Abstract
Genetic mutations on signaling pathways are found in patients with T-cell acute lymphoblastic leukemia (T-ALL) and act as markers of high-risk leukemia. Mutations in dynamin 2 (DNM2) have been reported in T-ALL, particularly in early T-cell precursor-ALL. In the present study, DNM2 mutations were screened by sequencing DNM2 exons obtained by polymerase chain reaction amplification and gel purification in adult T-ALL patients. A total of 4 novel DNM2 mutations were identified in adult T-ALL patients, with a mutation rate of 9.5%, and the DNM2 mutations were found to co-exist with NOTCH1 and PHD finger protein 6, and were also associated with high-risk leukemia. A high rate of silent mutation was also found in the patients, but no significant association was found between the silent mutations and patients' clinical features. The present findings suggested the DNM2 mutations may be involved in the oncogenesis of T-ALL.
Keywords: dynamin 2, adult, T-cell acute lymphoblastic leukemia
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in B cells, immature T lymphocytes or lymphoid progenitors (1). T-cell acute lymphoblastic leukemia (T-ALL) accounts for ~15 and 25% of ALL in pediatric and adult patients, respectively (2). Patients with T-ALL usually have high white blood cell (WBC) counts and may present with organomegaly, particularly mediastinal enlargement and central nervous system (CNS) involvement (3). The biological knowledge of T-ALL is limited. Previously, T-ALL was classified into five subgroups (pro-T, pre-T, cortical, mature T-ALL and ETP) based on the results of fluorescent in situ hybridization (FISH), molecular biology and gene expression profiling (4). With exome-sequencing and whole genome sequencing, genetic mutations on genes including NOTCH1, F-box and WD repeat domain containing 7 (FBXW7), Ras, PHD finger protein 6 (PHF6) and Janus kinase 1 (JAK1) have been found to be high-risk markers in patients with T-ALL (3,5,6).
The GTPase dynamin 2 (DNM2) is essential for intracellular vesicle formation and trafficking, cytokinesis and receptor endocytosis. DNM2 contains five domains, as follows: GTPase domain; intermediate domain (MD); pleckstrin homology domain (PH); GTPase effector domain (GED); and proline-arginine-rich domain (PRD). High DNM2 expression is observed in prostate cancer and associated with cancer progression (7). DNM2 potentiates invasive migration of pancreatic tumor cells (8). Inhibition of DNM2 induced cell death in 11 cancer cell lines (9). Previously, DNM2 genetic mutations were identified in a subtype of T-ALL, termed early T-cell precursor (ETP) ALL, which accounts for up to 15% of T-ALL and is associated with a high risk of treatment failure (6,10). However, to the best of our knowledge, no studies have investigated DNM2 genetic mutations in adult ALL. The present study sequenced the exons of DNM2 genes in 42 patients with T-ALL, and the clinical features in the patients with DNM2 mutations were analyzed.
Materials and methods
Patients and samples
Bone marrow (BM) samples from 42 patients with newly diagnosed T-ALL, consisting of 31 male patients with a median age of 26 years (range, 16–62 years) and 11 female patients with a median age of 29 years (range, 19–60 years), were collected between July 2010 and December 2014 at the First Affiliated Hospital of Nanjing Medical University (Nanjing, Jiangsu, China). The diagnosis of ALL was made according to the morphological, immunophenotypical, cytogenetic and molecular criteria of the 2008 World Health Organization Diagnosis and Classification of ALL (11). All patients provided written informed consent, in accordance with the Declaration of Helsinki, prior to enrollment in the study. The present study was approved by the Institutional Review Board of Nanjing Medical University.
