Abstract
Diffuse large B-cell lymphoma (DLBCL), the most common type of non-Hodgkin's lymphoma (NHL) in adults, accounts for approximately 30–40% of newly diagnosed lymphomas worldwide. Environmental factors, such as viruses and bacteria, may contribute to cancer development through chronic inflammation and the integration of oncogenes, and have previously been indicated in cervical cancer, hepatocellular carcinoma, gastric cancer and lymphoproliferative disorders. In the present study, the presence of microbial agents was analyzed in the lymphoma tissue of patients with activated B-cell like (ABC) DLBCL. The present study compared two groups of patients from geographically varied regions that possess a difference in the prevalence of viral and other microbial agents. The patient populations were from Sweden (a low endemic infectious disease region) and Egypt (a high endemic infectious disease region). A differential expression of several viruses in lymphoma tissues was noted when comparing Swedish and Egyptian patients. JC polyomavirus (JCV) was detected in Swedish and Egyptian patients and, uniquely, the complete hepatitis B virus (HBV) genome was detected only in Egyptian lymphoma patients. None of these viruses were detected in control lymph tissues from Sweden or Egypt. In total, 38% of the Egyptian patients were found to have HBV surface antigens (HBsAgs) in their serum; however, HBsAgs were not found in any of the Swedish patients. The percentage of serum HBsAgs in Egyptian patients with ABC DLBCL was significantly increased compared with the general Egyptian population (P<0.05). The present study may support a notion that viral agents, including JCV and HBV, may be involved in the tumorigenesis of DLBCL in regions of high infectious disease.
Keywords: molecular genetics, lymphoma and Hodgkin disease, virus, hepatitis B virus, gene array
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common type of non-Hodgkin's lymphoma (NHL) in adults, accounts for approximately 30–40% of newly diagnosed lymphomas worldwide (1). Using gene expression profiling, DLBCL can be divided into three distinguishable subtypes; the germinal center B-cell like (GBC), activated B-cell like (ABC) and primary mediastinal B-cell lymphoma subtypes (1–3). These subtypes differ in the expression of thousands of genes and arise at separate stages of B-cell differentiation. The ABC subgroup has a poor prognosis with a short survival time compared with the GCB subgroup (2–4). In addition, the frequency of the ABC subtype in Asian countries, such as Japan and China, is increased compared with the GCB subtype. The ABC subtype is also more common in certain Asian countries compared with in the western world (5,6).
Environmental factors, including viruses and bacteria, may contribute to the development of cancer, and have been previously indicated in cervical cancer, hepatocellular carcinoma, gastric cancer and lymphoproliferative disorders (7–10). Viruses may be involved in the development of cancer by activating the innate immune system, as well as activating intracellular signaling cascades that control viral infection and tumor cell growth (11). Bacteria, viruses and parasites may also directly contribute to carcinogenesis through the transfection and integration of oncogenes, which results in malignant cell transformation (12,13). Viruses involved in carcinogenesis may be targets for diagnosis, prevention and therapies (11).
Although the incidence of NHL has increased during the last three decades, the etiology of the most common types remains unclear (American Cancer Society, 2001). Chromosomal translocations are common in numerous NHL subtypes, which could result in the inactivation of tumor suppressor genes or activation of oncogenes (14). Viruses may induce genetic damage to the genome of lymphoid cells and dysregulate normal cell growth (15). Certain viruses have been specifically linked to particular types of lymphomas, including Epstein-Barr virus (EBV), human T-lymphotropic virus type 1, herpes virus-8 and hepatitis (16–18). Prevention of viral infections should reduce the number of individuals at risk of lymphoma development; thus recognizing the viruses involved in the tumorigenesis of NHL is important.
The virus genome has been shaped to interact with host cell regulatory and signaling networks. Viruses may alter host cell proteins in all steps of the viral cycle and utilize common host cell response pathways for pathogenic invasion (19,20). The Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway may be activated by viral infections that result in maintained cell growth (21–23). Replicating hepatitis B virus (HBV) modulates STAT signaling by the upregulation of STAT3, which contributes to immune suppression, viral persistence and malignant transformation (21). As a part of the JAK/STAT pathway, STAT3 has been shown to be upregulated in several types of malignancies, including B-cell lymphomas (24). STAT3 has been previously suggested as a therapeutic target of ABC DLBCL, since STAT3 and nuclear factor κ B activation may be associated with a poor survival in this subgroup of patients (24).
A previous study analyzed the gene and micro RNA (miRNA) expression patterns in the lymphoma tissues of patients from two geographically varied regions with differing loads of microbial infections, and found a difference in the expression of STAT3 (25). The patient populations were from Sweden (a low infectious disease region) and Egypt (a high endemic infectious disease region) (25). STAT3 was overexpressed in Swedish patients compared with Egyptian patients (26) and miR-1234 was shown to be a possible regulator of STAT3 expression (27).
