:633-6; PMID:15608277; http://dx.doi.org/ 10.1093/nar/gki052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 2005; 16:260-9; PMID:15496452; http://dx.doi.org/ 10.1091/mbc.E04-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 2006; 18:3218-34; PMID:17114353; http://dx.doi.org/ 10.1105/tpc.106.044529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan A, Garbelli A, Grossi S, Florentin A, Batelli G, Acuna T, Zolla G, Kaye Y, Paul LK, Zhu JK. et al.. The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J 2014; 79:28-43; PMID:24724701; http://dx.doi.org/ 10.1111/tpj.12533 [DOI] [PubMed] [Google Scholar]
- 23.Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JW, Shaw PJ. Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 2009; 21:1592-606; PMID:19435936; http://dx.doi.org/ 10.1105/tpc.108.060434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 2009; 21:2045-57; PMID:19602621; http://dx.doi.org/ 10.1105/tpc.109.067736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleiff E, Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol 2011; 12:48-59; PMID:21139638; http://dx.doi.org/ 10.1038/nrm3027 [DOI] [PubMed] [Google Scholar]
- 26.Ulrich T, Gross LE, Sommer MS, Schleiff E, Rapaport D. Chloroplast β-barrel proteins are assembled into the mitochondrial outer membrane in a process that depends on the TOM and TOB complexes. J Biol Chem 2012; 287:27467-79; PMID:22745120; http://dx.doi.org/ 10.1074/jbc.M112.382093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E. The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J 2014; 80:1043-56; PMID:25319368; http://dx.doi.org/ 10.1111/tpj.12703 [DOI] [PubMed] [Google Scholar]
- 28.Weis BL, Palm D, Missbach S, Bohnsack MT, Schleiff E. atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 2015; 21:415-25; PMID:25605960; http://dx.doi.org/ 10.1261/rna.047563.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missbach S, Weis BL, Martin R, Simm S, Bohnsack MT, Schleiff E. 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One 2013; 8:e54084; PMID:23382868; http://dx.doi.org/ 10.1371/journal.pone.0054084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barneche F, Steinmetz F, Echeverría M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem 2000; 275:27212-20; PMID:10829025; http://dx.doi.org/10.1074/jbc.M002996200 [DOI] [PubMed] [Google Scholar]
- 31.Stewart II, Thomson T, Figeys D. 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom 2001; 15:2456-65; PMID:11746917; http://dx.doi.org/ 10.1002/rcm.525 [DOI] [PubMed] [Google Scholar]
- 32.Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol 2013; 23:242-50; PMID:23375955; http://dx.doi.org/ 10.1016/j.tcb.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll AJ. The Arabidopsis cytosolic ribosomal proteome: From form to function. Front Plant Sci 2013; 4:32; PMID:23459595; http://dx.doi.org/10.3389/fpls.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 2006; 47:480-9; PMID:16813578; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02802.x [DOI] [PubMed] [Google Scholar]
- 35.Kerényi Z, Mérai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 2008; 27:1585-95; PMID:18451801; http://dx.doi.org/ 10.1038/emboj.2008.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K. Arabidopsis SMG7 protein is required for exit from meiosis. J Cell Sci 2008; 121:2208-16; PMID:18544632; http://dx.doi.org/ 10.1242/jcs.027862 [DOI] [PubMed] [Google Scholar]
- 37.Nyikó T, Kerényi F, Szabadkai L, Benkovics AH, Major P, Sonkoly B, Mérai Z, Barta E, Niemiec E, Kufel J. et al.. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res 2013; 41:6715-28; PMID:23666629; http://dx.doi.org/ 10.1093/nar/gkt366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013; 25:3657-83; PMID:24179125; http://dx.doi.org/ 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 2005; 18:601-607; PMID:15916966; http://dx.doi.org/ 10.1016/j.molcel.2005.04.021 [DOI] [PubMed] [Google Scholar]
- 40.Rao R, Fiskus W, Ganguly S, Kambhampati S, Bhalla KN. HDAC inhibitors and chaperone function. Adv Cancer Res 2012; 116:239-62; PMID:23088873; http://dx.doi.org/ 10.1016/B978-0-12-394387-3.00007-0 [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci U S A 2013; 110:6841-6; PMID:23569248; http://dx.doi.org/ 10.1073/pnas.1217692110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, Banks S, Deng J, Van Meter AJ, Geho DH, et al.. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol. Cell. Proteom 2008; 7:1998-2018; PMID:18667411; http://dx.doi.org/10.1074/mcp.M700596-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 2008; 7:347-69; PMID:17934214; http://dx.doi.org/ 10.1074/mcp.M700052-MCP200 [DOI] [PubMed] [Google Scholar]
- 44.Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Bagisnky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physol 2009; 150:889-903; PMID:19376835; http://dx.doi.org/ 10.1104/pp.109.138677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae MS, Cho EJ, Choi EY, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003; 36:652-63; PMID:14617066; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01907.x [DOI] [PubMed] [Google Scholar]
- 46.Calikowski TT, Meulia T, Meier I. A proteomic study of the arabidopsis nuclear matrix. J Cell Biochem. 2003; 90:361-78; PMID:14505352; http://dx.doi.org/ 10.1002/jcb.10624 [DOI] [PubMed] [Google Scholar]
