Abstract
Background:
Methicillin resistant Staphylococcus aureus (MRSA) is responsible for a wide spectrum of nosocomial and community associated infections worldwide. The aim of this study was to analyze MRSA strains from the general population in Canton Sarajevo, B&H.
Methods:
Our investigation including either phenotypic and genotypic markers such as antimicrobial resistance, pulsed-field gel electrophoresis (PFGE), SCC typing, and Panton-Valentine leukocidin (PVL) detection.
Results:
Antimicrobial susceptibility: all MRSA isolates were resistant to the β-lactam antibiotics tested, and all isolates were susceptible trimethoprim sulphamethoxazole, rifampicin, fusidic acid, linezolid and vancomycin. Sixty-eight per cent of the MRSA isolates were resistant to erythromycin, 5% to clindamycin, 5% to gentamicin and 4% to ciprofloxacin. After the PFGE analysis, the isolates were grouped into five similarity groups: A-E. The largest number of isolates belonged to one of two groups: C: 60 (60%) and D: 27 (27%). In both groups C and D, SCCmec type IV was predominant (60% and 88, 8%, respectively). A total of 24% of the isolates had positive expression of PVL genes, while 76% showed a statistically significantly greater negative expression of PVL genes.
Conclusion:
SCCmec type IV, together with the susceptibility profile and PFGE grouping, is considered to be typical of CA-MRSA
Keywords: infection, CA-MRSA, markers, antimicrobial susceptibility
1. INTRODUCTION
Methicillin resistant Staphylococcus aureus (MRSA) is one of the most important hospital pathogens, and since the end of the last century has become responsible for community infection in patients without previous health-care contact. It is mainly responsible for a wide spectrum of nosocomial and community associated infections worldwide and cause endemic and epidemic infections in many parts of the world (1). Methicillin resistance in S.aureus emerged via the acquisition of the mecA gene located on mobile genomic island designated staphylococcal chromosome cassette mec (SCCmec) by methicillin-susceptible S.aureus (2).
The mecA gene is responsible for the synthesis of a novel penicillin-binding protein known as penicillin-binding protein 2a, which has decreased binding affinity for penicillin and cephalosporins. Therefore, MRSA strains are resistant to all ß-lactam antibiotics. MRSA was initially susceptible to non-beta lactam antibiotics. However, from the late 1979s onwards, new strains of MRSA appeared that were resistant to multi-non- ß-lactam agents including aminoglycosides, with only vancomycin left as an antibiotic of last resort for treating MRSA infections. These strains, described as epidemic MRSA (EMRSA or HA-MRSA), also possessed the capacity to spread extensively and caused serious infections mostly among hospitalized patients worldwide (3). In the 1990s, a new type of MRSA causing infections in the community among healthy and younger people who had no history of hospital admission or medical treatment in the previous year was reported in Western Australia (4).
These types of MRSA strains were described as community-acquired, community-originated, community-associated, or community-onset MRSA (CA-MRSA). HA-MRSA and CA-MRSA belong to different genetic lineages. CA-MRSA strains are usually sensitive to antibiotics other than beta-lactams and contain staphylococcal cassette chromosome SCCmec type IV, V or VII, while HA-MRSA are generally multidrug-resistant and harbor larger SCCmec type I, II or III (5).
The pathogenicity of MRSA is related to an exhaustive arsenal of virulence factors and toxins. The most common and probably important is the Panton-Valentine leukocidin (PVL) toxin which is lethal to neutrophils and is associated with skin and soft tissue infections as well as severe necrotizing pneumonia. Numerous CA-MRSA clones have emerged on every continent (6). Importantly, these CA-MRSA strains that initially were associated with community-onset (CO) infections, have begun to enter into hospitals and may be replacing the conventional HA-MRSA strains with significant clinical and public health implications (7).
However, CA-MRSA penetration has not been thoroughly explored among a large number of hospitals and knowledge of the risk factors involved in nosocomial transmission of CA-MRSA compared with HA-MRSA remains largely undefined (8). The prevalence of in vitro resistance to non-β-lactam antimicrobial agents may be increasing among MRSA strains associated with community transmission.