The main aim of the present study was to analyze the presence of microbial agents in the lymphoma tissues of patients with ABC DLBCL, and to associate this with STAT3 and miR-1234 expression.
Materials and methods
Patients and controls
In total, 47 ABC DLBCL patient biopsy samples were obtained from the National Cancer Institute, Cairo, Egypt, between July 2003 and January 2008, and 14 ABC DLBCL patient biopsy samples were obtained from the Karolinska University Hospital, Stockholm, Sweden, between October 2005 and September 2011. The biopsy material was obtained at the time of diagnosis and embedded in paraffin. The median ages of the Swedish and Egyptian patients were 57 and 66 years, respectively. The gender distribution (% male/female) was 53/47 and 43/57 in Swedish and Egyptian patients, respectively. Reactive lymph nodes from teh same patients were collected as controls, from the National Cancer Institute, Cairo, Egypt (n=10) and from the Karolinska University Hospital, Stockholm, Sweden (n=10). The study was approved by the Regional Ethics Committee (www.epn.se) and written informed consent was obtained from all patients.
Histopathological diagnosis
Primary selection of DLBCL patients was performed using formalin-fixed (FF) paraffin-embedded (PE) material stained with eosin and hematoxylin and automated immunohistochemistry (IHC) for cluster of differentiation (CD)20, CD3, Ki67, B-cell lymphoma (Bcl)-2, Bcl-6, CD10 and melanoma associated antigen (mutated) 1, according to the algorithm described by Hans et al (28). This algorithm has recently been confirmed as one of two IHC algorithms that can predict the non-germinal center cell origin with a high accuracy, and has 86% concordance with gene expression profiling results (29). FFPE blocks classified as the non-GCB DLBCL subtype, as described previously (30), were selected for the study and were analyzed by the Department of Pathology.
Handling of patient and control material
The tissue blocks were cut into 20-µm thick sections, using sterile blades. All benches, instruments and pipettes were cleaned with RNaseZap solution (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA). A previous study reported that FFPE materials may be used for gene expression analysis, and are comparable with fresh frozen material (30).
RNA extraction
Extraction of RNA from PE material with the Ambion Recover All Total Nucleic Acid Isolation optimized for FFPE samples (Thermo Fisher Scientific, Inc.) was successfully performed following the manufacturer's instructions. RNA samples were stored at −80°C until required for amplification. In brief, 20-µm thick sections were treated with xylene, centrifuged at full speed (10,000 × g) and washed with 100% ethanol twice. The pellets were air dried for 15 min. Digestion buffer and protease were added, followed by 3 h incubation at 50°C. Isolation additive was added followed by 100% ethanol, and the samples were transferred to a filter cartridge. Two rounds of centrifugation with Wash 1 and Wash 2/3 was performed. DNase combined with 10X DNase buffer was added followed by 30 min incubation at room temperature. The membranes were then washed with Wash 1 and Wash 2/3. The RNA was eluted in 30 µl RNase free water preheated at 95°C for 30 min. Yield and quality was measured using the Nanodrop Technologies ND 1000 (Thermo Fisher Scientific, Inc.). Absorbance ratios (260/280 for DNA and 260/230 for RNA) of >1.8 were considered to be of high purity.
Complementary DNA (cDNA) preparation and amplification from PE DLBCL material
RNA (100 ng) was used for amplification according to the protocols of the WT-Ovation FFPE RNA Amplification System V2 (Nugen Technologies, Inc., San Carlos, CA, USA), with a Spike control. All incubations were performed using the 1294 Techne Progene Peltier Thermal cycler thermal cycler (Scientific support, Inc., Hayward, CA, USA). The purification step was performed using Agencourt RNA Clean purification beads (Beckman Coulter, Inc., Fullerton, CA, USA). The amplified cDNA was purified using DNA Clean & Concentrator kit (Zymo Research Corporation, Irvine, CA, USA). The yield and quality of the cDNA was measured using the Nanodrop Technologies ND 1000. cDNA to be used in the microarray analysis were pooled using the same concentration of each sample.
Microbial detection array (MDA)
Nucleic acid concentrations were determined using the Invitrogen Qubit Fluorometer (Thermo Fisher Scientific, Inc.). DNA and RNA were randomly amplified using a protocol described previously (31). Following amplification, the amplified samples were purified using the Qiagen QIAquick PCR Purification kit (Qiagen GmbH, Hilden, Germany). Amplified nucleic acids were fluorescently labeled using the Roche NimbleGen One-Color DNA Labeling kit (Roche Applied Science, Madison, WI, USA), according to the recommended protocols. DNA was purified subsequent to labeling, and hybridized using the NimbleGen Hybridization kit (Roche Applied Science) and the Lawrence Livermore Microbial Detection Array (LLMDA), according to manufacturers' instructions. Microarrays were allowed to hybridize for 17 h and washed using the NimbleGen Wash Buffer kit (Roche Applied Science), according to manufacturer's instructions. The samples were then scanned on an Axon GenePix 4000B 5 µM scanner from Molecular Devices, LLC (Sunnyvale, CA, USA). Signals on the LLMDA were analyzed using the maximum likelihood analysis method (32). In brief, probes were identified when binding intensity to the probes exceeded a threshold equal to the 99th percentile of intensities for the negative control probes. Targets in an internal database of viral sequences were screened against stringency criteria and a log-odd score was computed for each target. A selection algorithm was applied to find the collection of targets most likely to be present in the sample. At every forward selection step, a conditional log-odd score was computed for each remaining target. The target with the largest conditional log-odd score was selected and added to the collection. The conditional log-odd score is a value representing the probability of being the detected target. Higher log-odd scores have an increased probability of being the detected target compared with lower log-odd scores.
