Abstract
Aberrant smooth muscle cells (SMCs) play important roles in the formation of abdominal aortic aneurysm (AAA). Although the molecular mechanism of AAA formation has been investigated, there is a lack of understanding concerning the role of microRNAs (miRNAs) in AAA, which the current study aimed to address. Firstly, miRNA array analysis was performed in order to compare the miRNA profiles in a mouse model of AAA with those in normal control mice, and differentially expressed miRNAs were identified. miR-129-5p was selected for further analysis, and was used to transfect human SMCs. The results of an MTT assay revealed that miR-129-5p inhibited the proliferation of SMCs, and flow cytometry indicated that apoptosis was induced. Bioinformatic analysis predicted that Wnt5a was the potential target gene of miR-129-5p, and this was verified by luciferase assay. In summary, miR-129-5p inhibits cellular proliferation, induces apoptosis and modulates the Wnt5a signaling pathway in SMCs.
Keywords: abdominal aortic aneurysm, miRNA-129-5p, Wnt5a
Introduction
In the development and progression of vascular diseases such as abdominal aortic aneurysm (AAA), smooth muscle cells (SMCs) play very important roles (1–3). Due to lack of terminal differentiation, SMCs have the ability to proliferate, migrate and synthesize extracellular matrix (ECM) (4). In certain pathological conditions such as atherosclerosis, the proliferation and cell cycling of SMCs is considered to be dangerous (5). However, in other conditions, such as aneurysmal dilation, the regrowth of SMSs is considered to be beneficial (6). The microenvironment affects the proliferation of SMCs, and SMC growth is stimulated by a variety of extracellular and intracellular factors (7–9).
It has been identified that microRNAs (miRNAs) play a significant role in determining the fate and behavior of SMCs (10). miRNAs are small non-coding RNAs comprising ~22 nucleotides that can lead to the inhibition of target gene expression (11,12). Previous studies have demonstrated that miRNAs are involved in various cellular functions, including differentiation, growth and development in vascular-associated diseases (12,13). miRNAs regulate target gene expression in angiogenesis, endothelial cell function and vascular inflammation (14). However, little is known about how miRNAs function in SMCs.
In the present study, the aim was to investigate the roles of miRNAs in SMC pathobiology. An miRNA array was used to identify the significant miRNAs in a mouse model of AAA. The effect of one of the most differentially expressed miRNAs identified by the array on SMC proliferation and apoptosis was further investigated. Its target gene was also predicted and verified.
Materials and methods
Murine models of AAA
A total of 20 male ApoE−/− mice (10 weeks old; n=4 per group) were purchased from Better Biotechnology Co., Ltd. (Nanjing, China). The mice were maintained in cages with food and water ad libitum, and with a 12 h light/dark cycle (room temperature, 23–25°C). AAA models were established according to a previously reported procedure (15). Briefly, 10% hydrated chlorine aldehyde was used to anesthetize the mice (0.3 ml/100 g body weight). The infra-renal abdominal aortas of the mice were isolated and temporary ligatures were applied. Saline solution containing 4.5 U/ml type 1 porcine pancreatic elastase (E1250; Sigma-Aldrich, St. Louis, MO, USA) was instilled for 5 min at 100 mmHg. The aortotomy was closed with a 10-0 suture, and aortic flow was restored. Mice were sacrificed 7 days after modeling by peritoneal injection of pentobarbital (50 mg/kg). Aortas were harvested from the mice and immediately flash-frozen in liquid nitrogen. The SMCs were isolated from the aortas according to a previously described method (15). The study was approved by the Animal Experiments Ethics Committee of the Affiliated Hospital of Nanjing University Medical School (Nanjing, China).
