Abstract
The opportunistic bacteria of the Burkholderia cepacia complex (Bcc) are extremely pathogenic to cystic fibrosis (CF) patients, and acquisition of Bcc bacteria is associated with a significant increase in mortality. Treatment of Bcc infections is difficult because the bacteria are multidrug resistant and able to survive in biofilms. Short palate, lung, and nasal epithelial clone 1 (SPLUNC1) is an innate defense protein that is secreted by the upper airways and pharynx. While SPLUNC1 is known to have antimicrobial functions, its effects on Bcc strains are unclear. We therefore tested the hypothesis that SPLUNC1 is able to impair Bcc growth and biofilm formation. We found that SPLUNC1 exerted bacteriostatic effects against several Bcc clinical isolates, including B. cenocepacia strain J2315 (50% inhibitory concentration [IC50] = 0.28 μM), and reduced biofilm formation and attachment (IC50 = 0.11 μM). We then determined which domains of SPLUNC1 are responsible for its antimicrobial activity. Deletions of SPLUNC1's N terminus and α6 helix did not affect its function. However, deletion of the α4 helix attenuated antimicrobial activity, while the corresponding α4 peptide displayed antimicrobial activity. Chronic neutrophilia is a hallmark of CF lung disease, and neutrophil elastase (NE) cleaves SPLUNC1. However, we found that the ability of SPLUNC1 to disrupt biofilm formation was significantly potentiated by NE pretreatment. While the impact of CF on SPLUNC1-Bcc interactions is not currently known, our data suggest that understanding this interaction may have important implications for CF lung disease.
INTRODUCTION
The Burkholderia cepacia complex (Bcc) is comprised of 18 Gram-negative bacteria that, while phenotypically similar, are genetically distinct species (1–3). Although Bcc strains are commonly found in the environment, Bcc bacteria are a group of opportunistic pathogens associated with immunocompromised patients, such as those with cystic fibrosis (CF) (4, 5). Unlike Pseudomonas aeruginosa infections, which usually result in a relatively slow decline in CF lung function (6), Bcc infections are unusually virulent and are associated with a rapid decline in CF life expectancy (7, 8). Indeed, Bcc infections result in “cepacia syndrome,” which is characterized by pneumonia, deteriorating lung function, bacteremia, and increased mortality (9, 10). Treatment of Bcc infections is difficult because these pathogens are resistant to many antibiotics, including polymyxins, trimethoprim, quinolones, β-lactams, chloramphenicol, aminoglycosides, and antimicrobial peptides (11–13). Bcc infection is usually planktonic and invasive, and the bacteria survive intracellularly in pulmonary macrophages and respiratory epithelial cells (14, 15). Although biofilms are not typically observed, Bcc bacteria have been shown to form biofilms in vitro and to form mixed biofilms when cultured with P. aeruginosa (16–18). In addition, Bcc biofilms are more resistant to antibiotic cocktails than P. aeruginosa biofilms (19).
Short palate, lung, and nasal epithelial clone 1 (SPLUNC1) is a 25-kDa protein that is primarily secreted by the airways and nasopharynx (20). SPLUNC1, also known as PLUNC (palate, lung, and nasal epithelium clone), SPURT (secretory protein in upper respiratory tracts), LUNX (lung-specific X protein), NASG (nasopharyngeal carcinoma-related protein), and BPIFA1 (BPI fold-containing family A member 1), has also been found in saliva and nasal lavage fluids from healthy individuals, at concentrations ranging from 0.4 to 10 μM (21), and its expression levels increase greatly with inflammation (22, 23). SPLUNC1 is a multifunctional protein that regulates the epithelial sodium channel (ENaC) to modulate airway hydration levels (24, 25) as well as having surfactant-like properties and antimicrobial actions (21, 26). For example, SPLUNC1 is part of the bactericidal permeability-increasing (BPI) protein family (27) and has structural similarities to the BPI protein (28, 29). As part of the innate immune response to infections, SPLUNC1 has been shown to have antimicrobial and antibiofilm activities against many Gram-negative bacteria. Furthermore, SPLUNC1 knockout mice are more susceptible to Klebsiella pneumoniae and P. aeruginosa infections (26, 30). While SPLUNC1 has antimicrobial activity against P. aeruginosa, Haemophilus influenzae, and K. pneumoniae (22, 26, 30, 31), its effects against Bcc strains have only recently been examined and are not fully understood (32).
Airway epithelia utilize several host defense mechanisms, including mucociliary clearance (MCC), antimicrobial peptides, oxidative bursts, proteases, cytokines, and growth factors, to reduce bacterial invasion (15, 33–35). For example, Bcc infection stimulates inflammatory responses resulting in neutrophil influx into the lung (9, 36). CF airways have chronic inflammation and increased levels of cytokines, such as interleukin-1 (IL-1), IL-6, IL-8, and tumor necrosis factor alpha, as well as chronic neutrophilia and increased protease activity in the lung lumen, including increased neutrophil elastase (NE) activity (37, 38). Although NE is needed for killing of Gram-negative bacteria (39), increased levels of NE have been shown to cleave SPLUNC1 and have been proposed to impair airway epithelial defenses (25, 31). We recently showed that SPLUNC1 affects Burkholderia cenocepacia J2315 (32). However, little is known about SPLUNC1's ability to affect different Bcc strains. Since SPLUNC1 is the most abundantly expressed protein in the airways (29), we sought to fully understand its interaction with Bcc strains as a first step toward developing novel antibiotics against Bcc bacteria for the treatment of CF. In this study, we therefore tested SPLUNC1's antimicrobial activity against Bcc clinical isolates under planktonic and biofilm conditions. In addition, we tested the antimicrobial and antibiofilm effects of SPLUNC1 exposed to NE.
MATERIALS AND METHODS
Bacterial strains and media.
Bcc clinical isolates (Table 1) (obtained from John J. Lipuma, CFF Burkholderia cepacia Research Laboratory and Repository, University of Michigan Medical School), except for Bcc isolate K56-2 and the ΔhldE and ΔwbxE mutants (obtained from Miguel A. Valvano, Queens University, Belfast, Northern Ireland), P. aeruginosa PAO1, and Staphylococcus aureus CDL (obtained from Matthew Wolfgang, University of North Carolina at Chapel Hill) were grown in Luria broth (LB) at 37°C for 24 h with shaking at 300 rpm. The number of CFU per milliliter was determined by serial dilution plating on LB agar plates.
TABLE 1.
Burkholderia cepacia complex clinical isolates used in this study
| Species | Isolate |
|---|---|
| B. cenocepacia GIIIb | PHDC |
| AU19445 | |
| AU20454 | |
| B. cenocepacia GIIIa | AU21968 |
| J2315 | |
| K56-2 | |
| B. cepacia | AU25837 |
| AU25940 | |
| AU28001 | |
| PC763 | |
| B. multivorans | AU27629 |
| AU27847 | |
| AU28062 |
SPLUNC1 proteins.
A plasmid containing SPLUNC1 cDNA was transformed into BL21-Codon Plus competent cells (Agilent Technologies) and purified as previously described (40). After purification, all recombinant SPLUNC1 proteins were produced as described previously and stored at −80°C until required (40). The recombinant SPLUNC1 proteins included Δ19 SPLUNC1 (referred to as SPLUNC1), which lacks the cleavable N-terminal signal sequence (residues M1 to M19) but is otherwise full length; the S18 peptide, which corresponds to residues G22 to A39; the Δ44 mutant (residues T45 to V256), which lacks residues M1 to S43, including the S18/G22-to-A39 region; the α4 helix peptide (residues K77 to L101); the Δα4 mutant, which lacks the α4 helix (residues I76 to I105 and includes nonnative Gly-Ser-Gly-Ser linker to residues L75 to I106); the α6 helix peptide (residues I252 to V256); and the Δα6 mutant, which lacks the α6 helix (residues I242 to V256).