HBV antigen and antibody analyses of patient serum
DNA extracted from the serum of Swedish and Egyptian patients was used to analyze the presence of the HBVsAg by polymerase chain reaction (PCR), by detecting HBV core genes as previously described (33). Briefly, 100 µl of reaction mixture containing 10 µl of extracted DNA, 50 mM potassium chloride, 10 mM Tris-hydrochloric acid (pH 8.3), 2 mM magnesium chloride, 200 µM deoxyribonucleosides, 2.5 units Taq polymerase (PerkinElmer, Inc., Waltham, MA, USA), and 20 pmol of each of the oligonucleotide primers [C1 sense, CTGGGAGGAGTTGGGGGA (1730–1747) and C2 antisense, GTAGAAGAATAAAGCCC (2503–2487)] for the core genes. Amplification was performed for one cycle of 95°C for 5 min, followed by 35 cycles that consisted of denaturating for 1 min at 94°C, annealing for 1 min at 55°C and extension for 1.5 min at 72°C. Antibodies against HBV were analyzed in Swedish and Egyptian patient serum using a chemiluminescent microparticle immunoassay (Abbott Diagnostics, Lake Forest, IL, USA). In brief, serum sample was mixed with paramagnetic microparticles coated with recombinant HBsAg (rHBsAg; Abbott Diagnostics). Subsequent to washing, acridium-marked rHBsAg-conjugate (Abbott Diagnostics) was added. Following additional washing, pre-trigger and trigger solution were added. The chemiluminescence reaction was then measured and the concentration of anti-HBs was set by a pre-designed calibration curve (Abbott Diagnostics).
Relative expression of STAT3 in HBVsAg-positive (HBV+) and -negative (HBV-) Egyptian patients
To evaluate the relative expression of STAT3 and micro RNA (miR)-1234 in association with HBV, HBV+ and HBV− Egyptian and Swedish patients were analyzed individually using Applied Biosystems HT7900 quantitative PCR (Thermo Fisher Scientific, Inc.). For the STAT3 relative expression analysis, the High Capacity RNA to cDNA kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for generating cDNA, according to manufacturer's recommendations. cDNA was further amplified (250 ng) by applying the TaqMan PreAmp Master Mix kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to manufacturer's recommendations. The TaqMan assays designed for STAT3 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression of individual patient samples was calculated using the reference gene GAPDH. For the miR-1234 relative expression analysis, total RNA was poly-adenylated using yeast polyA polymerase (Affymetrix, Inc., Santa Clara, CA, USA) for 1 h at 37°C, followed by 10 min at 65°C, according to manufacturer's recommendations. The RNA was precipitated over night using sodium acetate-isopropanol at −20°C, washed in ethanol and diluted in H2O. The reverse transcription reaction was performed using the TaqMan miRNA reverse transcription kit (Thermo Fisher Scientific, Inc.) using 1.17 µM universal RT primer. The reaction was incubated at 72°C for 5 min and put on ice prior to adding the RT enzyme. The reaction was then incubated at 42°C for 45 min, followed by 5 min at 85°C. qPCR was performed using an Applied Biosystems HT7900 qPCR machine (Thermo Fisher Scientific, Inc.) with miR-1234 primers and TaqMan universal PCR master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression of miR-1234 was calculated relative to the expression of 5S rRNA. The mean Cq values and the relative expression for STAT3 and miR-1234 expression were calculated, as previously described (34).
Statistics
Statistical analysis was performed using GraphPad Prism version 5 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical calculation was performed by a one-way analysis of variance, followed by post-hoc Tukey's honest significant difference test or Student's t-test. P<0.05 was used to indicate a statistically significant difference.