Mutational analysis of DNM2
Mutational analysis of DNM2 exons 2–22 was performed. Genomic DNA was isolated using a Wizard® Genomic DNA Purification kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. DNA fragments spanning the aforementioned DNM2 exons were amplified by PCR using AmpliTaq Gold kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and exon-specific primers. The primers for PCR amplification of DNM2 exons were as follows: exon 2 forward, TGCAAGACAGAGTTGCTCCAC and reverse, TGTGTAAGTGTTCACTGAGCCG; exon 3 forward, CCAGCCTGGGTCATTACTTTC and reverse, ACACAGGCTCACCCATAGCAC; exon 4 forward, GTGGTTCAGGCAGAGTGTCAG and reverse, GACTTGGAACCAAGGATGCTG; exon 5 forward, CTGTGAGATCAGGGCTGTGAC and reverse, GGAGAAGCAATGACTTCCAGG; exon 6 forward, TACTTGAATCTTGCCCATCCC and reverse, CTGAAACAAGTGCCAGTGAGG; exon 7 forward, ATAGTGGCACCCTGGTGTTG and reverse, GTGGACGAGTGATGAGTGGTG; exon 8 forward, GTAAACCCTGGCTTGACTTGG and reverse, CTTGAGACCTTATTGCCTGGG; exon 9 forward, GTGTGAGCCACTGTATCTGGC and reverse, GGACTCAGAGGTGTGGGTGAC; exon 10 forward, CAACCTTCATTCCTTGTTGGG and reverse, CTGGGAGCCTGATACCAAACC; exon 12 forward, TCTTCTGCTCTTAGCTCCCAG and reverse, TGTCAGCATGCACAGAACAGT; exon 13 forward, TCTGTTGCCTATGAGGGTGTG and AATCCCAACTCAGTCACCTCC; exon 14 forward, CTACCTGTGGCTGCTCACTTG and reverse, TAGAGAGAGCAGATGGCCTGG; exon 16 forward, GGGCTGGAGGTGTCTCTATTG and reverse, GCAGTGACTGAGTTCTGCCC; exon 17 forward, TCATATACAGCAGCGACCAGC and reverse, GTGCTCAGTGCTCAGTGAAGG; exon 18 forward, CTAGAGCCCATTCCTCTCGG and reverse, CATGATTTCAGAGACTCCTGGC; exon 19 forward, TAGGGCAGATGGTTTCCAGAG and reverse, CTCCTTAGCTCGTGATCCGC; exon 20 forward, CCCGCCCTGTGAGAGATG and reverse, AGGACCCTGCAGGACACAC; exon 21 forward, CACCTCAGGTTCTGGCAGC and reverse, ACTGGGAGGAAGTGAGACAGG; and exon 22 forward, GAGTTGATGCCTAGGTTTGGC and reverse GAGCCTGGTCCCAGCATAG. Exons in NOTCH1, FBXW7, PHF6, phosphatase and tensin homolog (PTEN), JAK1 and interleukin (IL)-7R were also amplified as previously reported (11–15).
The PCR products of the DNM2 gene exons 2–22, NOTCH1 gene exons 26–28 and 34, FBXW7 gene exons 5–12, PHF6 gene exons 2–10, PTEN gene exons 1–9, JAK1 gene exons 13, 14, 16, 18 and 19 and IL-7R exons 2–8 were purified in 2% agarose gel and cloned into the vector by The Beijing Genomics Institute (BGI; Beijing, China) and sequenced by BGI or Shanghai Bojin Medical Instrument Co., Ltd. (Shanghai, China).
Cytogenetic and molecular analyses
Conventional cytogenetic analysis was performed at the time of diagnosis, using unstimulated short-term cultures, according to the recommendations of the International System for Human Cytogenetic Nomenclature (16). For each sample, at least 20 BM metaphase cells were analyzed.
Immunophenotypical analyses were performed by flow cytometry on fresh BM samples as described previously (17,18). The following antibody conjugates were used: anti-cluster of differentiation (CD)3-fluorescein isothiocyanate (FITC; catalog no., 555339), anti-CD2-allophycocyanin (APC; catalog no., 560642), anti-CD5-phycoerythrin (PE; catalog no., 555353), anti-CD7-brilliant violet 421 (BV421; catalog no., 562635), anti-CD19-FITC (catalog no., 555412), anti-CD20-APC (catalog no., 559776), anti-CD10-BV421 (catalog no., 562902), anti-CD34-PE (catalog no., 555822) and anti-CD33-BV605 (catalog no., 740400). All the antibodies were mouse anti-human and purchased from BD Biosciences (San Jose, CA, USA) and used for cell staining according to the manufacturer's protocol. FITC, PE, BV605 and BV421-conjugated antibodies were diluted 1:5; APC-conjugated antibodies were diluted 1:20. The stained cells were analyzed on a FACScalibur flow cytometer (BD Biosciences), which was equipped with red and blue lasers. Routine machine calibration was performed daily according to the standard operating procedures of our laboratory. Fluorescence compensation calibration was run at least once a week using standard fluorescence beads (Calibrite beads; BD Biosciences). Data analysis was performed using BD CellQuest™ Pro software version 6.0 (BD Biosciences), and lymphocytes were delineated using forward scatter/side scatter dot plots. Cell-surface antigens were defined as present when the fluorescence intensity of ≥20% of cells exceeded the fluorescence of the negative control.