Cell culture
Human SMCs (T/G HA-VSMC) obtained from X-Y Biotechnology (Shanghai, China) were cultured in SmGM-2 Smooth Muscle Growth Medium-2 (Lonza, Walkersville, MD, USA) with 5% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Cells were harvested for RNA or protein analysis at ~90% confluence.
miRNA array hybridization and analysis
The RNA from the SMCs was isolated following homogenization using a mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA), which was then used to conduct the RT-qPCR. RNA samples from normal (control) and AAA model mice were prepared for analysis using an Agilent Mouse miRNA 8×15K Array kit (Agilent Technologies, Inc., Santa Clara, CA, USA). This array consisted of probes for 135 mice miRNAs from the Sanger database, version 10.1. The analysis was conducted according to the manufacturer's protocol. Significant miRNAs were those that showed a >2.0-fold difference in expression from the control. The most differentially expressed miRNAs were confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR
Human SMCs were transfected with miR129-5p or miR control. Total RNA was extracted from the mouse or transfected SMCs using an miRNeasy Mini kit (Qiagen, Valencia, CA, USA). Total RNA was reverse transcribed to cDNA using M-MuLV reverse transcriptase (Promega Corporation, Madison, WI, USA). A Taqman® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) used the stem-loop method to detect the expression levels of mature miRs. Total RNA (10 ng) was reverse transcribed and mixed with specific stem-loop primers (Applied Biosystems; Thermo Fisher Scientific, Inc.), and then qPCR was performed using an ABI PRISM 7900HT Sequence Detection System (Thermo Fisher Scientific, Inc.). miRNA amplification was carried out using the GenoExplorer miRNA qRT-PCR kit (Genosensor Corporation, Tempe, AZ, USA). The primer sequences of miR-129-5p and miRNA control were as follows: miR-129-5p, forward: 5′-ACACTCCTTTTTGCGTCTGGGCTTGC-3′ and reverse: 5′-TGGTGTCGTGGAGTCG-3′; miRNA control, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′; Wnt5a forward: 5′-CTTGGTGGTCGCTAGGTATGAAT-3′ and reverse: 5′-GGTGTTATCCACAGAGTGCTGC-3′; β-actin forward: 5′-CCTGGAGAAGAGCTATGAGCTGCCTG-3′ and reverse: 5′-CGATCCACACAGAGTACTTGCGC-3′. Relative mRNA quantification was determined with respect to the mean of β-actin. Relative miR quantification was determined with respect to the mean of RNU48. The relative expression levels of the miRNAs were evaluated with the 2−ΔΔCq method. Experiments were conducted in triplicate.
Luciferase assay
Wnt5a was predicted as the target gene of miR-129-5p by TargetScan (http://www.targetscan.org and http://www.mirbase.org). The 3′ untranslated region (3′UTR) of Wnt5a containing a potential binding site for miR-129-5p was amplified and cloned into a pGL4 luciferase reporter vector (Promega Corporation, Madison, WI, USA). Site-directed mutagenesis of the miR-129-5p binding site in the Wnt5a 3′UTR was performed using the GeneTailor Site-Directed Mutagenesis System (Invitrogen; Thermo Fisher Scientific). The pGL4 vector contains a firefly luciferase reporter gene. SMCs were co-transfected with miR-129-5p, or miRNA control combined with Wnt5a 3′-UTR for 24 h and then luciferase activity was measured. Luciferase activity was measured 36 h after cell transfection using the Dual-Luciferase Reporter Assay System (Promega Corporation). Values were normalized with firefly luciferase activity.
Cell transfection
Lentivirus of miR-129-5p or its control were produced in 293T cells transfected with the miRNA vector. Human SMCs were seeded in triplicate into 6-well plates at a density of 1×105 cells/well, and were grown to 30–50% confluence at 37°C in an atmosphere containing 5% CO2. Turbofect transfection reagent (Thermo Fisher Scientific, Inc.) was used for transfection, according to the manufacturer's instructions. Human SMCs were transfected with the indicated plasmid or nucleotides using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific). After 24 h of transfection, the cells were seeded into 96-well culture plates at a density of 2×103 cells/well. The cell proliferation of the SMCs was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich) assay according to the manufacturer's protocol.