MRSA typing is an essential component of an effective surveillance system to describe epidemiological trends and infection control strategies. Current challenges for MRSA typing are focused on selecting the most appropriate technique in terms of efficiency, reliability, ease of performance and cost involved. The phenotypic methods in general are easier to perform, easier to interpret, cost effective and are widely available, however less discriminatory. The genotypic methods are expensive and technically demanding, however more discriminatory. Newer technologies involving sequencing of various genes are coming up as broadly applicable and high throughput typing systems. Still there is no consensus regarding the single best method for typing of MRSA strains (9). Pulsed-field gel electrophoresis (PFGE) has been the ‘gold standard’ genotyping method for MRSA for over a decade, and it has been used widely for local outbreak investigation, long-term surveillance of MRSA infections at regional and national levels and for international comparisons (10). In Europe, harmonization efforts have been made to standardize the PFGE typing protocol of MRSA and enable multicentric comparison of PFGE data. The macro restriction analysis of chromosomal DNA using PFGE is a reference method for Staphylococcus aureus typing and can be combined with other methods (10). Our aim was to use PFGE to determine the number and distribution of clones in MRSA isolates from the general population in Canton Sarajevo, Bosnia and Herzegovina. Also, our investigation including both phenotypic and genotypic markers such as antimicrobial resistance, Scene type, and PVL genes.
2. MATERIALS AND METHODS
Out of 2279 Staphylococcus aureus strains, 653 were methicillin-resistant Staphylococcus aureus. A total of 653 methicillin-resistant Staphylococcus aureus isolates were collected out of 4,341 patient samples admitted to the Laboratory of Institute of Public Health of Sarajevo Canton, Bosnia and Herzegovina (B&H) from January 2015 to August 2015.
The samples included nose swabs, throat, ear, eye, umbilicus swabs, wound and skin swabs. All collected MRSA isolates were identified using standard microbiological methods based on the demonstration of deoxyribonuclease, bound coagulase (rabbit plasma, bioMerieux, France) and free coagulase (Slidex Staph Plus, bioMerieux, France) (11). Antibiotic susceptibility was determined using the agar disk diffusion method according to the guidelines of the Clinical Laboratory Standards Institute (12, 13). We tested the susceptibility of the S. aureus isolates to 12 following antibiotics: oxacillin, cefoxitin, erythromycin, clindamycin, linezolid, ciprofloxacin, sulfamethoxazole-trimethoprim, tetracycline, fusidic acid, rifampicin, vancomycin and gentamicin. All MRSA strains were tested for the presence of the mecA gene by multiplex PCR. Molecular analysis of the SCCmec cassette was performed using a method previously described by Oliveira et al. (14), with certain modification described by Budimir et al. (15). Presence of the gene for PVL was detected using PCR primers previously described by Lina et al. (16).
All of isolates were analyzed for epidemiological relatedness by pulsed-field gel electrophoresis (PFGE). Macrorestriction of chromosomal DNA was analyzed using the restriction enzyme SmaI, according to the procedure described previously. DNA fragments were separated using a CHEF-DR III electrophoresis system (Bio-Rad laboratories, Hercules, California, USA) at 6.0 V/cm for 2O h, with pulse times ramped from 5 s to 40 s. Gels were stained with ethidium bromide and photographed under UV illumination. PFGE patterns were analyzed using GelCompare software (Applied Maths, Ghent, Belgium) according to the scheme of Tenover et al. (17). Isolates with indistinguishable band patterns were assigned to the same PFGE pattern type. Isolates that differed by ≤ three fragments were considered to be subtypes of a given clonal group (closely related). The dendrogram was produced using 0, 5% optimization, a band tolerance of 3% and the unweighted pair group method with arithmetic averages based on Dice coefficients.