Statistical analysis
For qualitative parameters, overall group differences were analyzed using a χ2 test. All statistical analyses were performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate statistical significance.
Results
Mutational analysis of DNM2
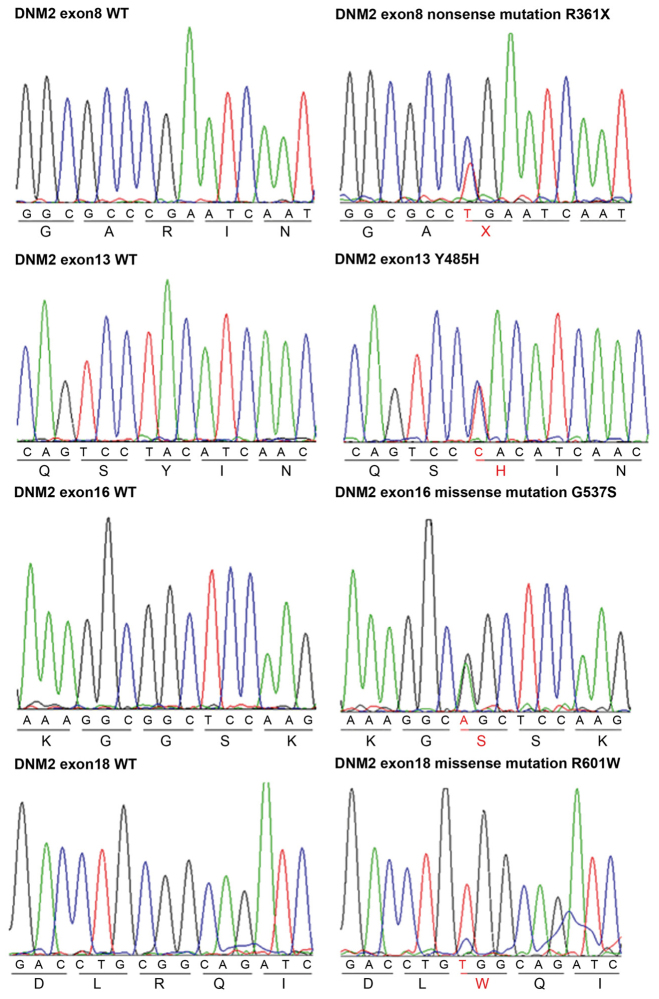
DNM2 mutations were identified in 4 out of 42 T-ALL patients, resulting in a mutation incidence of 9.5%. The 4 mutations were the point mutations c.1081C>T, c.1453T>C, c.1609G>A and c.1801C>T, which were located in exons 8, 13, 16 and 18, respectively. In the 4 mutations, 1 mutation was a nonsense mutation and 3 mutations were missense mutations. All 4 mutations led to amino acid changes (R361X, Y485H, G537S and R601W). The R361X and Y485H mutations are located within the DNM2 MD; G537S and R601W mutations are located within the DNM2 PH domain (Fig. 1).
Figure 1.
Representative DNA sequencing chromatograms of T-cell acute lymphoblastic leukemia genomic DNA samples showing mutations in exons of DNM2. DNM2, dynamin 2; WT, wild type.
DNM2 mutation in combination with NOTCH1 and PHF6 mutation
The present study found that in these 4 patients, DNM2 mutations co-existed with NOTCH1 mutations. In 3 of the 4 patients, the DNM2 mutation co-existed with two NOTCH1 mutations (point mutations and/or indel mutations) located in NOTCH1 exons 26, 27 and 34. In addition, 3 of the 4 patients with DNM2 mutations had PHF6 mutations located in PHF6 exons 4, 5 and 8 (Table I).
Table I.