Antimicrobial assay.
The antimicrobial activity of SPLUNC1 was tested by incubating Bcc strains in the presence of various concentrations of SPLUNC1 or SPLUNC1 mutants. The bacterial cultures were grown overnight at 37°C and 300 rpm. After 24 h, bacteria at 106 CFU/ml were added to round-bottomed 96-well plates (Corning Incorporated) with increasing doses of SPLUNC1. Plates were incubated at 37°C for 24 h, and bacterial growth was measured by determining the optical density at 600 nm (OD600) by using a Tecan Sunrise plate reader. Samples were also collected at 24 h, serially diluted in Ringer's solution, and plated on LB agar plates to determine the number of CFU per milliliter. Percent inhibition was then determined using the following equation: % inhibition = [(CFU/ml from vehicle − CFU/ml from SPLUNC1 present)/(CFU/ml from vehicle)] × 100.
Antibiofilm assay.
The antibiofilm activity of SPLUNC1 was tested by incubating Bcc strains in the presence of increasing concentrations of SPLUNC1 or SPLUNC1 mutants. For biofilm inhibition, SPLUNC1 was coincubated with 106 CFU/ml Bcc strains in flat-bottomed 96-well plates for 24 h. For disruption of biofilms, bacteria at 106 CFU/ml were added to flat-bottomed 96-well plates and incubated for 24 h for biofilm formation, and 0.4 μM SPLUNC1 was then added for a further 1 or 24 h. Plates were incubated at 37°C and then washed. Biofilms were fixed with methanol and stained with 1% crystal violet. After rinsing with distilled water, the stained biofilms were resolubilized with 33% acetic acid. Biofilm formation was measured by determining the OD590 by using a Tecan Sunrise plate reader.
Attachment assay.
Bcc cultures were grown overnight, adjusted to an OD600 of 1.0, and added to flat-bottomed 96-well plates. SPLUNC1 was added at 0.4 μM and incubated for 1 to 3 h. Attachment was measured by 1% crystal violet staining and determination of the OD590 as previously described (41).
Cleavage of SPLUNC1 by proteases.
SPLUNC1 (40 μM) was incubated alone or with 1 μM neutrophil elastase (NE; Elastin Product Company) at 37°C for up to 24 h. NE alone (1 μM) was used as a control. To stop NE activity, 1 μM sivelestat (NE inhibitor ONO5046; Sigma) was added, and the samples were placed immediately on ice. An aliquot of the sample was denatured and run in a 4 to 15% Mini-Protean TGX SDS-PAGE gel (Bio-Rad). Gels were stained by Coomassie brilliant blue R-250 (Thermo Scientific) and visualized by a Bio-Rad Chemidoc instrument. To test for antimicrobial and antibiofilm activities, samples were then diluted 1:100, to a final concentration of 0.4 μM, and incubated with 106 CFU/ml B. cenocepacia J2315 for 24 h at 37°C. Bacterial growth and biofilm formation were measured as described above.
Neutrophil elastase activity assay.
Inhibition of NE activity by sivelestat was confirmed by incubating 1 μM NE and 10 μM Suc-Ala-Ala-Ala-MCA substrate (MAA-3133-v; Peptides International) with or without 1 μM sivelestat for 90 min at 37°C. Substrate fluorescence was measured every 5 min as an indicator of NE activity at excitation/emission wavelengths of 380/460 nm, using a Tecan Infinite M1000 multiplate reader.
Multiangle static light scattering.
SPLUNC1 (5 mg/ml) was treated with 5 μM NE in 50 mM HEPES, 150 mM NaCl, pH 7.4, and 0.02% azide for up to 24 h at 37°C, and the treatment was stopped with 1.25 μM sivelestat. Samples were injected onto a GE Superdex S200 size-exclusion column connected to a multiangle light scattering instrument (Dawn EOS; Wyatt Technologies) and a refractometer (Optilab T-rEX; Wyatt Technologies). The molecular weight of the sample eluting for each peak was calculated based on light scattering and refractive index data by using the ASTRA 6 software package (Wyatt Technologies). A dn/dc value of 0.185 was assumed.
Circular dichroism.
SPLUNC1 was placed in circular dichroism (CD) buffer containing 10 mM potassium phosphate (pH 7.4) and 150 mM potassium fluoride. SPLUNC1 (40 μM) was treated with 1 μM NE for various times, and then NE was inhibited with 1 μM sivelestat. Samples were diluted to 10 μM in the buffer described above and loaded into 1-mm cuvettes. Using a Chirascan-Plus instrument (Applied Photophysis Limited), spectra were recorded from 185 to 280 nm at 20 ± 1.0°C. Measurements were corrected for the background signal by using CD buffer containing 1 μM NE and 1 μM sivelestat without SPLUNC1.
Statistical analysis.
All data are shown as means ± standard errors. Data were analyzed using Prism software (GraphPad Software, Inc.). Nonparametric one-way analysis of variance (ANOVA; Kruskal Wallis) was used to compare multiple groups. P values of <0.05 were considered statistically significant. All experiments were performed a minimum of three times.
RESULTS
Antimicrobial activity of SPLUNC1 against Bcc strains.