Detection of cell apoptosis
Human SMCs were transfected with miR-129-5p or miR control. After 48 h of transfection, the cells were washed with phosphate-buffered saline and then stained with 5 µl Annexin V and 5 µl propidium iodide for 15 min at room temperature in the dark in accordance with the manufacturer's protocol (BD Biosciences, San Jose, CA, USA). The apoptosis was analyzed using flow cytometry.
Western blot analysis
Transfected SMCs were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.) containing 1X protease inhibitor cocktail, and protein concentrations were determined using the Bradford assay (Bio-Rad Laboratories, Inc., Philadelphia, PA, USA). Protein lysates (50 µg) were loaded onto 10% SDS-PAGE gels, fractionated, and electroblotted onto nitrocellulose membranes (Millipore, Bedford, MA, USA) at 80 V for 2 h. After blocking in 5% non-fat dry milk, the membranes were incubated with primary antibodies (1:1,000) overnight at 4°C and then incubated with goat anti-rabbit (1:2,000; cat. no. sc-2301; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and goat anti-mouse (1:3,000; cat. no. sc-2031; Santa Cruz Biotechnology, Inc.) secondary antibodies conjugated with horseradish peroxidase for 1 h at room temperature. The following primary antibodies were used: Mouse monoclonal Wnt-5a antibody (A-5; sc-365370), rabbit polyclonal Rac 1 antibody (C-11; sc-95), rabbit polyclonal Rho A antibody (119; sc-179) and mouse monoclonal glucreraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (G-9; sc-365062) were purchased from Santa Cruz Biotechnology, Inc. Protein bands were visualized by enhanced chemiluminescence using LumiGLO Chemiluminescent Substrate (KPL Inc., Gaithersburg, MD, USA).
Statistical analysis
The study data were analyzed using SPSS software, version 13.0 (SPSS, Inc., Chicago, IL, USA). Results are presented as the mean ± standard error of the mean of at least three independent experiments. Comparisons of two independent groups were analyzed using the two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA profile of SMCs from AAA models
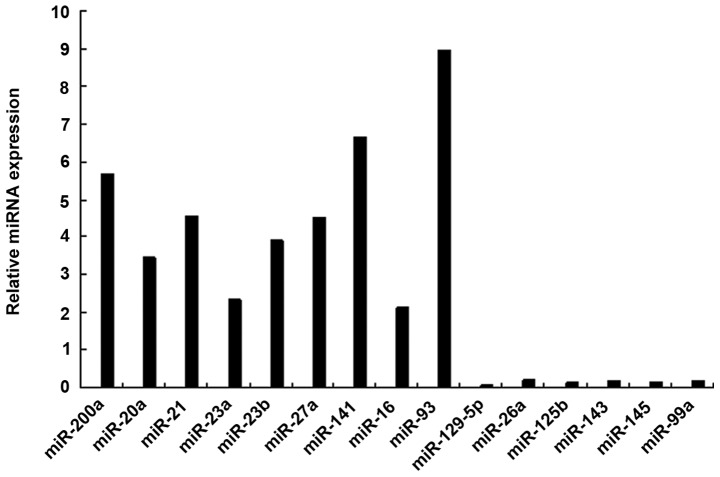
To identify the miRNAs that differed in expression between AAA model and normal mice, a customized miRNA microarray that contained 135 mouse miRNAs was used to analyze miRNA expression levels. miRNAs were considered differentially expressed if their M values (log2-fold change values) were >1.0 or <-1.0. There were 15 miRNAs that showed significant changes in ≥12 of the 20 AAA models (Table I). The upregulated miRNAs in the AAA model were miR-21, miR-20a, miR-93, miR-200a, miR-23a, miR-23b, miR-27a, miR-141 and miR-16, and the downregulated miRNAs were miR-129-5p, miR-26a, miR-125b, miR-143, miR-145 and miR-99a. Of these miRNAs, the most downregulated miRNA in the SMCs was miR-1259-5p. The results were confirmed by RT-qPCR analysis (Fig. 1).