Co-existence of DNM2, NOTCH1 and PHF6 mutations in adult patients with T-cell acute lymphoblastic leukemia.
| DNM2 | NOTCH1 | PHF6 | ||||
|---|---|---|---|---|---|---|
| Patient ID | Mutation | Location | Mutation | Location | Mutation | Location |
| Mu1# | c.1081C>T, | exon 8 | c.4732_4734delGTG, | exon 26 | c.820C>T, | exon 8 |
| p.R361X | p.V1578delV, | p.R274X | ||||
| c.5094C>T, p.D1698D | exon 27 | |||||
| Mu2# | c.1453T>C, | exon 13 | c.5033T>C, p.L1678P | exon 27 | c.346C>T, | exon 4 |
| p.Y485H | c.7400C>A, p.S2467* | exon 34 | p.R116X | |||
| Mu3# | c.1609G>A, | exon 16 | c.4721T>C, p.L1574P | exon 26 | c.385C>T, | exon 5 |
| p.G537S | p.R129X | |||||
| Mu4# | c.1801C>T, | exon 18 | c.7427G>C, p.V7427L | exon 34 | ||
| p.R601W | c.7171delinsTTTT, | |||||
| p.Q2391fs*3 | ||||||
DNM2, dynamin 2; PHF6, PHD finger protein 6.
Clinical characteristics of the patients with DNM2 mutations
The clinical characteristics of the patients with DNM2 mutations are listed on Table II. No significant differences in clinical characteristics were observed between the patients with DNM2 mutations and those without mutations in terms of age (P=0.094), sex (P=1.000), T/B subtype diagnosis (P=1.000), initial WBC (P=0.453), HGB (P=0.602), platelets (P=0.950), lactate dehydrogenase (P=0.317), blasts in bone marrow (P=0.939), blasts in peripheral blood (P=0.900), immune phenotype CD34+ (P=0.098), CD10+ (P=0.866), CD19+ (P=1.000), CD20+ (P=1.000), CD33+ (P=1.000), CD2+ (P=0.525), CD3+ (P=0.535), CD5+ (P=0.777) and CD7+ (P=1.000), frequency of extramedullary infiltration on liver (P=0.888), spleen (P=0.204) and lymph nodes (P=1.000), and days for complete remission (P=0.234). However, it was found that 1 patient experienced relapse, 1 patient had a high WBC count, and 2 patients had complex karyotypes and lymph node infiltration. Notably, the time taken to reach complete remission was >4 weeks in all 4 patients. These data suggest that the patients with DNM2 mutation exhibit high-risk leukemia and possess a poor prognosis.
Table II.
Clinical characteristics of T-ALL patients with DNM2 mutations.
| Mutation | |||||
|---|---|---|---|---|---|
| Characteristics | Mu1# | Mu2# | Mu3# | Mu4# | No mutation, median (range) |
| Age, years | 27 | 26 | 26 | 14 | 30.0 (14.0–70.0) |
| Gender | F | M | M | M | F/M |
| Diagnosis | T-ALL | T-ALL | T-ALL | T-ALL | T-ALL |
| DNM2 mutations | R361X | Y485H | G537S | R601W | No |
| WBC, nx109/l | 2.9 | 106.7 | 28.9 | 54.4 | 44.2 (1.0–546.0) |
| HGB, g/l | 117 | 124 | 92 | 114 | 115.5 (56.0–171.0) |
| PLT, nx109/l | 85 | 76 | 46 | 56 | 58.5 (17.0–267.0) |
| LDH, U/l | 934 | 731 | 861 | 588 | 1,144.0 (131.0–8601.0) |
| Blasts in bone marrow, % | 56.4 | 95.6 | 90.2 | 72.4 | 76.0 (20.0–100.0) |
| Blasts in peripheral blood, % | 6.0 | 68.0 | 88.0 | 89.0 | 64.0 (0.0–100.0) |
| Immune phenotypea | |||||
| CD34+ | Negative | Negative | Negative | Negative | |
| CD10+ | Negative | Negative | Positive | Negative | |
| CD19+ | Negative | Negative | Negative | Negative | |
| CD20+ | Negative | Negative | Negative | Negative | |
| CD33+ | Negative | Positive | Negative | Negative | |
| CD2+ | Negative | Positive | Negative | Positive | |
| CD3+ | Negative | Positive | Positive | Negative | |
| CD5+ | Positive | Positive | Negative | Positive | |
| CD7+ | Positive | Positive | Positive | Positive | |
| Extramedullary infiltration | |||||
| Liver | No | No | No | No | |
| Spleen | No | No | No | No | |
| Lymph node metastasis | No | No | No | Yes | |
| Karyotype | 46, XX[20] | 46, XY, der (1), 9p-, 14q+[3]/47, XY, t(6:11;) (p10;p10), +?8,9p-[1]/46, XY[6] | 46, XY[20] +[3][inc]/46, XY[1] 2013.1.7: 47–48, XY, −1,2q-, −4,4q-, −5, +8,9p-, der (9), +11,11q-*2,12q+, 16q+, 17q-, +2mar[7cp]/46, XY[3] | 46, XY, 11q | |
| Rearrangement | TCR/TCR | TCR/TCR | TCR/TCR | TCR/TCR | |
| Time to complete remission, days | 61 | 43 | 60 | 65 | 28 (9–169) |
Cell-surface antigens were defined as present when the fluorescence intensity of ≥20% of cells exceeded the fluorescence of the negative control. F, female; M, male; T-ALL, T-cell acute lymphoblastic leukemia; DNM2, dynamin 2; WBC, white blood cell; HGB, hemoglobin; PLT, platelet; LDH, lactate dehydrogenase; CD, cluster of differentiation; TCR, T cell receptor.