Results
Microbial detection in Swedish and Egyptian patients using MDA analysis
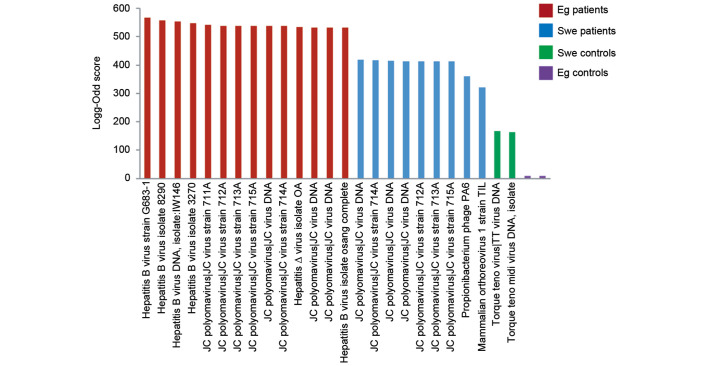
Viruses were detected by the MDA, a high-density oligonucleotide array for the detection and discovery of viruses and bacteria. Viruses that were found in all groups (controls and patients) were excluded, leaving exclusively those found in each patient group. Among excluded viruses were influenza virus A and human endogenous retroviruses found in the patient and control groups. Human immunodeficiency virus (HIV) was excluded as it was found in Swedish patients and controls. Clinically irrelevant viruses, including plant and animal viruses that have not been reported to infect human cells (for example faba bean necrotic yellow virus, porcine circovirus and melon necrotic spot virus) were also excluded. The log-odd scores were plotted in a bar graph (Fig. 1). The HBV had the highest log-odd scores, including the complete genome, and was only found in Egyptian patients. JC polyomavirus (JCV) was found in Egyptian and Swedish patients, with a higher log-odd score in the Egyptian patients, but not in Swedish and Egyptian controls. Generally, the highest log-odd scores were found in Egyptian patients rather than in Swedish patients. Few viruses were found in the control groups. To visualize the differences in log-odd scores between the groups, a mean log-odd score was calculated for each group. A statistically significant difference was found between the mean log-odd scores for each patient and control group (P=0.032 for Swedish controls versus Egyptian patients; P=0.041 for Swedish patients versus Egyptian patients; Fig. 2).
Figure 1.
Bar graph showing detected viruses from the microbial detection array. Log-odd scores are indicated on the y-axis. Red bars indicate Egyptian patients, blue bars indicate Swedish patients, purple bars indicate Egyptian control samples and green bars indicate Swedish control samples. Each bar indicates the detection of a virus, including the complete genome, strain or isolate. Strains and isolate numbers are indicated. Eg, Egyptian; Swe, Swedish.
Figure 2.
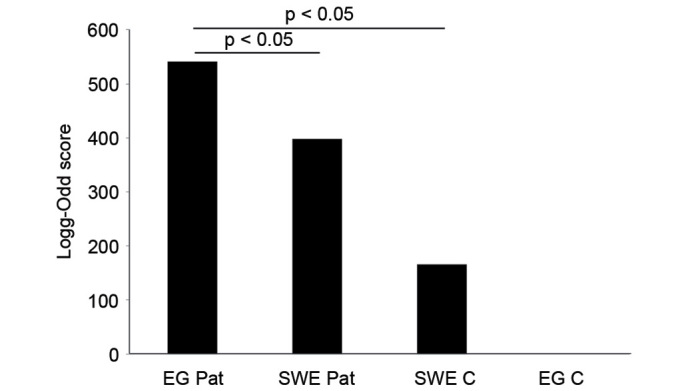
Bar graph summarizing log-odd scores for each patient or control group. Statistical analysis (Student's t-test) was performed and indicates statistical difference between the groups. No significant difference was shown between the controls sample groups. EG, Egyptian; SWE, swedish; C, control; Pat, patient.
Serum HBVsAgs and antibody responses
HBV showed the highest log-odd score and the presence of the complete genome, but only in Egyptian patients using lymphoma tissues. HBVsAgs could be detected in the serum of 38% of Egyptian patients, but not in any of the Swedish patients. According to previous literature, the prevalence in the general population of Egypt for the HBsAg is 4% (35), and in Swedish patients <1% (36). The difference, with regard to the presence of HBVsAg, between the Egyptian patients and the general Egyptian population was statistically significant (P<0.05) (35). HBV antibodies could be detected in 100% of the Egyptian patients, but in none of the Swedish patients.
STAT3 and miR-1234 in association with serum HBVsAg positivity
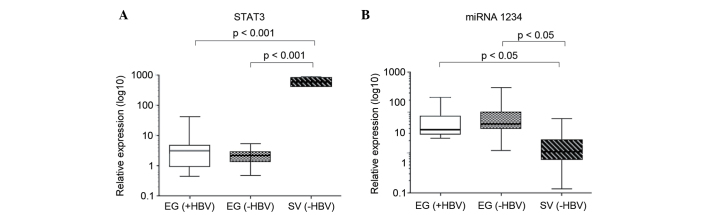
Based on the presence of the HBVsAgs in serum, Egyptian patients were divided into a HBV+ and HBV− groups. All Swedish patients were HBV-. The relative expression of STAT3 did not differ between HBV+ and HBV-Egyptian patients, but was significantly increased in the Swedish patients compared with the total Egyptian patient population (P=0.00098 versus HB+ and P=0.00093 versus HB− Egyptian patients; Fig. 3A). A study has recently shown that miR-1234 may regulate STAT3, and that the relative expression of miR-1234 is associated with HBV status (data to be published). In the present study, there was no difference in the relative expression of miR-1234 when comparing HBV+ and HBV− Egyptian patients; however, HBV− Swedish patients showed a significantly lower relative expression of miR-1234 compared with the total Egyptian patient population (P=0.044 versus HB+ and P=0.042 versus HB− Egyptian patients; Fig. 3B). miR-1234 expression was inversely associated with the expression patterns of STAT3.