3. RESULTS
A total of 653 MRSA isolates were collected from various samples of outpatients in the microbiological laboratory of Institute of Public Health of the Canton Sarajevo, B&H between January and August 2015. We selected a representative sample of 100 strains, eliminating the “copy” strains (strains from the same patient at different places and those that are repeated). Among one hundred MRSA isolates 49 (49%) were from male and 51 (51%) female. There was no significant sex difference in MRSA distribution. The mean age of examinees was 10.9±18.7 years. Of the total number of isolates largest number of isolates included in our study was isolated from nasal swabs or nasopharyngeal (41%), 35.0% isolates were from skin swabs, 7% from pustule, 5% from wound infections, 3% from throat swabs, 3% from ear swabs and 6% from various other body sites.
Antimicrobial Susceptibility Test
All MRSA isolates were resistant to the β-lactam antibiotics tested, i.e. penicillin, oxacillin, cefoxitin, and all isolates were susceptible trimethoprim sulphamethoxazole, rifampicin, fusidic acid, linezolid and vancomycin. Sixty-eight per cent of the MRSA isolates were resistant to erythromycin, 5% to clindamycin, 5% to gentamicin and 4% to ciprofloxacin. All isolates were identified as MRSA strains by the determination of methicillin resistance using oxacillin and cefoxitin disk diffusion method. The minimum inhibitory concentration (MIC) of oxacillin was between 4 µg/ml and >256 µg/ml (AB BioMérieux Swede). Staphylococcus aureus ATCC 25923 served for quality control in both methods. The presence of the mecA gene was tested by PCR as described elsewhere (Anonymous, 2008). Primers used for confirming the presence of the mecA gene were MecA-1 and MecA-2. All 100 MRSA isolates of contained the gene mecA.
PFGE analysis
One of the 100 isolates could not be typed using PFGE, either owing to difficulties with lysis of the bacteria or the digestion of the DNA. A total of 5 similarity groups (A to E) were found and two of these, C and D, were classified as major similarity groups (Figure 1). Similarity group C harbored 60 of the 100 isolates, whereas similarity group D harbored 27 of the 100 isolates. In both groups C and D, SCCmec type IV was predominant (i.e. 60% and 88, 8%, respectively). The group A comprised three isolates harboring SCCmec IV. Group B included the four isolates, where three isolates were SCCmec type I, while one was SCCmec type IV. Group E included 3 isolates, SCCmec type I, IV and V. Three isolates were singletons, i.e., not similar to any of the other strains. A total of 24% of the isolates had positive expression of PVL genes, while 76% showed a statistically significantly greater negative expression of PVL genes. Of the 24 PVL positive isolates, 16 were recovered from samples skin changes swabs, while 8 were from nasal swabs. The most prevalent SCCmec element was type IV (86, 0%), followed by SCCmec I (4%) and SCCmec type V (1%). Non-typeable strains were also found in 9% of cases. No isolates with SCCmec III was found during this analysis.
Figure 1.
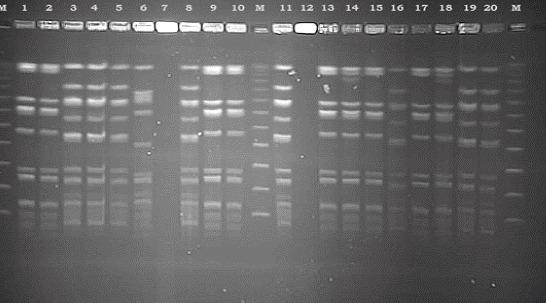
Representative PFGE patterns of MRSA strains isolated from population included in the study, after splitting enzyme SmaI and separation in an electric field and staining with ethidium bromide.
Figure 2.