DNM2 synonymous amino acid mutations
DNM2 synonymous amino acid mutations were identified in exon 4 (1/42 patients; 2.38%), exon 6 (1/42 patients; 2.38%), exon 22 (1/42 patients; 2.38%), and exon 20 (34/42 patients; 80.95%) in the T-ALL patients. There were two synonymous amino acid mutations, Ala713Ala and Asp720Asp, in exon 20, and the former mutation was found in 31 patients (31/42 patients; 73.81%) and the latter in 3 patients (3/42 patients; 7.14%). The coexistence of the two mutations was identified in 1 case (1/42 patients; 2.38%) (Table III). The clinical features of the patients with DNM2 synonymous amino acid mutations were also observed, but these synonymous amino acid mutations were not significantly associated with any clinical features observed (data not shown).
Table III.
Mutations of synonymous amino acids in exons of DNM2.
| Mutation, Exon | Mutation, Cases, n | nucleotide | amino acid |
|---|---|---|---|
| 4 | 1 | c.450A>G | p.P150P |
| 6 | 1 | c.789G>A | p.P263P |
| 20 | 31 | c.2139T>C | p.A713A |
| 20 | 3 | c.2160C>T | p.D720D |
| 20 | 1 | c.2139T>C+ | p.A713A+ |
| c.2160C>T | p.D720D | ||
| 22 | 1 | c.2571G>A | p.R857R |
DNM2, dynamin 2.
Discussion
The DNM2 protein is involved in a wide range of cellular functions, including phagocytosis, phagosome formation of actin and microtubule interactions, cytokinesis, cell migration and regulation of apoptosis (19). It has been reported that DNM2 gene mutation is an important factor for patients with autosomal dominant centronuclear myopathy (20) and Alzheimer's disease (21,22).
Recurrent DNM2 mutations have also been identified in patients with ETP-ALL by whole-exome sequencing (6,10,23). These mutations include: E78fs in the Ras-like GTPase domain; L3354P, R364C, K382E, T404N and E468* in the dynamin MD domain; S528fs, E544fs and K557_K558>K in the PH domain; S698L in the GTPase effector domain; and K770*, P791T, L789fs and I805fs in the C-terminus. In addition, it has been shown that certain mutations appear in patients that experience relapse with induction failure. The present study identified 4 novel DNM2 mutations in 42 adult T-ALL patients, with a mutation rate of 9.5%. These mutations also mainly appear in the dynamin central region (MD domain), PH domain and GTPase effector domains. It was also found that the patients with DNM2 mutations were more likely to demonstrate high-risk factors, such as a high WBC count, complex karyotype, lymph node infiltration and difficulty achieving complete remission. These data indicated that patients with T-ALL with DNM2 mutations have a poor outcome.
Patient Mu1# was diagnosed with a type of hypoproliferative leukemia termed ‘hypocellular leukemia’, with less tolerance to chemotherapy and poor clinical outcome compared to individual's with normal/increased WBC counts. Hypocelluar leukemia is most commonly observed in cases of acute myeloid leukemia, and certain cases are secondary leukemia resulting from myelodysplastic syndrome, indicating high-risk leukemia (24,25). The mechanism of hypoproliferative leukemia is not fully understood. The present study identified that 5 immunotypes were present in patient Mu2#, but only 2–3 immunotypes were present in the other 3 patients with detected mutations. Only CD33 was observed in patient Mu2#, suggesting this immunotype may lead to a poorer outcome compared with those observed in the other 3 patients. In addition, the association of the mutations with survival of the patients requires clarification with more patients with the mutations in future.