Table I.
miRNA profile of AAA model mice.
| miRNA symbol | Upregulated/downregulated | Fold expressiona |
|---|---|---|
| has-miR-200a | Upregulated | 3.42 |
| has-miR-20a | Upregulated | 4.72 |
| has-miR-21 | Upregulated | 9.51 |
| has-miR-23a | Upregulated | 2.52 |
| has-miR-23b | Upregulated | 4.65 |
| has-miR-27a | Upregulated | 3.45 |
| has-miR-141 | Upregulated | 7.88 |
| has-miR-16 | Upregulated | 2.23 |
| has-miR-93 | Upregulated | 10.43 |
| has-miR-129-5p | Downregulated | 0.12 |
| has-miR-26a | Downregulated | 0.34 |
| has-miR-125b | Downregulated | 0.29 |
| has-miR-143 | Downregulated | 0.32 |
| has-miR-145 | Downregulated | 0.24 |
| has-miR-99a | Downregulated | 0.15 |
miRNA, microRNA; AAA, abdominal aortic aneurysm.
Fold expression AAA model vs. normal mice.
Figure 1.
miRNA profile in the SMCs of AAA models. miRNAs found to be changed by microarray were confirmed by reverse transcription-quantitative polymerase chain reaction. miRNA/miR, microRNA; SMC, smooth muscle cell; AAA, abdominal aortic aneurysm.
miR-129-5p inhibits SMC proliferation and induces apoptosis
Following the identification of miR-129-5p as a frequently downregulated miRNA in SMCs, its role in AAA was investigated. SMCs were infected with miR-129-5p using a lentiviral vector, and the expression of miR-129-5p was confirmed to be increased (Fig. 2A). Cell proliferation was evaluated by MTT assay. The results showed that miR-129-5p inhibited the proliferation of SMCs (Fig. 2B). Furthermore, flow cytometry data indicated that apoptosis was increased in the SMCs with miR-129-5p overexpression compared with that in the control (Fig. 2C).
Figure 2.

miR-129-5p inhibits SMC proliferation and induces apoptosis. (A) The expression level of miR-129-5p was tested in SMCs transfected with LV-miRNA-129-5p (miR-129-5p) and control (miR-control) by reverse transcription-quantitative polymerase chain reaction at 48 h after transfection. (B) The effect of transient transfection with miR-129-5p for 48 h on the growth of the SMCs was examined by MTT assay. (C) Flow cytometric analysis indicated that transfection with miR-129-5p induced apoptosis of the SMCs. Data are represented as the mean ± standard deviation. **P<0.01 vs. miR-control. miRNA/miR, microRNA; SMC, smooth muscle cell; LV, lentivirus; PI, propidium iodide; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Wnt5a is a target gene of miR-129-5p in SMCs
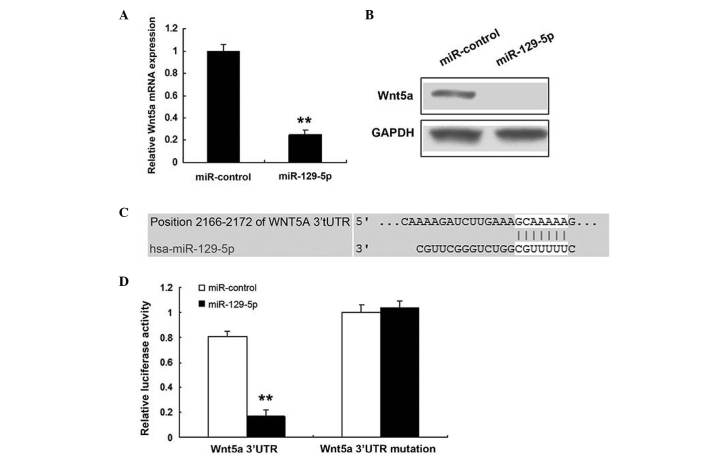
The prediction that miR-129-5p regulates endogenous Wnt5a expression in SMCs was investigated. Endogenous Wn5a mRNA levels (Fig. 3A) were downregulated when human SMCs were transfected with miR-129-5p, compared with those in the control. Western blotting demonstrated that the Wnt5a protein level was low in the cells transfected with miR-129-5p (Fig. 3B). Bioinformatic analysis indicated that Wnt5a would be directly suppressed by binding to miR-129-5p (Fig. 3C). As shown in Fig. 3D, the luciferase activity of Wnt5a in SMCs transfected with miR-129-5p was much lower than that in control cells. The luciferase activity of mutant Wnt5a was not affected by miR-129-5p.