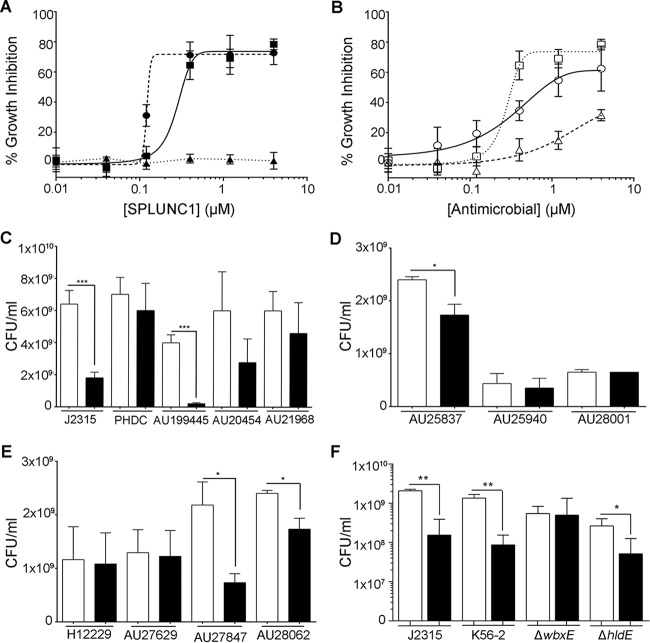
The Bcc epidemic Edinburg-Toronto (ET)-12 strain B. cenocepacia J2315 is known to cause cepacia syndrome (42). Therefore, SPLUNC1's antimicrobial effects were initially tested against this strain in a dose-dependent manner. SPLUNC1 was found to have antimicrobial activity against J2315, with a 50% inhibitory concentration (IC50 = 0.28 μM) similar to that observed for P. aeruginosa (IC50 = 0.12 μM) (Fig. 1A). We next tested SPLUNC1's ability to affect a Gram-positive bacterium (S. aureus). Consistent with previous reports that SPLUNC1 affects only Gram-negative bacteria (22, 30), S. aureus was insensitive to SPLUNC1. We also found that SPLUNC1 was more potent than tobramycin (IC50 = 0.33 μM) to reduce growth of J2315, while the antibiotic polymyxin B, to which B. cenocepacia is resistant, had relatively little effect (Fig. 1B) (43). To test SPLUNC1's ability to affect the growth of other Bcc clinical isolates, a physiological concentration of 0.4 μM SPLUNC1, which is comparable to that found in nasal and saliva lavage fluids from healthy humans (21), was added along with Bcc clinical isolates at time zero, and bacterial growth was determined 24 h later. The data indicate that SPLUNC1 significantly affected the growth of many, but not all, of the Bcc clinical isolates that were tested (Fig. 1C to E). Increasing concentrations of up to 4 μM SPLUNC1 were also tested against the Bcc clinical isolates that were not initially susceptible, and these isolates remained insensitive to SPLUNC1 (n = 3) (data not shown). These data suggest that susceptibility to SPLUNC1 is strain dependent. SPLUNC1 has previously been shown to bind to bacterial lipopolysaccharide (LPS) (22, 32). The LPS composition varies between the different Bcc strains, which may alter its susceptibility to SPLUNC1. To determine if the different LPS structures of Bcc strains play a role in their susceptibility to SPLUNC1, we used the B. cenocepacia K56-2 strain and its ΔhldE and ΔwbxE LPS mutants. The hldE gene codes for a heptokinase and is required for the assembly of the inner core oligosaccharide region of LPS, while the wbxE gene encodes a glycosyltransferase that mediates assembly of the O-antigen subunits (44). SPLUNC1 (0.4 μM) reduced the bacterial growth of wild-type K56-2 to a degree similar to that for J2315 (Fig. 1F). While the ΔwbxE mutant was susceptible to 0.4 μM SPLUNC1, growth of the ΔhldE mutant was unaffected by SPLUNC1, suggesting that different regions of the LPS structure play a role in SPLUNC1 susceptibility.
FIG 1.
SPLUNC1 has antimicrobial activity against B. cepacia complex clinical isolates. (A) SPLUNC1 was coincubated with 106 CFU/ml B. cenocepacia J2315 (■), P. aeruginosa PAO1 (●), or S. aureus CDL (▲) for 24 h, and growth was measured. The number of CFU per milliliter was determined, and inhibition was calculated as follows: % inhibition = [(CFU/ml from vehicle − CFU/ml from SPLUNC1 present)/(CFU/ml from vehicle)] × 100. (B) SPLUNC1 (□), tobramycin (○), and polymyxin B (△) were incubated with 106 CFU/ml J2315 for 24 h, and growth was measured. (C to E) SPLUNC1 (0.4 μM) was incubated for 24 h with 106 CFU/ml of B. cenocepacia (C), B. cepacia (D), and Burkholderia multivorans (E) isolates, and growth was measured. (F) SPLUNC1 (0.4 μM) was incubated for 24 h with 106 CFU/ml of B. cenocepacia K56-2 and its ΔwbxE and ΔhldE LPS mutants, and growth was measured. Open bars, vehicle; closed bars, 0.4 μM SPLUNC1. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (n = 4 for all panels).
SPLUNC1 has bacteriostatic activity against B. cenocepacia J2315.
To better understand SPLUNC1's effects on Bcc strains, we subsequently focused on its effects on the epidemic strain J2315 (42). As growth was still seen after 24 h, even with higher concentrations of SPLUNC1 (Fig. 1A), we next determined whether SPLUNC1 also exerted bacteriostatic and/or bactericidal activity against J2315. We therefore incubated this strain with or without 0.4 μM SPLUNC1 for 2 h. Bacteria were then washed with 0.1% Triton X-100 to remove SPLUNC1 and grown for an additional 9 h. In the presence of SPLUNC1, bacterial growth was inhibited at 2 h. However, after removal of SPLUNC1, bacterial growth resumed, reaching levels similar to those of nontreated J2315 (Fig. 2). In addition, after 24 h, some J2315 bacteria were still present in the medium (Fig. 1B), suggesting that SPLUNC1 has bacteriostatic rather than bactericidal activity.
FIG 2.
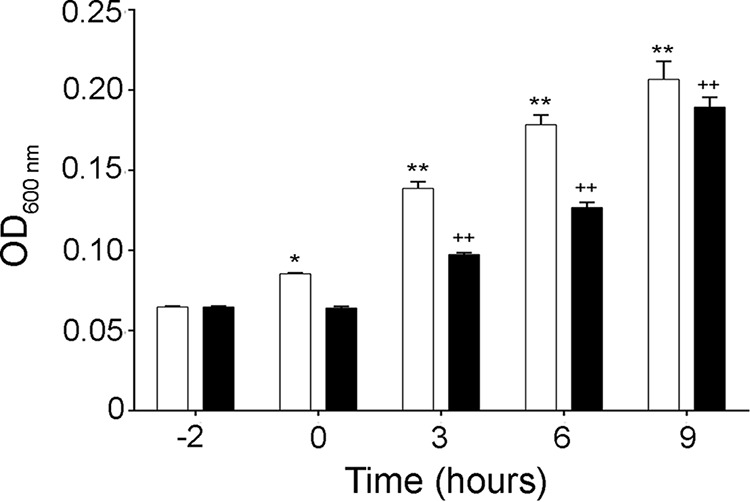
SPLUNC1 is bacteriostatic, not bactericidal. B. cenocepacia J2315 was incubated with or without 0.4 μM SPLUNC1 for 2 h (starting at −2 h). Bacteria were then washed at 0 h with 0.1% Triton X-100 to remove SPLUNC1, grown for an additional 9 h, and measured by determining the OD600 every 3 h. Open bars, vehicle; closed bars, 0.4 μM SPLUNC1. *, P < 0.05 compared to vehicle at −2 h; **, P < 0.01 compared to vehicle at −2 h; ++, P < 0.01 compared to 0.4 μM SPLUNC1 at −2 h (n = 3).
SPLUNC1 has antibiofilm activity against Bcc strains.