DNM2 can bind to membrane phospholipids through PH domains and the endogenous GED can activate GTPase activity. In addition, high DNM2 expression is associated with a poor prognosis and high rate of metastasis in patients with solid cancer. Inhibition of DNM2 induces the apoptosis of cancer cells (8–10). Therefore, the present authors hypothesize that the DNM2 mutations identified in the present study may be gain-of-function mutations. The effects of these newly-identified DNM2 mutations on the proliferation of leukemia cells in the current study may be examined in future studies.
Notably, a high rate of DNM2 synonymous amino acid mutations (also termed silent mutations) was identified in the patients. Silent mutations are the evolutionary substitution of one base for another in an exon of a gene coding for a protein, such that the produced amino acid sequence is not modified. However, mutations do not always result in silent mutations (26–28). The point mutation may affect transcription, splicing, mRNA transport and translation, any of which may alter the phenotype, rendering the synonymous mutation non-silent (26–28).
In the present study, DNM2 expression was not observed in the patients with DNM2 silent mutations, and no significant changes in DNM2 expression were found in the patients with silent mutations compared to patients without mutations. In addition, no significant association between the DNM2 silent mutations and any clinical features was observed. These data indicated that the sites with silent mutations may only be the hot spots and the nucleotides in these sites are easily changed, but the nucleotide changes may also be quickly corrected. The clinical relevance of DNM2 silent mutations requires additional clarification. It was hypothesized in the present study that silent mutations may affect DNM2 translation in patients (26–28).
In summary, the present study identified 4 novel DNM2 mutations in T-ALL and their associations with high-risk leukemia. The current findings suggest the DNM2 mutations may be involved in the oncogenesis of T-ALL.
Acknowledgements
This study was supported by the following organizations: The National Natural Science Foundation of China (grant nos. 81270613 and 30973376); Jiangsu Province Key Medical Talents (grant no. RC2011077); The Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (39th); China Postdoctoral Science Foundation (grant no. 20090461134); Special Grade of the Financial Support from China Postdoctoral Science Foundation (grant no. 201003598); The Six Great Talent Peak Plan of Jiangsu (grant no. 2010-WS-024); The Scientific Research Foundation for the Returned Overseas Chinese Scholars, Nanjing Municipal Bureau of Personnel (2009); and Project of National Key Clinical Specialty, and Project funded by Jiangsu Provincial Special Program of Medical Science (grant no. BL2014086). The study was also supported partially by the Four Diamond Foundation of Pennsylvania State University.
References
- 1.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 2.Mody R, Li S, Dover DC, Sallan S, Leisenring W, Oeffinger KC, Yasui Y, Robison LL, Neglia JP. Twenty-five-year follow-up among survivors of childhood acute lymphoblastic leukemia: A report from the childhood cancer survivor study. Blood. 2008;111:5515–5523. doi: 10.1182/blood-2007-10-117150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiaretti S, Foà R. T-cell acute lymphoblastic leukemia. Haematologica. 2009;94:160–162. doi: 10.3324/haematol.2008.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 5.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: Insights and treatment implications. Nat Rev Clin Oncol. 2015;12:344–357. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M, Vosberg S, Schlee C, Heesch S, Schwartz S, Gökbuget N, Hoelzer D, Graf A, Krebs S, Bartram I, et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu B, Teng LH, Silva SD, Bijian K, Al Bashir S, Jie S, Dolph M, Alaoui-Jamali MA, Bismar TA. The significance of dynamin 2 expression for prostate cancer progression, prognostication, and therapeutic targeting. Cancer Med. 2014;3:14–24. doi: 10.1002/cam4.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razidlo GL, Wang Y, Chen J, Krueger EW, Billadeau DD, McNiven MA. Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1. Dev Cell. 2013;24:573–585. doi: 10.1016/j.devcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chircop M, Perera S, Mariana A, Lau H, Ma MP, Gilbert J, Jones NC, Gordon CP, Young KA, Morokoff A, et al. Inhibition of dynamin by dynole 34-2 induces cell death following cytokinesis failure in cancer cells. Mol Cancer Ther. 2011;10:1553–1562. doi: 10.1158/1535-7163.MCT-11-0067. [DOI] [PubMed] [Google Scholar]