Figure 3.
Box plot showing relative expression of (A) STAT3 and (B) miRNA in EG (+HBV) and EG (−HBV) patients and SV (−HBV) patients. Statistical analysis (analysis of variance) shows a significant difference between SV and EG patients. There were no statistical differences between EG (+HBV) and EG (−HBV) patients. EG, Egyptian, SV, Swedish; HBV, hepatitis B virus; STAT3, signal transducers and activators of transcription 3; miRNA, micro RNA.
Discussion
Virus-linked human cancers may be responsible for 15% of all cancers, which makes viruses the second most important risk factor for cancer development after tobacco (37,38). The present study compared the presence of microbial agents in the lymphoma tissues of two groups of ABC DLBCL patients from a high (Egypt) and low (Sweden) infectious disease region (39). The LLMDA system used in the present study was developed at Lawrence Livermore National Laboratory, Livermore, CA, USA, and contains probes to detect any RNA and DNA of sequenced viruses and bacteria (32,40). Several viruses were found and most the prevalent were HBV and JCV. HBV was not detected in the Swedish patients or in either control group. JCV was also present in Egyptian and Swedish patients, but not in the controls. Overall, the Egyptian group showed higher virus log-odd scores compared with Swedish patients. The HBVsAg could be found in 38% of the Egyptian patients. There was no difference in STAT3 expression compared between the HBV+ and HBV− groups of Egyptian patients. STAT3 has previously been reported to be dysregulated by HBV, resulting in viral persistence and malignant transformation (21,41).
Viral infection of host cells may induce cell proliferation and immune suppression, which are mechanisms that may contribute to the viral-induced development of human tumors (21). HIV infected patients have an increased risk of developing, for example, Bcl and Kaposi sarcoma (38). Adenovirus and poxvirus have cell-transforming properties, but none of these have been found in human tumor cells (38). EBV has been recognized as a viral agent involved in the development of certain Bcls. Barzon et al (42) reported that patients with primary adrenal Bcl harbored EBV and JCV together, and suggested that these viruses may act as cofactors in the development of lymphomas (42).
Hepatitis B and C have been associated with hepatocellular carcinoma and dermatological, hematological, endocrinological and autoimmune disorders (43–45). HBV reactivation may be observed in patients during treatment with rituximab (46); however, no conclusive evidence of an association between NHL and hepatitis virus infection has been reported (47). Two studies showed that HBV infection was associated with an increased risk of DLBCL in Korean populations (48,49). An association between HBV infection and NHL was suggested in patients with persistent HBsAg (50). A study of a Korean population reported that 12% of NHL patients were HBV+ compared with 6% of the control population (51). According to the Centers for Disease Control and Prevention, 2–7% of the Egyptian population are HBV+ (52). These numbers were confirmed by a study reporting that 4% of Egyptian blood donors were HBV+ (35). The results of the present study show that lymphoma tissues of the Egyptian patients were HBV+ and that 38% of the patients were HBV+. None of the Swedish patients or any of the controls were HBV+. The results indicate that Egyptian patients have an increased viral load and that HBV was the most common virus. Since none of the Swedish patients were HBV+, the virus is unlikely to be associated with the development of DLBCL originating in Sweden. However, these results may be of importance for treatment strategies in Egyptian patients, since a previous study has demonstrated the reactivation of HBV in patients from high endemic infectious regions during treatment with rituximab (46).
The first human polyomaviruses (PyV), BK polyomavirus (BKV) and JCV were isolated in 1971 (53,54). Human PyVs are common in the general population but rarely cause clinically apparent disease (55). Sources of infection that have been suggested include virus-bearing fluids, contaminated food and water (56,57). The association of JCV with human malignancies is controversial; however, several studies have shown JCV to be associated with human cancers as colorectal-, gastric-, lung− and brain cancers (58–62). In a study by Flægstad et al (63), BKV was reported to be expressed in the tissue samples of neuroblastomas, but not found in control tissues. This suggests that BKV may be important in the development of neuroblastomas through the inactivation of cell cycle regulation and apoptotic effects, which contribute to malignant transformation (63).