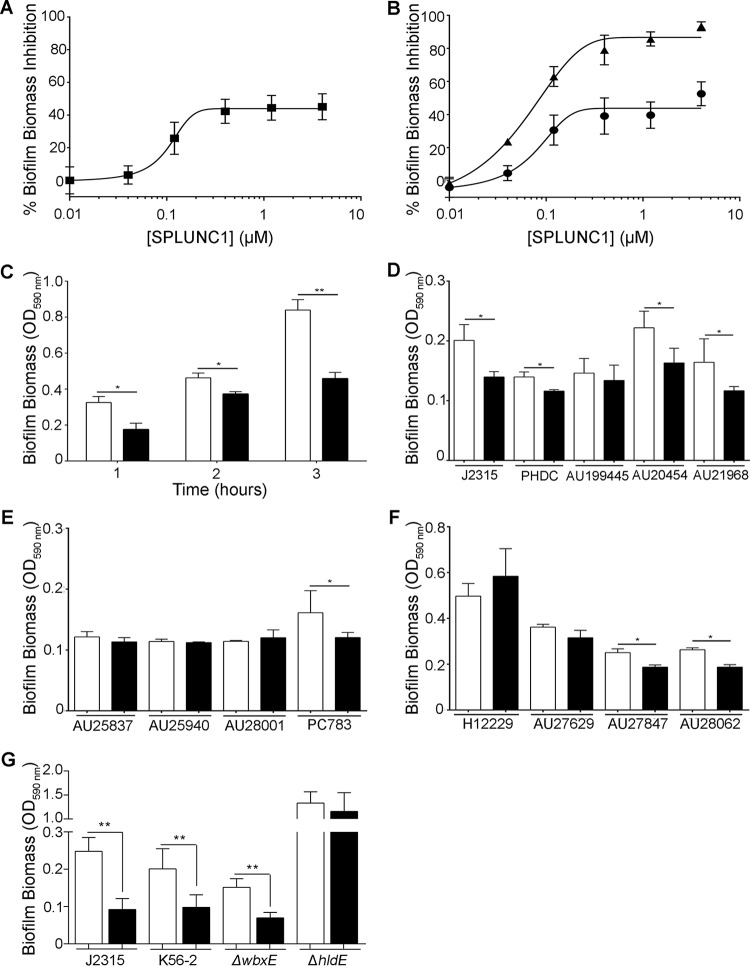
SPLUNC1 has previously been shown to exhibit antibiofilm activity against Gram-negative bacteria (22, 30, 32). To determine whether SPLUNC1 exerts antibiofilm activity against B. cenocepacia, increasing concentrations of SPLUNC1 were coincubated with 106 CFU/ml J2315 for 24 h. Biofilm biomass was then measured by crystal violet staining. Our data indicated that SPLUNC1 prevented J2315 biofilm formation, with an IC50 of 0.10 μM (Fig. 3A). To determine if SPLUNC1 also affected preformed biofilms, J2315 was grown for 24 h to allow for biofilm formation, and SPLUNC1 was added over a range of concentrations to the preformed biofilms and incubated for 1 h and 24 h. We found that SPLUNC1 significantly reduced preformed J2315 biofilms after both 1 h and 24 h (Fig. 3B), therefore suggesting that SPLUNC1 can exert antibiofilm activity.
FIG 3.
SPLUNC1 has antibiofilm activity against Bcc strains. (A) Dose-response curve for SPLUNC1 coincubated with 106 CFU/ml B. cenocepacia J2315 for 24 h. (B) Preformed (24 h) J2315 biofilms were incubated with increasing concentrations of SPLUNC1 for 1 h (●) or 24 h (▲). (C) Attachment assay for J2315 coincubated with 0.4 μM SPLUNC1 for up to 3 h. (D to F) SPLUNC1 (0.4 μM) was coincubated for 24 h with B. cenocepacia (D), B. cepacia (E), and B. multivorans (F) Bcc clinical isolates. (G) SPLUNC1 (0.4 μM) was incubated for 24 h with 106 CFU/ml of B. cenocepacia K56-2 and its ΔwbxE and ΔhldE LPS mutants. Bcc strains were stained with 1% crystal violet and measured by determining the OD590, and inhibition was calculated as follows: % inhibition = [(CFU/ml from vehicle − CFU/ml from SPLUNC1 present)/(CFU/ml from vehicle)] × 100. Open bars, vehicle; closed bars, 0.4 μM SPLUNC1. *, P < 0.05; **, P < 0.01 (n = 4 for all panels).
Bacterial attachment is the first step in biofilm formation; we therefore tested whether SPLUNC1 affected biofilm attachment. In addition to reducing biofilm formation, 0.4 μM SPLUNC1 also inhibited initial J2315 attachment for up to 3 h (Fig. 3C), suggesting an additional role for SPLUNC1 in biofilm inhibition.
Since SPLUNC1's antimicrobial activities against the various Bcc clinical strains differed, its antibiofilm activities against these strains were also tested. At 0.4 μM, SPLUNC1 reduced some but not all Bcc biofilm biomass (Fig. 3D to F). Increasing concentrations of up to 4 μM SPLUNC1 were also tested against the Bcc clinical isolates that were not initially susceptible. However, these strains remained insensitive to SPLUNC1, suggesting that SPLUNC1's antibiofilm activity is also strain specific (n = 3) (data not shown). Additionally, while B. cenocepacia strains PHDC, AU20454, and AU21968 were resistant to SPLUNC1's antimicrobial activity (Fig. 1C), they were susceptible to SPLUNC1's antibiofilm activity (Fig. 3D). Since LPS plays a role in Bcc strain susceptibility to SPLUNC1's antimicrobial activity (Fig. 1F), it may also be involved in susceptibility to SPLUNC1's antibiofilm activity. While 0.4 μM SPLUNC1 reduced the K56-2 biofilm biomass to levels similar to those for J2315, its effects on the ΔhldE and ΔwbxE LPS mutants varied (Fig. 3E). However, in contrast to the antimicrobial activity results, SPLUNC1 reduced ΔhldE biofilm biomass but did not affect ΔwbxE biofilm biomass, suggesting that different regions of LPS are involved in Bcc strain susceptibility to SPLUNC1's antibiofilm activity.
SPLUNC1 mutants reduce growth and biofilm formation of B. cenocepacia J2315.
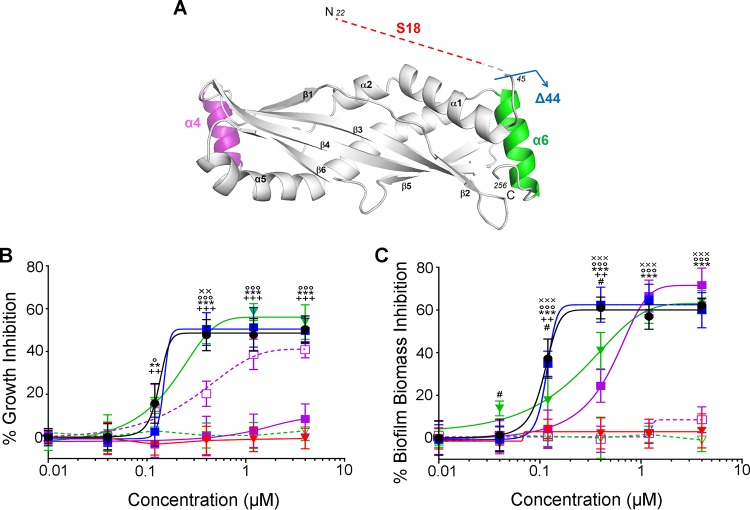
Since SPLUNC1 was effective against J2315, we then sought to determine which domains of SPLUNC1 were responsible for its antimicrobial activity. SPLUNC1's N-terminal S18 region is responsible for regulating ENaC (25), and we recently showed that deletion of the α4 helix resulted in attenuated antimicrobial activity (32). However, the following additional peptides and mutants of SPLUNC1 were generated to test whether the antimicrobial and antibiofilm activities were localized to a specific region of SPLUNC1: (i) the S18 peptide (residues G22 to A39), an N-terminal region containing only the ENaC inhibitory domain; (ii) the Δ44 mutant (residues T45 to V256), which lacks the SPLUNC1 N terminus, including the S18 region; (iii) the Δα4 mutant, which lacks the α4 helix (residues I76 to I105 and includes nonnative Gly-Ser-Gly-Ser linker to L75 to I106); (iv) a peptide corresponding to the α4 helix (residues K77 to L101); (v) the Δα6 mutant, in which the α6 helix (residues I242 to V256) is absent; and (vi) the α6 helix peptide (residues I242 to V256) (Fig. 4A). The Δα4 and Δα6 mutants were chosen because (i) α4 and α6 are two novel helixes that were present in our SPLUNC1 crystal structure and do not share homology with BPI (40) and (ii) our previous studies demonstrated that deletion of the α4 helix reduced SPLUNC1's antimicrobial effects (32). SPLUNC1 mutants were coincubated with 106 CFU/ml J2315 for 24 h, and bacterial growth and biofilm biomass were determined (Fig. 4B and C). The S18 peptide exhibited neither antimicrobial nor antibiofilm activity against J2315. Consistent with this observation, the Δ44 mutant retained full antimicrobial activity (IC50 = 0.14 μM) and antibiofilm activity (IC50 = 0.12 μM). The helix mutants varied in their effects against J2315. The Δα4 mutant lost antimicrobial activity against J2315 and also had significantly diminished antibiofilm activity (IC50 = 0.46 μM). However, the α4 peptide possessed antimicrobial activity against J2315 (IC50 = 0.36 μM) but did not have antibiofilm activity (Fig. 4B and C). The Δα6 mutant retained its antimicrobial activity (IC50 = 0.15 μM) but had significantly reduced antibiofilm activity (IC50 = 0.24 μM), while the α6 peptide had neither antimicrobial nor antibiofilm activity against J2315 (Fig. 4B and C).