- 10.Neumann M, Heesch S, Schlee C, Schwartz S, Gökbuget N, Hoelzer D, Konstandin NP, Ksienzyk B, Vosberg S, Graf A, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 11.Guo X, Zhang R, Liu J, Li M, Song C, Dovat S, Li J, Ge Z. Characterization of LEF1 high expression and novel mutations in adult acute lymphoblastic leukemia. PLoS One. 2015;10:e0125429. doi: 10.1371/journal.pone.0125429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin ZK, Zhang R, Ge Z, Liu J, Guo X, Qiao C, Wu YJ, Qiu HR, Zhang JF, Li JY. Characteristics of NOTCH1 mutation in adult T-cell acute lymphoblastic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21:1403–1408. doi: 10.7534/j.issn.1009-2137.2013.06.008. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Zhang R, Ge Z, Xu JY, Li M, Qiao C, Qiu HR, Li JY. Mutations of FBXW7 in adult T-cell acute lymphocytic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23:612–618. doi: 10.7534/j.issn.1009-2137.2015.03.002. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Qiu H, Jiang H, Wu L, Dong S, Pan J, Wang W, Ping N, Xia J, Sun A, et al. Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica. 2011;96:1808–1814. doi: 10.3324/haematol.2011.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer LG, Slovak ML, Campbell LJ, editors. Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. Karger; Basel, Switzerland: 2009. An International System for Human Cytogenetic Nomenclature. [Google Scholar]
- 17.Chen HY, Ge Z, Wu YJ, Wu LY, Sun M, Tian T, Qiou HR, Liu P, Li JY. Immunophenotypic analysis of Philadelphia chromosome positive acute lymphoblastic leukaemia in adults. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18:714–717. (In Chinese) [PubMed] [Google Scholar]
- 18.Miao Y, Fan L, Wu YJ, Xia Y, Qiao C, Wang Y, Wang L, Hong M, Zhu HY, Xu W, Li JY. Low expression of CD200 predicts shorter time-to-treatment in chronic lymphocytic leukemia. Oncotarget. 2016;7:13551–13562. doi: 10.18632/oncotarget.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippé J, González-García S, Toribio ML, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bitoun M, Maugenre S, Jeannet PY, Lacène E, Ferrer X, Laforêt P, Martin JJ, Laporte J, Lochmüller H, Beggs AH, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 22.Aidaralieva NJ, Kamino K, Kimura R, Yamamoto M, Morihara T, Kazui H, Hashimoto R, Tanaka T, Kudo T, Kida T, et al. Dynamin 2 gene is a novel susceptibility gene for late-onset Alzheimer disease in non-APOE-epsilon4 carriers. J Hum Genet. 2008;53:296–302. doi: 10.1007/s10038-008-0251-9. [DOI] [PubMed] [Google Scholar]
- 23.Koutsopoulos OS, Kretz C, Weller CM, Roux A, Mojzisova H, Böhm J, Koch C, Toussaint A, Heckel E, Stemkens D, et al. Dynamin 2 homozygous mutation in humans with a lethal congenital syndrome. Eur J Hum Genet. 2013;21:637–642. doi: 10.1038/ejhg.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai K, Kohno T, Chen YX, Tsushima H, Mori H, Nakamura H, Jinnai I, Matsuo T, Kuriyama K, Tomonaga M, Bennett JM. Diagnostic criteria for hypocellular acute leukemia: A clinical entity distinct from overt acute leukemia and myelodysplastic syndrome. Leuk Res. 1996;20:563–574. doi: 10.1016/0145-2126(95)00136-0. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Fu W, Wang L, Gao L, Lü S, Xi H, Qiu H, Chen L, Chen J, Ni X, et al. HAG regimen improves survival in adult patients with hypocellular acute myeloid leukemia. Oncotarget. 2016;7:3623–3634. doi: 10.18632/oncotarget.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 27.Goymer P. Synonymous mutations break their silence. Nat Rev Genet. 2007;8:92. doi: 10.1038/nrg2056. [DOI] [Google Scholar]
- 28.Zhou T, Ko EA, Gu W, Lim I, Bang H, Ko JH. Non-silent story on synonymous sites in voltage-gated ion channel genes. PLoS One. 2012;7:e48541. doi: 10.1371/journal.pone.0048541. [DOI] [PMC free article] [PubMed] [Google Scholar]