BKV and JCV were recently classified as ‘possibly carcinogenic to humans’ by the WHO international cancer research working committee (64). The contribution of PyVs to cancer progression can be challenged in three ways First, the PyV may establish chromosomal instability at an early stage of infection that contributes to cancer progression, but PyV may not be detectable until full progression to malignancy. Secondly, PyVs may find favorable conditions in an already transformed cell, but be neither necessary for nor contribute to the oncogenic characteristics. Thirdly, PyV may be detected in an anatomically connected compartment, but be unrelated to the malignancy (55). JCV may establish a latent infection and e reactivated upon immunosuppression; one a site of latency has been shown to be lymphoid cells (65). The results of the present study showed that JCV may be detected in Swedish and Egyptian patients, but not in Swedish and Egyptian control groups. These data are notable as it may suggest a role for JCV in DLBCL.
Previous studies on Swedish and Egyptian patients showed that STAT3 was overexpressed in Swedish patients compared with Egyptian patients (26), and that miR-1234 may be a possible regulator of STAT3 (data to be published). No association between STAT3 and HBV expression was detected in the present study. However, the regulation of STAT3 by miR-1234 may also be associated with other environmental factors and to the increased viral load as noted in the Egyptian patients.
The current study showed that Egyptian and Swedish patients had detectable JCV, which was not detected in Swedish and Egyptian controls, and HBV was only detected in Egyptian patients. Extended studies on the viral involvement in DLBCL are warranted in order to analyze whether specific infections are associated with the development of the disease. Such studies may be of value for the development of novel prevention and treatment strategies.
References
- 1.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma, corp-author. The Non-Hodgkin's Lymphoma Classification Project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu YH, Xu FP, Zhuang HG, Lai KC, Xie D, Luo DL, Li L, Luo XL, Xu J, Zhang MH, et al. Clinicopathologic significance of immunophenotypic profiles related to germinal center and activation B-cell differentiation in diffuse large B-cell lymphoma from Chinese patients. Hum Pathol. 2008;39:875–884. doi: 10.1016/j.humpath.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi A, Fujita S, Ikeda J, Nakamichi I, Fukuhara S, Hino M, Kanakura Y, Ogawa H, Sugiyama H, Kanamaru A, Aozasa K. Diffuse large B-cell lymphoma in the young in Japan: A study by the Osaka Lymphoma Study Group. Am J Hematol. 2007;82:893–897. doi: 10.1002/ajh.20968. [DOI] [PubMed] [Google Scholar]
- 7.Jarrett RF. Viruses and lymphoma/leukaemia. J Pathol. 2006;208:176–186. doi: 10.1002/path.1905. [DOI] [PubMed] [Google Scholar]
- 8.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, Lai KC, Hu WH, Yuen ST, Leung SY, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: A randomized controlled trial. JAMA. 2004;291:187–194. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 10.Zhang JY, Dai M, Wang X, Lu WQ, Li DS, Zhang MX, Wang KJ, Dai LP, Han SG, Zhou YF, Zhuang H. A case-control study of hepatitis B and C virus infection as risk factors for hepatocellular carcinoma in Henan, China. Int J Epidemiol. 1998;27:574–578. doi: 10.1093/ije/27.4.574. [DOI] [PubMed] [Google Scholar]
- 11.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.zurHausen H. Oncogenic DNA viruses. Oncogene. 2001;20:7820–7823. doi: 10.1038/sj.onc.1204958. [DOI] [PubMed] [Google Scholar]
- 13.Parsonnet J. Microbes and malignancy: Infection as a cause of human cancers. Oxford University Press; New York, NY: 1999. [Google Scholar]
- 14.Nogai H, Dörken B, Lenz G. Pathogenesis of non-Hodgkin's lymphoma. J Clin Oncol. 2011;29:1803–1811. doi: 10.1200/JCO.2010.33.3252. [DOI] [PubMed] [Google Scholar]
- 15.Capello D, Scandurra M, Poretti G, Rancoita PM, Mian M, Gloghini A, Deambrogi C, Martini M, Rossi D, Greiner TC, et al. Genome wide DNA-profiling of HIV-related B-cell lymphomas. Br J Haematol. 2010;148:245–255. doi: 10.1111/j.1365-2141.2009.07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engels EA, Chatterjee N, Cerhan JR, Davis S, Cozen W, Severson RK, Whitby D, Colt JS, Hartge P. Hepatitis C virus infection and non-Hodgkin lymphoma: Results of the NCI-SEER multi-center case-control study. Int J Cancer. 2004;111:76–80. doi: 10.1002/ijc.20021. [DOI] [PubMed] [Google Scholar]
- 17.Fisher SG, Fisher RI. The epidemiology of non-Hodgkin's lymphoma. Oncogene. 2004;23:6524–6534. doi: 10.1038/sj.onc.1207843. [DOI] [PubMed] [Google Scholar]