FIG 4.
The α4 helix is required for SPLUNC1's antimicrobial activity against B. cenocepacia J2315. (A) Three-dimensional rendering of SPLUNC1 structure with the intrinsically disordered S18 region appended (labeled in red). Also indicated are Δ44 SPLUNC1, which lacks the S18 region (blue arrow); the α4 region (labeled in purple), which is absent in the Δα4 mutant; and the α6 region (labeled in green), which is absent in the Δα6 mutant. Increasing concentrations of SPLUNC1 (black closed circles), the Δ44 (blue closed squares), Δα4 (purple closed squares), and Δα6 (green closed triangles) SPLUNC1 mutants, and the α4 (purple open squares), α6 (green open triangles), and S18 (red closed triangles) peptides were coincubated with 106 CFU/ml J2315 for 24 h and measured for antimicrobial activity by CFU counts and calculation of % growth inhibition as previously described (B) and for antibiofilm activity by 1% crystal violet staining, OD590 reading, and calculation of % biofilm biomass inhibition as previously described (C). **, P < 0.001 for S18 peptide compared to SPLUNC1; ***, P < 0.0001 for S18 peptide compared to SPLUNC1; ++, P < 0.001 for Δα4 mutant compared to SPLUNC1; +++, P < 0.0001 for Δα4 mutant compared to SPLUNC1; #, P < 0.01 for Δα6 mutant compared to SPLUNC1; ××, P < 0.05 for α4 peptide compared to SPLUNC1; ×××, P < 0.001 for α4 peptide compared to SPLUNC1; °, P < 0.05 for α6 peptide compared to SPLUNC1; °°°, P < 0.001 for α6 peptide compared to SPLUNC1 (n = 3 for all panels).
Neutrophil elastase maintains SPLUNC1's antibiofilm activity.
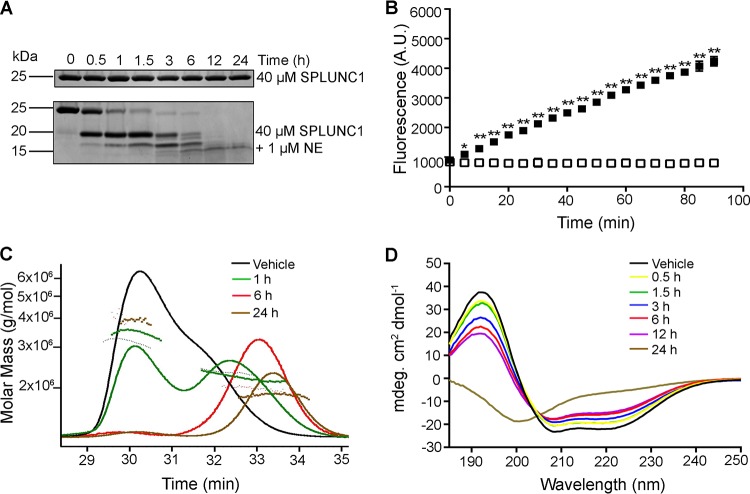
Chronic neutrophilia is a hallmark of CF lung disease and leads to elevated levels of NE in the lung lumen (45). SPLUNC1 is known to be a substrate for NE, and in some cases, NE may alter SPLUNC1's activity (25, 31, 46). To determine the effects of NE cleavage on SPLUNC1's antimicrobial and antibiofilm activities, SPLUNC1 was exposed to 1 μM NE for defined periods, after which NE activity was inhibited by sivelestat as previously reported (47). SPLUNC1 was extensively cleaved by NE, as shown by SDS-PAGE followed by Coomassie blue staining (Fig. 5A). Inhibition of NE by sivelestat was then confirmed by measuring the ability of NE to cleave the fluorogenic substrate Suc-Ala-Ala-Ala-MCA (Fig. 5B). SDS-PAGE fully denatures SPLUNC1, allowing individual fragments to be separated by size. However, under the nondenaturing conditions seen in the airways, NE cleavage of SPLUNC1 may not result in its dissociation, and SPLUNC1 may remain cohesive in the airway surface liquid (ASL) after cleavage. To determine if SPLUNC1 remained intact after NE exposure, we measured SPLUNC1's molecular size in a physiological solution after NE exposure by multiangle static light scattering. Molecular mass was determined by measuring the intensity of scattered light against SPLUNC1. SPLUNC1's initial (0 h) molecular mass was 23.5 kDa (Fig. 5C). After a 1-h exposure to NE, SPLUNC1's molecular mass was 22.3 kDa. There was a slight reduction in molecular mass within the initial 6 h after NE exposure, to 19.6 kDa. After 24 h of exposure to NE, SPLUNC1's molecular mass further decreased, to 18.6 kDa. However, these sizes were still greater than those of the individual fragments detected by SDS-PAGE (∼15 to 17 kDa) (Fig. 5A). To further examine changes in SPLUNC1's structure after NE exposure, cleaved SPLUNC1 was analyzed by circular dichroism (CD) spectroscopy in the far-UV spectral region (190 to 250 nm) to observe SPLUNC1's secondary structures. SPLUNC1 initially had a secondary alpha-helical structure, as indicated by a positive signal at 194 nm and two small negative signals, at 208 and 222 nm (Fig. 5D). Despite being cleaved by NE, SPLUNC1 retained its secondary alpha-helical structure for up to 12 h, but it lost this structure after 24 h of incubation with NE, as shown by a random coiling effect, with a negative signal at 200 nm and an increasing signal at 210 nm.
FIG 5.
SPLUNC1 does not dissociate and retains secondary structure after cleavage with NE. (A) Time course showing cleavage of 40 μM SPLUNC1 by 1 μM NE by SDS-PAGE with Coomassie blue staining. (B) Inhibition of 1 μM NE activity without (■) or with (□) 1 μM sivelestat and 10 μM substrate (Suc-Ala-Ala-Ala-MCA protein) (error bars are obscured by the symbols). A.U., arbitrary units. (C) Static light scattering of SPLUNC1 before and at timed intervals after exposure to NE and sivelestat. (D) Circular dichroism analysis of SPLUNC1 before and at timed intervals after exposure to NE and sivelestat. *, P < 0.01; **, P < 0.001 (n = 3 for all panels).