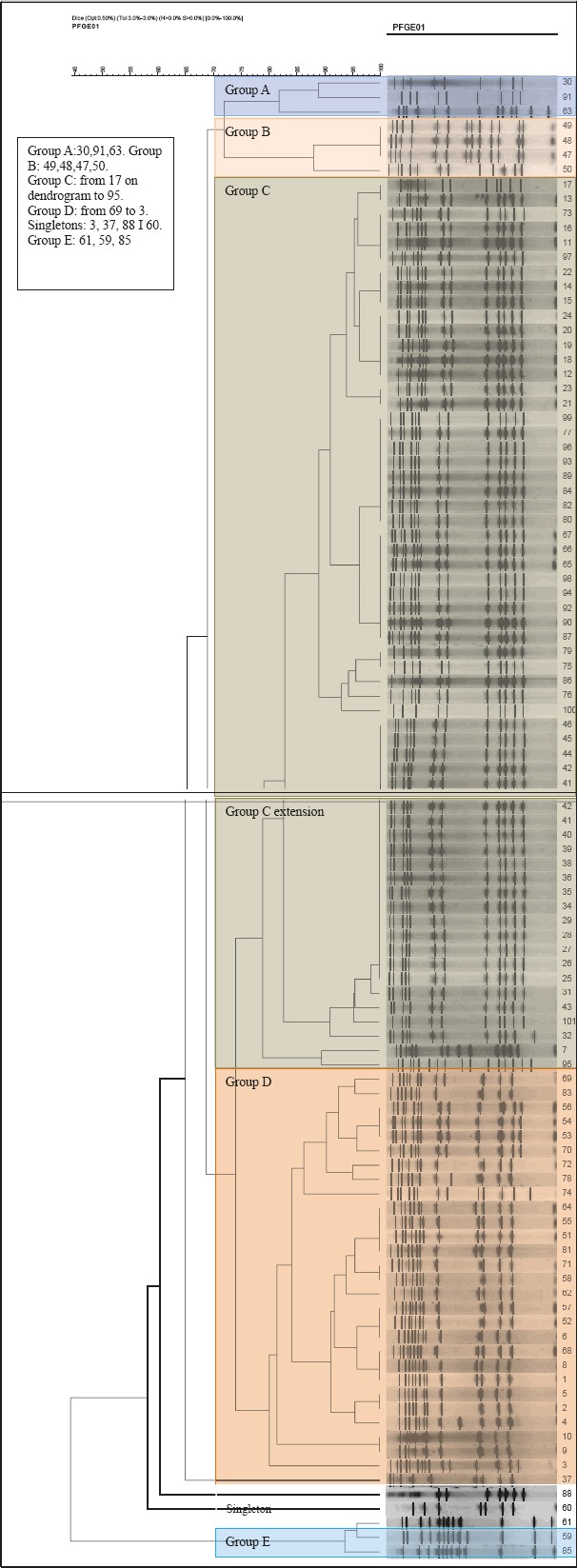
PFGE dendrogram of methicillin-resistant Staphylococcus aureus isolates included in the study
Figure 3.
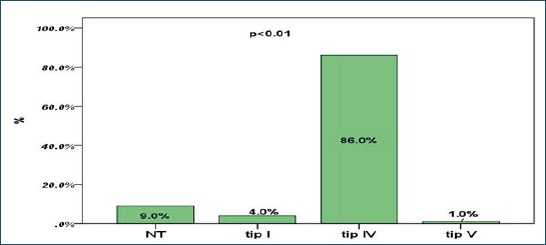
SCCmec types distribution among MRSA isolates Note: NT- non-typeable.
4. DISCUSSION
Methicillin-resistant Staphylococcus aureus is an increasing problem throughout the world, both in hospitals and in the community. Infections due to MRSA are becoming a major health issue in South East Europe, yet questions on the epidemiology and infection remain unanswered. This is the first study on the geographical area focused on PFGE analysis of S. aureus samples obtained from outpatients setting. Comparison grouping by PFGE and SCCmec typing was used to determine whether our strains belonged to HA or CA MRSA strains. The majority of isolates in PFGE groups C and D had SCCmec type IV, and fewer had SCCmec types I and V. In groups C and D prevail children in the first year of life, suggesting a very complex epidemiological situation and the region hospitals. The data presented here provide an important baseline for future surveillance of MRSA in our country. The group A comprised three isolates harboring SCCmec IV. One of these strains showed multiresistant. It was resistant to penicillin, oxacillin, cefoxitin, erythromycin, clindamycin, ciprofloxacin and gentamicin. From the epidemiological data to see that this isolate originated from an elderly patient who had been in contact with medical facilities.