Figure 3.
Wnt5a is the direct target of miR-129-5p in SMCs. (A) Effects of miR-125-5p on the expression of endogenous Wnt5a mRNA, as tested by reverse transcription-quantitative polymerase chain reaction. (B) Western blot analysis was used for monitoring Wnt5a expression in SMCs 48 h after transfection with miR-125-5p. (C) Binding between miR-125-5p and the miR-125-5p-binding site in the 3′UTR of Wnt5a. (D) Luciferase reporter assay with cotransfection of wild-type or mutant 3′UTR and miR-125-5p or miR-control in SMCs. Firefly luciferase activity of each sample was normalized against Renilla luciferase activity. Data from three separate experiments are presented as mean ± standard deviation. **P<0.01 vs. miR-control. miR, microRNA; SMC, smooth muscle cell; 3′UTR, 3′ untranslated region.
miR-129-5p inhibits SMC proliferation by targeting the Wnt5a signaling pathway
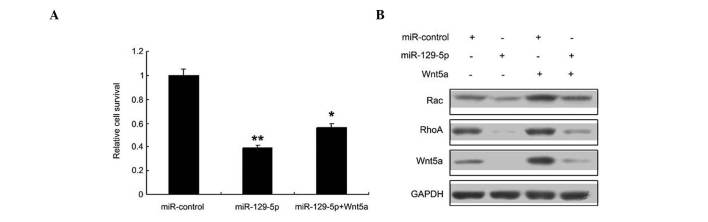
Whether miR-129-5p inhibits proliferation by targeting Wnt5a in SMCs was then investigated; it was found that miR-129-5p inhibited SMC proliferation, and its inhibitory effect was reduced when Wnt5a was overexpressed (Fig. 4A). Since the Wnt signaling pathway is an important pathway involved in SMC regulation, the levels of Wnt5a and downstream proteins in the Wnt signaling pathway, namely RhoA and Rac 1, were evaluated by western blotting (Fig. 4B). The results indicated that the Wnt5a signal pathway was inhibited in the SMCs transfected with miR-129-5p.
Figure 4.
miR-129-5p inhibits SMC proliferation by targeting the Wnt5a signaling pathway. (A) miR-129-5p inhibited the proliferation of SMCs, with and without Wnt5a overexpression. (B) Western blotting indicated that miRNA-129-5p inhibited the Wnt5a signaling pathway in SMCs. miR, microRNA; SMC, smooth muscle cell; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. *P<0.05, vs. the control and **P<0.01, vs. the control.