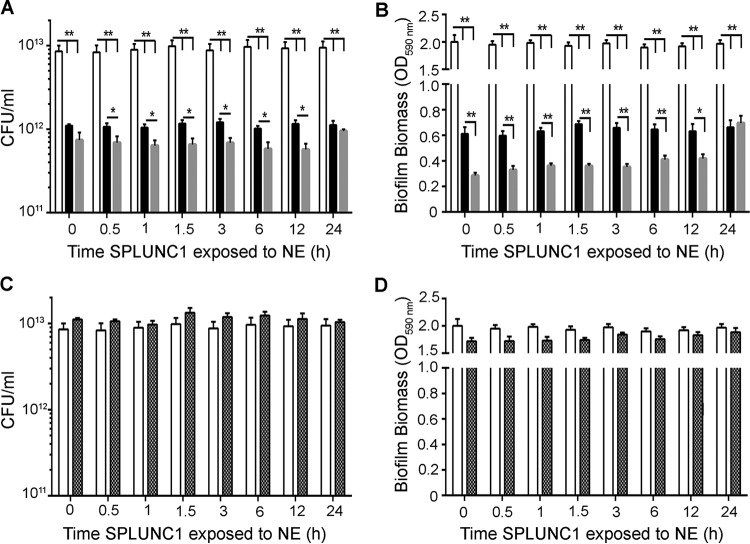
We next tested cleaved SPLUNC1, created by timed incubations with NE, for antimicrobial and antibiofilm activities by coincubation with 106 CFU/ml J2315 for 24 h. NE-exposed SPLUNC1 had increased antimicrobial and antibiofilm activities against J2315 compared to those of SPLUNC1 alone (Fig. 6A and B). However, after 24 h, NE-exposed SPLUNC1 had levels of antimicrobial activity similar to those of SPLUNC1 alone. Surprisingly, the effect of SPLUNC1 to disrupt biofilm formation was significantly potentiated by NE pretreatment for up to 12 h (n = 3; P < 0.01) (Fig. 6B). Importantly, these effects were not due to active NE, since NE activity had been halted by sivelestat (Fig. 5B). As a control, we tested the effects of NE plus sivelestat against J2315 growth, and these compounds had no antimicrobial or antibiofilm activity (Fig. 6C and D).
FIG 6.
Cleaved wild-type SPLUNC1 exerts larger effects on B. cenocepacia J2315 growth and biofilm formation than those seen with whole SPLUNC1. Aliquots of SPLUNC1 were exposed to NE for timed intervals, and NE activity was then halted with sivelestat. NE-cleaved SPLUNC1 was then incubated for 24 h with 106 CFU/ml J2315. (A) CFU counts to show antimicrobial activity after incubation with whole versus cleaved SPLUNC1. (B) Inhibition of biofilm formation as measured by crystal violet staining followed by OD590 readings. NE (1 μM) plus sivelestat (1 μM) alone had neither antimicrobial activity (C) nor antibiofilm activity (D). White bars, vehicle; black bars, 0.4 μM SPLUNC1; gray bars, 0.4 μM NE-cleaved SPLUNC1; hatched bars, 1 μM NE plus 1 μM sivelestat (control). *, P < 0.05; **, P < 0.01 (n = 5 for all panels).
DISCUSSION
The airways contain many antimicrobial agents, including peptides, such as cathelicidins and β-defensins, and proteins, including SPLUNC1, as part of the first line of innate defense against pathogens (27, 33). Previous reports showed that knockout of SPLUNC1 in mice led to increases in bacterial infections by P. aeruginosa, K. pneumoniae, and H. influenzae (26, 30, 31), suggesting that SPLUNC1 plays an important role in reducing bacterial infections. Furthermore, 0.4 μM SPLUNC1 reduced P. aeruginosa growth by 80% in vitro (48). While Bcc growth is not affected by cathelicidins or β-defensins (49), our results show that 0.4 μM SPLUNC1, which is within the physiological range of SPLUNC1 in the ASL (0.4 to 1 μM), also reduces J2315 growth (Fig. 1A and B) (22). SPLUNC1 is thought to exert its antimicrobial activity against P. aeruginosa by formation of pores in the bacterial cell wall, thus increasing cell wall permeability (22). SPLUNC1 shares structural homology with BPI and binds through hydrophobic interactions with the LPS of Gram-negative bacteria, such as K. pneumoniae and P. aeruginosa (50). Despite being structurally smaller than BPI, SPLUNC1 is thought to have similar mechanisms of interaction with P. aeruginosa (22). Although J2315 was susceptible to SPLUNC1, SPLUNC1's antimicrobial activity varied among the different Bcc species (Fig. 1C to E). This variation in susceptibility among the different Bcc species may be due to the unusual composition of the LPS structure, which differs among the Bcc species (51). Indeed, susceptibility to other antibiotics has been reported to vary among the Bcc members (11, 52). Our results demonstrate that changes to Bcc strains' LPS structure alter their susceptibility to SPLUNC1 (Fig. 1F and 3E). As LPS plays an important role in bacterial sensitivity to antimicrobial agents, more studies will be needed to compare the LPS structures of these strains to determine their interaction with SPLUNC1.
Researchers have proposed that the ASL is bacteriostatic rather than bactericidal (53–55) and must act in concert with functional mucociliary clearance (MCC) to remove bacteria. That is, as bacterial growth is impaired, MCC removes the bacteria in the airways, preventing bacterial colonization (56). SPLUNC1 has been shown to coat P. aeruginosa to inhibit growth rather than killing bacteria (22, 32). Indeed, our results revealed that when SPLUNC1 was removed, J2315 growth was restored to levels similar to those of the controls (Fig. 2), confirming that SPLUNC1 also has bacteriostatic activity against Bcc strains.
In order to determine which domain of SPLUNC1 is required for its antimicrobial activity, SPLUNC1 mutants and peptides were tested. The S18 peptide exerted neither antimicrobial nor antibiofilm activity against J2315. The Δ44 mutant, which lacks the S18 region, had antimicrobial and antibiofilm activities comparable to those of full-length SPLUNC1. Although the S18 region does not exert antimicrobial or antibiofilm activity against J2315, since this region is required to regulate ENaC (25), it still plays a role in mechanically clearing bacteria via the mucociliary escalator in vivo. Deletion of the α4 but not α6 helix resulted in a loss of antimicrobial activity. Consistent with this observation, the α4 peptide restored antimicrobial activity, suggesting that this region of SPLUNC1 is absolutely required for SPLUNC1's antimicrobial activity. In addition, both the Δα4 and Δα6 mutants had reduced biofilm activity, but here it was less clear, since the α4 deletion exerted a much stronger effect than the α6 deletion. In addition, the α4 and α6 peptides alone did not exert antibiofilm activity. However, it is likely that these helixes both play roles in SPLUNC1's antibiofilm activity while present in the SPLUNC1 protein (Fig. 4). While the Δ44 mutant retained antimicrobial activity (Fig. 4B), other SPLUNC1 fragments formed by NE activity may further expose the domains for antimicrobial and antibiofilm activities of SPLUNC1 for enhancement of the reduction of J2315 growth and biofilm formation.