Based on these data and in accordance with the definition of CA-MRSA by the CDC, we can assume that in this case a HA-MRSA. Group B included the four isolates, where three isolates were SCCmec type I, while one was SCCmec type IV. Strains with SCCmec type I originated from older patients who have had surgical interventions in the past six months. And these MRSA isolates showed multiresistant and did not have the PVL gene. And based on all the data we can conclude that this is a HA-MRSA, also. Group E included 3 isolates, SCCmec type I, IV and V. The epidemiological and molecular features of MRSA are evolving such that characteristics that initially distinguished CA-MRSA from HA-MRSA appear to be changing. For example, several reports have described transmission in healthcare settings of MRSA strains indistinguishable from those associated with community transmission. Eventually, these strains could become the predominant strains in both community and healthcare settings. In addition, the prevalence of in vitro resistance to non-betalactam antimicrobial agents may be increasing among MRSA strains associated with community transmission.
According to our results, MRSA represents a significant causative agent of infections and patient colonization in outpatient setting. A particularly important finding is the fact that most of our strains isolated from skin swabs obtained from the pediatric population (mean age 10.9 overall). Indeed, pediatrics patients are highly susceptible to MRSA colonization. CA-MRSA strains have been reported as a cause of colonization and infection in pediatric intensive units in many countries. It is interesting to note that newborns and young children, due to their immature immune systems, are easily infected by MRSA. Several similar studies reported that the majority of patients with MRSA infections were young children (18, 19). For all strains antimicrobial resistance showed that they were resistant to betal-lactams and susceptible to non beta-lactams. However, the resistance to erythromycin has been also observed. Molecular characterization of MRSA isolates by the identification of SCCmec type revealed that majority strains harbored the SCCmec type IV and PVL genes. According to these results, our strains have been classified as community-acquired strains. Ben Neima et al. described SCCmec IV and PVL genes as markers for CA-MRSA (20). On the other side, Tenover et al. (21) has been reported that the cassett type for IV and PVL genes have been found in some nosocomial MRSA strains. PFGE has been recommended as a „gold standard” for typing of MRSA strains however it is important to follow uniform standard protocols to achieve the internationally comparable results. The technique has been compared to other techniques in various studies and has been found extremely useful in the characterization of outbreak strains (22). In contrast, some workers have reported that the stability of PFGE is not sufficient for reliable application in long term epidemiological surveillance due to long evolutionary history of pandemic clones (23, 24). PFGE analysis showed that CA-MRSA strains belonged to the same clone according to criteria of Tenover et al. (21).
In our study, PFGE played a vital role in typing the MRSA isolates obtained from various samples and also in understanding the most predominant and potential strains of this region. HA-MRSA is considered as multi drug resistant while CA-MRSA in general shows resistance only to beta lactam antibiotics such as penicillin and cephalosporin groups. This is attributed to the low number of drug resistance gene harbored by the short SCCmec (21-25 kb) element of CA-MRSA compared to the large cassettes (34-67 kb) of HA-MRSA. The resistance pattern of all the current study isolates are characteristic of CA-MRSA type. Emergence of CA-MRSA as a health care associated pathogen is a challenging issue. This was even pointed out by Seybold et al., in one of their reports. A recent report by Dhawan et al., (25) adds more strength to this alarming fact as they noticed the emergence of SCCmec type IV and V clones of MRSA in an Indian hospital. Boucher et al. (26) in their review article exemplified a case of multi-drug resistant CA-MRSA infection. Major consequence of such infections is the treatment failure. A soothing fact to be noted is the absence of such multi-drug resistant MRSA in their study settings. This indicates that neither HA-MRSA has invaded into community settings nor CA-MRSA has acquired multi-drug resistance genes (27).
Of particular importance is the future impact of CA-MRSA in our community or, indeed, elsewhere, should this organism become more prevalent. These MRSA strains can complicate management of pediatric infection because they occur in unexpected circumstances. The true prevalence of CA-MRSA remains unknown, however, because no systematic, population-based study of community isolates of S. aureus exists. Furthermore, significant regional variation is likely. Thus, a significant increase in incidence would necessitate changes to the prescribing guidelines for CA-MRSA infections. Although most CA-MRSA isolates were susceptible to several antimicrobial agents, selection of alternative agents must remain dependent on local susceptibility patterns (28).