- 18.Mele A, Pulsoni A, Bianco E, Musto P, Szklo A, Sanpaolo MG, Iannitto E, De Renzo A, Martino B, Liso V, et al. Hepatitis C virus and B-cell non-Hodgkin lymphomas: An Italian multicenter case-control study. Blood. 2003;102:996–999. doi: 10.1182/blood-2002-10-3230. [DOI] [PubMed] [Google Scholar]
- 19.Davey NE, Travé G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Smith SB, Dampier W, Tozeren A, Brown JR, Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One. 2012;7:e33174. doi: 10.1371/journal.pone.0033174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeberlein B, zur Hausen A, Bektas N, Zentgraf H, Chin R, Nguyen LT, Kandolf R, Torresi J, Bock CT. Hepatitis B virus overexpresses suppressor of cytokine signaling-3 (SOCS3) thereby contributing to severity of inflammation in the liver. Virus Res. 2010;148:51–59. doi: 10.1016/j.virusres.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 23.Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, Staudt LM. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 26.Hogfeldt T, Bahnassy AA, Kwiecinska A, Osterborg A, Tamm KP, Porwit A, Zekri AR, Lundahl J, Khaled HM, Mellstedt H, Moshfegh A. Patients with activated B-cell like diffuse large B-cell lymphoma in high and low infectious disease areas have different inflammatory gene signatures. Leuk Lymphoma. 2013;54:996–1003. doi: 10.3109/10428194.2012.738365. [DOI] [PubMed] [Google Scholar]
- 27.Högfeldt T, Johnsson P, Grandér D, Bahnassy AA, Porwit A, Eid S, Österborg A, Zekri AR, Lundahl J, Khaled MH, et al. Expression of microRNA-1234 related signal transducer and activator of transcription 3 in patients with diffuse large B-cell lymphoma of activated B-cell like type from high and low infectious disease areas. Leuk Lymphoma. 2014;55:1158–1165. doi: 10.3109/10428194.2013.824077. [DOI] [PubMed] [Google Scholar]
- 28.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 29.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, Ott G, Rosenwald A, Braziel RM, Campo E, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson TA, Lundahl J, Mellstedt H, Moshfegh A. Gene expression analysis using long-term preserved formalin-fixed and paraffin-embedded tissue of non-small cell lung cancer. Int J Oncol. 2011;38:1075–1081. doi: 10.3892/ijo.2011.936. [DOI] [PubMed] [Google Scholar]
- 31.Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL. Viral nucleic acids in live-attenuated vaccines: Detection of minority variants and an adventitious virus. J Virol. 2010;84:6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner SN, Jaing CJ, McLoughlin KS, Slezak TR. A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics. 2010;11:668. doi: 10.1186/1471-2164-11-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: Comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.El-Gilany AH, El-Fedawy S. Bloodborne infections among student voluntary blood donors in Mansoura University, Egypt. East Mediterr Health J. 2006;12:742–748. [PubMed] [Google Scholar]
- 36.European Centre for Disease Prevention and Control, corp-author. Hepatitis B and C in the EU neighborhood, ECDC. http://ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx?ID=300&List=4f55ad51-4aed-4d32-b960-af70113dbb90. Technical Report. 2010 Accessed June 15, 2016.
- 37.zurHausen H. Intracellular surveillance of persisting viral infections. Human genital cancer results from deficient cellular control of papillomavirus gene expression. Lancet. 1986;2:489–491. doi: 10.1016/s0140-6736(86)90360-0. [DOI] [PubMed] [Google Scholar]
- 38.zurHausen H. Viruses in human cancers. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organisation, corp-author. http://www.who.int/mediacentre/factsheets/fs204/en/ Media Centre, Hepatitis B. 2016 Jun 15; Accessed.
- 40.Erlandsson L, Rosenstierne MW, McLoughlin K, Jaing C, Fomsgaard A. The microbial detection array combined with random Phi29-amplification used as a diagnostic tool for virus detection in clinical samples. PLoS One. 2011;6:e22631. doi: 10.1371/journal.pone.0022631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K, Kim KH, Cheong J. Hepatitis B virus X protein impairs hepatic insulin signaling through degradation of IRS1 and induction of SOCS3. PLoS One. 2010;5:e8649. doi: 10.1371/journal.pone.0008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barzon L, Trevisan M, Marino F, Guzzardo V, Palù G. Primary bilateral adrenal B-cell lymphoma associated with EBV and JCV infection. Infect Agent Cancer. 2009;4:1. doi: 10.1186/1750-9378-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anzola M. Hepatocellular carcinoma: Role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383–393. doi: 10.1111/j.1365-2893.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 44.Dammacco F, Sansonno D, Piccoli C, Racanelli V, D'Amore FP, Lauletta G. The lymphoid system in hepatitis C virus infection: Autoimmunity, mixed cryoglobulinemia, and Overt B-cell malignancy. Semin Liver Dis. 2000;20:143–157. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- 45.Gumber SC, Chopra S. Hepatitis C: A multifaceted disease. Review of extrahepatic manifestations. Ann Intern Med. 1995;123:615–620. doi: 10.7326/0003-4819-123-8-199510150-00008. [DOI] [PubMed] [Google Scholar]