Although biofilms in CF patients are rare, Bcc strains have been shown to form biofilms in vitro and to form thick biofilms in sputa of CF patients (57–59). Biofilms increase the bacterium's antibiotic resistance. However, we found that 0.4 μM SPLUNC1 both prevents biofilm formation and reduces preformed J2315 biofilms (Fig. 3A and B). Surfactants change flagellar development, leading to altered bacterial attachment and altered biofilm formation (30, 60). SPLUNC1 has surfactant activity (21, 32), which may play a role in antibiofilm activity. Indeed, our results have shown that SPLUNC1 reduces J2315 attachment (Fig. 3C), and we speculate that SPLUNC1's surfactant activities may play a role in antibiofilm activity against Bcc strains. Additionally, SPLUNC1's antibiofilm activity varied among the Bcc clinical isolates (Fig. 3D and E), as was seen with SPLUNC1's antimicrobial activity (Fig. 1C to E), which may be due to differences in the LPS or flagellar proteins of Bcc clinical isolates.
Persistent bacterial infection in CF lungs leads to airway inflammation, chronic neutrophilia, increased protease activity, and subsequent lung damage (23, 37, 61). NE readily cleaves SPLUNC1. However, we previously showed that the S18 peptide, which is analogous to SPLUNC1's ENaC inhibitory domain, remains as an intact and functional peptide capable of regulating ENaC even after NE exposure (25). Conversely, Jiang et al. reported that addition of NE to normal human tracheobronchial epithelia impaired their antimicrobial activity against Mycoplasma pneumoniae and H. influenzae (31). Here we found that NE-cleaved SPLUNC1 maintains antimicrobial/antibiofilm activity for up to 24 h against J2315 and that this activity is significantly enhanced compared to that of whole SPLUNC1 for a limited period (Fig. 6A and B). During the early stages of infection, both SPLUNC1 and NE expression levels increase (22, 23, 37), which may serve to potentiate SPLUNC1's antimicrobial activities, leading to a quicker resolution of the infection. However, in CF airways, SPLUNC1 is inactive due to the acidic environment, leading to a failure to regulate ENaC and to clear mucus, and chronic neutrophilia occurs, resulting in increased NE levels (29, 37, 40). We noted that the beneficial effects of NE on SPLUNC1 were eventually abolished (Fig. 6). Chronically increased NE levels may therefore lead to an altered ASL milieu that contributes to SPLUNC1's degradation and further impairment of SPLUNC1's antimicrobial activities (31). Indeed, we previously reported that SPLUNC1 is differentially cleaved in CF versus normal sputum (25). Our data indicate that as NE initially cleaves SPLUNC1, SPLUNC1 initially retains its secondary alpha-helical structure (Fig. 5D), which may allow the protein to continue to exert antimicrobial activity against J2315 as well as releasing the S18 peptide, which can help to flush out the airways by inhibiting ENaC and increasing hydration and MCC.
In conclusion, Bcc strain resistance to many antibiotics poses a problem for immunocompromised individuals (62, 63). For example, as Bcc strains colonize CF lungs in the later stages of the disease, patients exhibit a greater decline in pulmonary function and an increase in mortality (64). Our data have shown that SPLUNC1 affects J2315 by (i) bacteriostatic effects to reduce growth and (ii) antibiofilm activity to prevent and reduce biofilm formation via its α4 and α6 helixes. Further investigation into these helixes may provide novel therapies for treating Bcc infections. While the impact of CF lung disease on SPLUNC1-Bcc interactions is not currently known, our data suggest that understanding this phenomenon may have important implications for CF lung disease.
ACKNOWLEDGMENTS
We thank John LiPuma from the University of Michigan for sending the Bcc isolates, Miguel Valvano (Queens University Belfast) for the Bcc K56-2 strain and LPS mutants, and Colin Bingle (University of Sheffield, United Kingdom) for the wild-type SPLUNC1 construct.
This work was funded by NIH grant R01 HL108927 and by an INOVCF grant from the UK Cystic Fibrosis Trust.
Robert Tarran has equity in Spyryx Biosciences Inc. No other conflicts of interest exist.
REFERENCES
- 1.Mahenthiralingam E, Baldwin A, Vandamme P. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J Med Microbiol 51:533–538. doi: 10.1099/0022-1317-51-7-533. [DOI] [PubMed] [Google Scholar]
- 2.Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 58:1580–1590. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- 3.Dedeckova K, Fila L, Skalicka V, Bartosova J, Kucerova T, Vavrova V, Zemkova D, Kalferstova L, Melter O, Cinek O, Drevinek P. 2012. PCR detection of Burkholderia cepacia complex as one of key factors to handle a long-term outbreak. J Cyst Fibros 11:440–445. doi: 10.1016/j.jcf.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Saldias MS, Valvano MA. 2009. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology 155:2809–2817. doi: 10.1099/mic.0.031344-0. [DOI] [PubMed] [Google Scholar]
- 5.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 78:4088–4100. doi: 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin R, Lam M, Dupuis A, Ratjen F. 2011. The effect of early Pseudomonas aeruginosa treatment on lung function in pediatric cystic fibrosis. Pediatr Pulmonol 46:554–558. doi: 10.1002/ppul.21417. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho CP, Dos Santos SC, Madeira A, Mira NP, Moreira AS, Sa-Correia I. 2011. Long-term colonization of the cystic fibrosis lung by Burkholderia cepacia complex bacteria: epidemiology, clonal variation, and genome-wide expression alterations. Front Cell Infect Microbiol 1:12. doi: 10.3389/fcimb.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 9.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr 104:206–210. doi: 10.1016/S0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 10.Valvano MA, Keith KE, Cardona ST. 2005. Survival and persistence of opportunistic Burkholderia species in host cells. Curr Opin Microbiol 8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Nzula S, Vandamme P, Govan JR. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J Antimicrob Chemother 50:265–269. doi: 10.1093/jac/dkf137. [DOI] [PubMed] [Google Scholar]
- 12.Sousa SA, Ramos CG, Almeida F, Meirinhos-Soares L, Wopperer J, Schwager S, Eberl L, Leitao JH. 2008. Burkholderia cenocepacia J2315 acyl carrier protein: a potential target for antimicrobials' development? Microb Pathog 45:331–336. doi: 10.1016/j.micpath.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Leitao JH, Sousa SA, Cunha MV, Salgado MJ, Melo-Cristino J, Barreto MC, Sa-Correia I. 2008. Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. Eur J Clin Microbiol Infect Dis 27:1101–1111. doi: 10.1007/s10096-008-0552-0. [DOI] [PubMed] [Google Scholar]
- 14.Burns JL, Jonas M, Chi EY, Clark DK, Berger A, Griffith A. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect Immun 64:4054–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DW, Mohr CD. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect Immun 68:24–29. doi: 10.1128/IAI.68.1.24-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwab U, Abdullah LH, Perlmutt OS, Albert D, Davis CW, Arnold RR, Yankaskas JR, Gilligan P, Neubauer H, Randell SH, Boucher RC. 2014. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect Immun 82:4729–4745. doi: 10.1128/IAI.01876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forier K, Messiaen AS, Raemdonck K, Nelis H, De Smedt S, Demeester J, Coenye T, Braeckmans K. 2014. Probing the size limit for nanomedicine penetration into Burkholderia multivorans and Pseudomonas aeruginosa biofilms. J Control Release 195:21–28. doi: 10.1016/j.jconrel.2014.07.061. [DOI] [PubMed] [Google Scholar]