Our study has some limitations. Only one regional public institute participated in collection of data on CA-MRSA, and among the non -participants were the two largest regions. The prevalence of CA-MRSA in B&H is therefore difficult to assess. In addition, in hospitalized patients, important data on the time between admittance to hospital and isolation of the MRSA were missing, if that period is < 48 h, the strain is definitely regarded as being community acquired. Lastly unfortunately, B&H does not participate in the European Antimicrobial Resistance Surveillance Systems and therefore our data insufficient. Methicillin-resistant Staphylococcus aureus is a major cause of both nosocomial and community-acquired infections, with reported high level of morbidity and mortality within infected patients, especially in newborns and children. Outbreaks of infection caused by MRSA have been reported even in neonatal units. MRSA Pediatric clone expressing ermC plus lnuA genes causing nosocomial transmission and healthcare workers colonization in a neonatal intensive care unit (29). The combination of molecular typing methods (PFGE, mecA, Tn554 probes, SCCmec, and MLST) with epidemiological and clinical information allows the detection of MRSA clusters and outbreaks and therefore provides rationale for appropriate infection control intervention.
5. CONCLUSION
With global transmission of MRSA, our local epidemiology may be changing due to the introduction of new strains. We emphasize the importance of identification and therapeutic practice in infections associated with S. aureus, particularly those caused by methicillin-resistant isolates. These findings suggest the need to reassess the choice of empirical treatments for infections in areas where MRSA is prevalent in the community. However, the prevalence of CA-MRSA in the B&H is unknown, but this report indicate that it is likely increasing. SCCmec type IV, together with the susceptibility profile and PFGE grouping, is considered to be typical of CA-MRSA. Surveillance programs should be implemented by public health laboratories to determine the extent of CA-MRSA dissemination. Until the scope of this problem is better defined, clinicians should consider the use of non-β-lactam antistaphylococcal antibiotics in the empirical treatment of CA-MRSA.
Footnotes
• Conflict of Interests: The authors declare that they have no conflict of interest.
REFERENCES
- 1.Stryjewski ME, Corey GR. Methicillin-Resistant Staphylococcus aureus. An Evolving Pathogen. Clin Infect Dis. 2014;58(1):10–19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 2.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 3.Udo EE. Community-acquired Methicillin-resistant Staphylococcus aureus: The New Face of an Old Foe? Med Princ Pract. 2013;22(1):20–9. doi: 10.1159/000354201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 5.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated methicillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Curr Opin Microbiol. 2012;15(5):588–95. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Otter JA, Yezli S, French GL. Impact of the suspending medium on susceptibility of meticillin-resistant Staphylococcus aureus to hydrogen peroxide vapour decontamination. J Hosp Infect. 2012;82(3):213–5. doi: 10.1016/j.jhin.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Popoola BO, Denloye OO, Iyun OI. Influence of parental socioeconomic status on caries prevalence among children seen at the university college hospital, Ibadan. Ann Ib Postgrad Med. 2013;11(2):81–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Mehndiratta PL, Bhalla P. Typing of Methicillin resistant Staphylococcus aureus: a technical review. Indian J Med Microbiol. 2012;30(1):16–23. doi: 10.4103/0255-0857.93015. [DOI] [PubMed] [Google Scholar]
- 10.Struelens MJ, Hawkey PM, French GL, Witte W, Tacconelli E. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect. 2009;15(2):112–9. doi: 10.1111/j.1469-0691.2009.02698.x. [DOI] [PubMed] [Google Scholar]