Discussion
Previous studies have indicated that several miRNAs play a role in vascular physiology, including miRNA-145, miRNA-21 and miRNA-221 (15,16). However, the mechanism of regulation of SMC function by miRNAs in AAA remains unknown. In the present study, the aim was to clarify this by performing miRNA microarray analysis of vascular SMCs from AAA models. Mouse AAA models were established and SMCs were isolated from the animals. Through the miRNA array assay, 15 miRNAs that were significantly upregulated or downregulated were identified. Further experiments were conducted to investigate the most notable miRNA, namely miR-129-5p, including its target genes and function.
miR-129-5p was selected as it was downregulated to a greater extent than other miRNAs such as miR-26a, miR-125b, miR-99a and miR-145. It was hypothesized that miR-129-5p could be a key regulator of SMC physiology. Despite its downregulation during SMC differentiation, it was found that the overexpression of miR-129-5p inhibits SMC proliferation. It was then postulated that miR-129-5p may be an inducer of cell apoptosis in SMCs. The results of flow cytometric analysis indicated that miR-129-5p serves as a stimulator of SMC apoptosis. The results of the present study identified that Wnt5a was a direct target gene of miR-129-5p. Previous reports demonstrated that Wnt5a-induced Wnt1-inducible secreted protein-1 suppresses vascular smooth muscle cell apoptosis induced by oxidative stress (16). Wnt5a is induced by TGF-β/Smad3 in rat aortic SMC and promotes its proliferation (17). Wnt5a is associated with the differentiation of bone marrow mesenchymal stem cells in vascular calcification by connecting with various receptors (18). Curculigoside A induces cell proliferation and angiogenesis via Wnt5a/β-catenin and VEGF/CREB/Egr-3/VCAM-1 signaling, and promotes maturation and stability of new blood vessels by increasing Ang1 and Tie-2 expression (19). Wnts are expressed during the differentiation of MSCs during calcification. MSCs are able to differentiate into various cell phenotypes when there is direct cell-cell contact with VSMCs or calcified VSMCs, and the Wnt5a/Ror2 pathway may be associated with the determination of differentiation of MSCs during this process (20).
In a conclusion, numerous significantly regulated miRNAs in a mouse model of AAA were identified using a microarray. Notably, miR-129-5p was suggested to be a potentially critical regulator of AAA development, an inhibitor of cell proliferation and inducer of apoptosis in SMCs. Wnt5a was identified as a target gene of miR-129-5p, and transfection with miR-129-5p downregulated Wnt5a mRNA and protein expression in SMCs. Further studies are required to investigate other biological effects of miR-129-5p in AAA.
Acknowledgements
The authors thank the Natural Science Foundation of Jiangsu Province, China for support (No: BK2009035).
References
- 1.Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H. Prevention of abdominal aortic aneurysm by anti-microRNA-712 or anti-microRNA-205 in angiotensin II-infused mice. Arterioscler Thromb Vasc Biol. 2014;34:1412–1421. doi: 10.1161/ATVBAHA.113.303134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biros E, Moran CS, Wang Y, Walker PJ, Cardinal J, Golledge J. MicroRNA profiling in patients with abdominal aortic aneurysms: The significance of miR-155. Clin Sci (Lond) 2014;126:795–803. doi: 10.1042/CS20130599. [DOI] [PubMed] [Google Scholar]
- 3.Maegdefessel L, Dalman RL, Tsao PS. Pathogenesis of abdominal aortic aneurysms: MicroRNAs, proteases, genetic associations. Annu Rev Med. 2014;65:49–62. doi: 10.1146/annurev-med-101712-174206. [DOI] [PubMed] [Google Scholar]