- 46.Niitsu N, Hagiwara Y, Tanae K, Kohri M, Takahashi N. Prospective analysis of hepatitis B virus reactivation in patients with diffuse large B-cell lymphoma after rituximab combination chemotherapy. J Clin Oncol. 2010;28:5097–5100. doi: 10.1200/JCO.2010.29.7531. [DOI] [PubMed] [Google Scholar]
- 47.Okan V, Yilmaz M, Bayram A, Kis C, Cifci S, Buyukhatipoglu H, Pehlivan M. Prevalence of hepatitis B and C viruses in patients with lymphoproliferative disorders. Int J Hematol. 2008;88:403–408. doi: 10.1007/s12185-008-0175-3. [DOI] [PubMed] [Google Scholar]
- 48.Engels EA, Cho ER, Jee SH. Hepatitis B virus infection and risk of non-Hodgkin lymphoma in South Korea: A cohort study. Lancet Oncol. 2010;11:827–834. doi: 10.1016/S1470-2045(10)70167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang J, Cho JH, Suh CW, Lee DH, Oh HB, Sohn YH, Chi HS, Park CJ, Jang SS, Lee KH, et al. High prevalence of hepatitis B and hepatitis C virus infections in Korean patients with hematopoietic malignancies. Ann Hematol. 2011;90:159–164. doi: 10.1007/s00277-010-1055-5. [DOI] [PubMed] [Google Scholar]
- 50.Marcucci F, Spada E, Mele A, Caserta CA, Pulsoni A. The association of hepatitis B virus infection with B-cell non-Hodgkin lymphoma-a review. Am J Blood Res. 2012;2:18–28. [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Bang YJ, Park BJ, Yoo T, Kim CW, Kim TY, Heo DS, Lee HS, Kim NK. Hepatitis B virus infection and B-cell non-Hodgkin's lymphoma in a hepatitis B endemic area: A case-control study. Jpn J Cancer Res. 2002;93:471–477. doi: 10.1111/j.1349-7006.2002.tb01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Centers for Disease Control and Prevention, corp-author. http://www.cdc.gov/hepatitis/hbv/ Viral Hepatitis - Hepatitis B Information. 2016 Jun 29; Accessed.
- 53.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/S0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 54.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/S0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 55.Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology. 2013;437:63–72. doi: 10.1016/j.virol.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Bofill-Mas S, Formiga-Cruz M, Clemente-Casares P, Calafell F, Girones R. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J Virol. 2001;75:10290–10299. doi: 10.1128/JVI.75.21.10290-10299.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66:238–245. doi: 10.1128/AEM.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boldorini R, Pagani E, Car PG, Omodeo-Zorini E, Borghi E, Tarantini L, Bellotti C, Ferrante P, Monga G. Molecular characterisation of JC virus strains detected in human brain tumours. Pathology. 2003;35:248–253. doi: 10.1080/0031302031000123245. [DOI] [PubMed] [Google Scholar]
- 59.Delbue S, Pagani E, Guerini FR, Agliardi C, Mancuso R, Borghi E, Rossi F, Boldorini R, Veggiani C, Car PG, Ferrante P. Distribution, characterization and significance of polyomavirus genomic sequences in tumors of the brain and its covering. J Med Virol. 2005;77:447–454. doi: 10.1002/jmv.20474. [DOI] [PubMed] [Google Scholar]
- 60.Laghi L, Randolph AE, Chauhan DP, Marra G, Major EO, Neel JV, Boland CR. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci USA. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin PY, Fung CY, Chang FP, Huang WS, Chen WC, Wang JY, Chang D. Prevalence and genotype identification of human JC virus in colon cancer in Taiwan. J Med Virol. 2008;80:1828–1834. doi: 10.1002/jmv.21296. [DOI] [PubMed] [Google Scholar]
- 62.Zheng H, Aziz HO Abdel, Nakanishi Y, Masuda S, Saito H, Tsuneyama K, Takano Y. Oncogenic role of JC virus in lung cancer. J Pathol. 2007;212:306–315. doi: 10.1002/path.2188. [DOI] [PubMed] [Google Scholar]
- 63.Flaegstad T, Andresen PA, Johnsen JI, Asomani SK, Jorgensen GE, Vignarajan S, Kjuul A, Kogner P, Traavik T. A possible contributory role of BK virus infection in neuroblastoma development. Cancer Res. 1999;59:1160–1163. [PubMed] [Google Scholar]
- 64.Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K. WHO International Agency for Research on Cancer Monograph Working Group: Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012;13:339–340. doi: 10.1016/S1470-2045(12)70125-0. [DOI] [PubMed] [Google Scholar]
- 65.Lafon ME, Dutronc H, Dubois V, Pellegrin I, Barbeau P, Ragnaud JM, Pellegrin JL, Fleury HJ. JC virus remains latent in peripheral blood B lymphocytes but replicates actively in urine from AIDS patients. J Infect Dis. 1998;177:1502–1505. doi: 10.1086/515305. [DOI] [PubMed] [Google Scholar]