- 11.Murray PR. Manual of Clinical Microbiology. 8th ed. Washington, D. C: American Society for Microbiology Press; 2003. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing. Sixteenth informational supplement. CLSI. 2006;27(1):M100–S16 (M2). [Google Scholar]
- 13.Villanova PA. Susceptibility to mupirocin and fusidic acid was determined using methods recommended by the British Society for Antimicrobial therapy British Society for Antimicrobial Chemotherapy. BSAC methods for antimicrobial susceptibility testing. Version 5. 2006 [Google Scholar]
- 14.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–61. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budimir A, Deurenberg RH, Plecko V, Vink C, Kalenic S, Stobberingh EE. Molecular characterization of methicillin-resistant Staphylococcus aureus bloodstream isolates from Croatia. J Antimicrob Chemother. 2006;57(2):331–4. doi: 10.1093/jac/dki452. [DOI] [PubMed] [Google Scholar]
- 16.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 17.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan SI, Hulten KG, Gonzales BE, et al. Three year surveillance of community-acquired methicillin-resistant Staphylococcus aureus infections in children. Cl Infect Dis. 2005;12(40):1785–91. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 19.Lee MC, Rios AM, Aten MF, Mejias A, Cavuoti D, McCracken GH, Jr, Hardy RD. Management and outcome of children with skin and soft tissue abscesses caused by community-acquired methicillin-resistant Staphylococcus aureus. Pediatr Infect Dis J. 2004;23(2):123–7. doi: 10.1097/01.inf.0000109288.06912.21. [DOI] [PubMed] [Google Scholar]
- 20.Ben Nejma M, Mastouri M, Bel Hadj Jrad B, Nour M. Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn Microbiol Infect Dis. 2013;77(1):20–4. doi: 10.1016/j.diagmicrobio.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Tenover FC. Rapid detection and identification of bacterial pathogens using novel molecular technologies: infection control and beyond. Clin Infect Dis. 2007;44(3):418–23. doi: 10.1086/510684. [DOI] [PubMed] [Google Scholar]
- 22.Mehndiratta PL, Bhalla P. Typing of Methicillin resistant Staphylococcus aureus: a technical review. Indian J Med Microbiol. 2012;30(1):16–23. doi: 10.4103/0255-0857.93015. [DOI] [PubMed] [Google Scholar]
- 23.Hallin M, Deplano A, Denis O, De Mendonca R, De Ryck R, Struelens MJ. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of staphylococcus aureus Infections. J Clin Microbiol. 2007;45:127–33. doi: 10.1128/JCM.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melles DC, van Leeuwen WB, Sniders SV, Horst-Kreft D, Peeters JK, Verbrugh HA, et al. Compariosn of multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP) for genetic typing of Staphylococcus aureus. J Microbiol Methods. 2007;69:371–5. doi: 10.1016/j.mimet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Dhawan B, Rao C, Udo EE, Gadepalli R, Vishnubhatla S, Kapil A. Dissemination of methicillin-resistant Staphylococcus aureus SCCmec type IV and SCCmec type V epidemic clones in a tertiary hospital: challenge to infection control. Epidemiol Infect. 2015;143(2):343–53. doi: 10.1017/S095026881400065X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2010;15(Suppl 2):S183–97. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- 27.Govindan S, Maroli A S, Ciraj A M, Bairy I. Molecular epidemiology of methicillin resistant staphylococcus aureus colonizing the anterior Nares of school children of Udupi Taluk. Indian J Med Microbiol. 2015;33(Suppl S1):129–33. doi: 10.4103/0255-0857.150919. [DOI] [PubMed] [Google Scholar]
- 28.Wang CC1, Lo WT, Chu ML, Siu LK. Epidemiological typing of community-acquired methicillin-resistant Staphylococcus aureus isolates from children in Taiwan. Clin Infect Dis. 2004;39(4):481–7. doi: 10.1086/422642. [DOI] [PubMed] [Google Scholar]
- 29.Faccone D, Togneri AM, Podesta L, Perez M, Gagetti P, Sanchez S, Romero G, Corso A. MRSA Pediatric clone expressing ermC plus lnuA genes causing nosocomial transmission and healthcare workers colonization in a neonatal intensive care unit. Infect Genet Evol. 2014;25:78–80. doi: 10.1016/j.meegid.2014.04.005. [DOI] [PubMed] [Google Scholar]