- 4.Maegdefessel L, Azuma J, Tsao PS. MicroRNA-29b regulation of abdominal aortic aneurysm development. Trends Cardiovasc Med. 2014;24:1–6. doi: 10.1016/j.tcm.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maegdefessel L, Spin JM, Adam M, Raaz U, Toh R, Nakagami F, Tsao PS. Micromanaging abdominal aortic aneurysms. Int J Mol Sci. 2013;14:14374–14394. doi: 10.3390/ijms140714374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheuk BLY, Cheng SWK. Identification and characterization of microRNAs in vascular smooth muscle cells from patients with abdominal aortic aneurysms. J Vasc Surg. 2014;59:20–209. doi: 10.1016/j.jvs.2013.02.244. [DOI] [PubMed] [Google Scholar]
- 7.Adam M, Raaz U, Spin JM, Tsao PS. MicroRNAs in abdominal aortic aneurysm. Curr Vasc Pharmacol. 2015;13:280–290. doi: 10.2174/15701611113119990015. [DOI] [PubMed] [Google Scholar]
- 8.Lee HJ, Yi JS, Lee HJ, Lee IW, Park KC, Yang JH. Dysregulated expression profiles of MicroRNAs of experimentally induced cerebral aneurysms in rats. J sKorean Neurosurg Soc. 2013;53:72–76. doi: 10.3340/jkns.2013.53.2.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golledge J, Kuivaniemi H. Genetics of abdominal aortic aneurysm. Curr Opin Cardiol. 2013;28:290–296. doi: 10.1097/HCO.0b013e32835f0d55. [DOI] [PubMed] [Google Scholar]
- 10.Kin K, Miyagawa S, Fukushima S, Shirakawa Y, Torikai K, Shimamura K, Daimon T, Kawahara Y, Kuratani T, Sawa Y. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc. 2012;1:e000745. doi: 10.1161/JAHA.112.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pahl MC, Derr K, Gäbel G, Hinterseher I, Elmore JR, Schworer CM, Peeler TC, Franklin DP, Gray JL, Carey DJ, et al. MicroRNA expression signature in human abdominal aortic aneurysms. BMC Med Genomics. 2012;5:25. doi: 10.1186/1755-8794-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, Leeper NJ, Raaz U, Schoelmerich AM, McConnell MV, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. 2012;4:122ra22. doi: 10.1126/scitranslmed.3003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ, et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. 2012;122:497–506. doi: 10.1172/JCI61598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milewicz DM. MicroRNAs, fibrotic remodeling and aortic aneurysms. J Clin Invest. 2012;122:490–493. doi: 10.1172/JCI62204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G, Huang Y, Lu X, Lu M, Huang X, Li W, Jiang M. Identification and characteristics of microRNAs with altered expression patterns in a rat model of abdominal aortic aneurysms. Tohoku J Exp Med. 2010;222:187–193. doi: 10.1620/tjem.222.187. [DOI] [PubMed] [Google Scholar]
- 16.Mill C, Monk BA, Williams H, Simmonds SJ, Jeremy JY, Johnson JL, George SJ. Wnt5a-induced Wnt1-inducible secreted protein-1 suppresses vascular smooth muscle cell apoptosis induced by oxidative stress. Arterioscler Thromb Vasc Biol. 2014;34:2449–2456. doi: 10.1161/ATVBAHA.114.303922. [DOI] [PubMed] [Google Scholar]
- 17.Jin Y, Wang W, Chai S, Liu J, Yang T, Wang J. Wnt5a attenuates hypoxia-induced pulmonary arteriolar remodeling and right ventricular hypertrophy in mice. Exp Biol Med (Maywood) 2015;240:1742–1751. doi: 10.1177/1535370215584889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan S, Wang Z, Xin F, Xin H. Wnt5a is associated with the differentiation of bone marrow mesenchymal stem cells in vascular calcification by connecting with different receptors. Mol Med Rep. 2014;10:1985–1991. doi: 10.3892/mmr.2014.2449. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, He J, Ye L, Lin F, Hou J, Zhong Y, Jiang W. Mechanisms of angiogenesis in a Curculigoside A-treated rat model of cerebral ischemia and reperfusion injury. Toxicol Appl Pharmacol. 2015;288:313–321. doi: 10.1016/j.taap.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Guan S, Wang Z, Xin F, Xin H. Wnt5a is associated with the differentiation of bone marrow mesenchymal stem cells in vascular calcification by connecting with different receptors. Mol Med Rep. 2014;10:1985–1991. doi: 10.3892/mmr.2014.2449. [DOI] [PubMed] [Google Scholar]