- 47.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol 2004; 7:631-7; PMID:15556036; http://dx.doi.org/ 10.1016/j.mib.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 48.Karbstein K. Inside the 40S ribosome assembly machinery. Curr Opin Chem Biol 2011; 15:657-63; PMID:21862385; http://dx.doi.org/ 10.1016/j.cbpa.2011.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weis BL, Kovacevic J, Missbach S, Schleiff E. Plant-Specific Features of Ribosome Biogenesis. Trends Plant Sci. 2015; 20:729-40; PMID:26459664; http://dx.doi.org/ 10.1016/j.tplants.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 50.Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL Jr. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 2014; 28:198-210; PMID:24449272; http://dx.doi.org/ 10.1101/gad.228825.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamita M, Kimura Y, Ino Y, Kamp RM, Polevoda B, Sherman F, Hirano H. N(α)-Acetylation of yeast ribosomal proteins and its effect on protein synthesis. J Proteom 2011; 74:431-41; PMID:21184851; http://dx.doi.org/ 10.1016/j.jprot.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 52.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010; 38:4487-502; PMID:20338880; http://dx.doi.org/ 10.1093/nar/gkq172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA. 2015; 6:129-39; PMID:25176256; http://dx.doi.org/ 10.1002/wrna.1263 [DOI] [PubMed] [Google Scholar]
- 54.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA. 2015; 6:191-209; PMID:25327757; http://dx.doi.org/ 10.1002/wrna.1267 [DOI] [PubMed] [Google Scholar]
- 55.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014; 65:473-503; PMID:24579988; http://dx.doi.org/ 10.1146/annurev-arplant-050213-035728 [DOI] [PubMed] [Google Scholar]
- 56.Comella P, Pontvianne F, Lahmy S, Vignols F, Barbezier N, Debures A, Jobet E, Brugidou E, Echeverria M, Sáez-Vásquez J. Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3'ETS in Arabidopsis. Nucleic Acids Res. 2008; 36:1163-75; PMID:18158302; http://dx.doi.org/ 10.1093/nar/gkm1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiyota E, Okada R, Kondo N, Hiraguri A, Moriyama H, Fukuhara T. An Arabidopsis RNase III-like protein, AtRTL2, cleaves double-stranded RNA in vitro. J Plant Res. 2011; 124:405-414; PMID:20978817; http://dx.doi.org/ 10.1007/s10265-010-0382-x [DOI] [PubMed] [Google Scholar]
- 58.Patel RV, Nahal HK, Breit R, Provart NJ. BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J. 2012. 71:1038-50; PMID:22607031; http://dx.doi.org/ 10.1111/j.1365-313X.2012.05055.x [DOI] [PubMed] [Google Scholar]
- 59.Scharf K-D, Rose S, Zott W, Schöff F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J 1990; 12:4495-501; PMID:2148291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sommer M, Rudolf M, Tillmann B, Tripp J, Sommer MS, Schleiff E. Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ 2013; 36:970-83; PMID:23131143; http://dx.doi.org/ 10.1111/pce.12030 [DOI] [PubMed] [Google Scholar]
- 61.Sommer MS, Daum B, Gross LE, Weis BL, Mirus O, Abram L, Maier UG, Kühlbrandt W, Schleiff E. Chloroplast Omp85 proteins change orientation during evolution. Proc Natl Acad Sci USA 2011; 108:13841-6; PMID:21825140; http://dx.doi.org/ 10.1073/pnas.1108626108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popov N., Schmitt M., Schulzeck S., Matthies H. Reliable micromethod for determination of the protein content in tissue homogenates. Acta Biol Med Ger 1975; 34:1441-6; PMID:1221733 [PubMed] [Google Scholar]
- 63.Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 1970; 227:680-5; PMID:5432063; http://dx.doi.org/ 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 64.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979; 76:4350-4; PMID:388439; http://dx.doi.org/ 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 66.Menck K, Scharf C, Bleckmann A, Dyck L, Rost U, Wenzel D, Dhople VM, Siam L, Pukrop T, Binder C, et al.. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J Mol Cell Biol 2014; 7:143-53; PMID:25503107; http://dx.doi.org/ 10.1093/jmcb/mju047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al.. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 2012; 40:1202-10; PMID:22140109; http://dx.doi.org/ 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 2002; 74:5383-92; PMID:12403597; http://dx.doi.org/ 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 69.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003; 75:4646-58; PMID:14632076; http://dx.doi.org/ 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 70.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010; 10:1265-9; PMID:20077414; http://dx.doi.org/ 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- 71.Ebersberger I, Strauss S, von Haeseler A. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 2009; 9:157; PMID:19586527; http://dx.doi.org/ 10.1186/1471-2148-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006; 34:D363-8; PMID:16381887; http://dx.doi.org/ 10.1093/nar/gkj123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sonnhammer EL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234-9; PMID:25429972; http://dx.doi.org/ 10.1093/nar/gku1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paul P, Simm S, Blaumeiser A, Scharf KD, Fragkostefanakis S, Mirus O, Schleiff E. The protein translocation systems in plants - composition and variability on the example of Solanum lycopersicum. BMC Genomics 2013; 14:189; PMID:23506162; http://dx.doi.org/ 10.1186/1471-2164-14-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res 2014; 42:222-30; PMID:24288371; http://dx.doi.org/17688688 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007; 8:298; PMID:17688688; http://dx.doi.org/ 10.1186/1471-2105-8-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29-W37; PMID:21593126; http://dx.doi.org/ 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simm S, Keller M, Selymesi M, Schleiff E. The composition of the global and feature specific cyanobacterial core-genomes. Front Microbiol 2015; 6:219; PMID:25852675; http://dx.doi.org/ 10.3389/fmicb.2015.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.