- 18.Conway BA, Venu V, Speert DP. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J Bacteriol 184:5678–5685. doi: 10.1128/JB.184.20.5678-5685.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dales L, Ferris W, Vandemheen K, Aaron SD. 2009. Combination antibiotic susceptibility of biofilm-grown Burkholderia cepacia and Pseudomonas aeruginosa isolated from patients with pulmonary exacerbations of cystic fibrosis. Eur J Clin Microbiol Infect Dis 28:1275–1279. doi: 10.1007/s10096-009-0774-9. [DOI] [PubMed] [Google Scholar]
- 20.Bingle CD, Bingle L. 2000. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta 1493:363–367. doi: 10.1016/S0167-4781(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 21.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PB Jr. 2010. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One 5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayeed S, Nistico L, St Croix C, Di YP. 2013. Multifunctional role of human SPLUNC1 in Pseudomonas aeruginosa infection. Infect Immun 81:285–291. doi: 10.1128/IAI.00500-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bingle L, Barnes FA, Cross SS, Rassl D, Wallace WA, Campos MA, Bingle CD. 2007. Differential epithelial expression of the putative innate immune molecule SPLUNC1 in cystic fibrosis. Respir Res 8:79. doi: 10.1186/1465-9921-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, Stutts MJ, Tarran R. 2009. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci U S A 106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs CA, Blanchard MG, Alijevic O, Tan CD, Kellenberger S, Bencharit S, Cao R, Kesimer M, Walton WG, Henderson AG, Redinbo MR, Stutts MJ, Tarran R. 2013. Identification of the SPLUNC1 ENaC-inhibitory domain yields novel strategies to treat sodium hyperabsorption in cystic fibrosis airway epithelial cultures. Am J Physiol Lung Cell Mol Physiol 305:L990–L1001. doi: 10.1152/ajplung.00103.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Di ME, Chu HW, Liu X, Wang L, Wenzel S, Di YP. 2013. Increased susceptibility to pulmonary Pseudomonas infection in Splunc1 knockout mice. J Immunol 191:4259–4268. doi: 10.4049/jimmunol.1202340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di YP. 2011. Functional roles of SPLUNC1 in the innate immune response against Gram-negative bacteria. Biochem Soc Trans 39:1051–1055. doi: 10.1042/BST0391051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britto CJ, Liu Q, Curran DR, Patham B, Dela Cruz CS, Cohn L. 2013. Short palate, lung, and nasal epithelial clone-1 is a tightly regulated airway sensor in innate and adaptive immunity. Am J Respir Cell Mol Biol 48:717–724. doi: 10.1165/rcmb.2012-0072OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarran R, Redinbo MR. 2014. Mammalian short palate lung and nasal epithelial clone 1 (SPLUNC1) in pH-dependent airway hydration. Int J Biochem Cell Biol 52:130–135. doi: 10.1016/j.biocel.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Bartlett JA, Di ME, Bomberger JM, Chan YR, Gakhar L, Mallampalli RK, McCray PB Jr, Di YP. 2013. SPLUNC1/BPIFA1 contributes to pulmonary host defense against Klebsiella pneumoniae respiratory infection. Am J Pathol 182:1519–1531. doi: 10.1016/j.ajpath.2013.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang D, Wenzel SE, Wu Q, Bowler RP, Schnell C, Chu HW. 2013. Human neutrophil elastase degrades SPLUNC1 and impairs airway epithelial defense against bacteria. PLoS One 8:e64689. doi: 10.1371/journal.pone.0064689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton WG, Ahmad S, Little MS, Kim CS, Tyrrell J, Lin Q, Di YP, Tarran R, Redinbo MR. 2016. Structural features essential to the antimicrobial functions of human SPLUNC1. Biochemistry 55:2979–2991. doi: 10.1021/acs.biochem.6b00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bals R, Weiner DJ, Wilson JM. 1999. The innate immune system in cystic fibrosis lung disease. J Clin Invest 103:303–307. doi: 10.1172/JCI6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiemstra PS, McCray PB Jr, Bals R. 2015. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur Respir J 45:1150–1162. doi: 10.1183/09031936.00141514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker D, Prince A. 2011. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elborn JS, Cordon SM, Parker D, Delamere FM, Shale DJ. 1993. The host inflammatory response prior to death in patients with cystic fibrosis and chronic Pseudomonas aeruginosa infection. Respir Med 87:603–607. doi: 10.1016/S0954-6111(05)80263-X. [DOI] [PubMed] [Google Scholar]
- 37.Voynow JA, Fischer BM, Zheng S. 2008. Proteases and cystic fibrosis. Int J Biochem Cell Biol 40:1238–1245. doi: 10.1016/j.biocel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richman-Eisenstat J. 1996. Cytokine soup: making sense of inflammation in cystic fibrosis. Pediatr Pulmonol 21:3–5. doi:. [DOI] [PubMed] [Google Scholar]
- 39.Belaaouaj A. 2002. Neutrophil elastase-mediated killing of bacteria: lessons from targeted mutagenesis. Microbes Infect 4:1259–1264. doi: 10.1016/S1286-4579(02)01654-4. [DOI] [PubMed] [Google Scholar]
- 40.Garland AL, Walton WG, Coakley RD, Tan CD, Gilmore RC, Hobbs CA, Tripathy A, Clunes LA, Bencharit S, Stutts MJ, Betts L, Redinbo MR, Tarran R. 2013. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc Natl Acad Sci U S A 110:15973–15978. doi: 10.1073/pnas.1311999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margolis JJ, El-Etr S, Joubert LM, Moore E, Robison R, Rasley A, Spormann AM, Monack DM. 2010. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl Environ Microbiol 76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, LiPuma JJ, Govan JR. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res Microbiol 154:91–96. doi: 10.1016/S0923-2508(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 43.Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Cell Infect Microbiol 1:6. doi: 10.3389/fcimb.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol 188:2073–2080. doi: 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conese M, Copreni E, Di Gioia S, De Rinaldis P, Fumarulo R. 2003. Neutrophil recruitment and airway epithelial cell involvement in chronic cystic fibrosis lung disease. J Cyst Fibros 2:129–135. doi: 10.1016/S1569-1993(03)00063-8. [DOI] [PubMed] [Google Scholar]
- 46.Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, Frasch SC, Michels NM, Case SR, Chu HW. 2011. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol 178:2159–2167. doi: 10.1016/j.ajpath.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawabata K, Suzuki M, Sugitani M, Imaki K, Toda M, Miyamoto T. 1991. ONO-5046, a novel inhibitor of human neutrophil elastase. Biochem Biophys Res Commun 177:814–820. doi: 10.1016/0006-291X(91)91862-7. [DOI] [PubMed] [Google Scholar]
- 48.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. 2008. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem 309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 49.Baird RM, Brown H, Smith AW, Watson ML. 1999. Burkholderia cepacia is resistant to the antimicrobial activity of airway epithelial cells. Immunopharmacology 44:267–272. doi: 10.1016/S0162-3109(99)00122-8. [DOI] [PubMed] [Google Scholar]
- 50.Meszaros K, Parent JB, Gazzano-Santoro H, Little R, Horwitz A, Parsons T, Theofan G, Grinna L, Weickmann J, Elsbach P, Weiss J, Conlon PJ. 1993. A recombinant amino terminal fragment of bactericidal/permeability-increasing protein inhibits the induction of leukocyte responses by LPS. J Leukoc Biol 54:558–563. [DOI] [PubMed] [Google Scholar]
- 51.De Soyza A, Silipo A, Lanzetta R, Govan JR, Molinaro A. 2008. Chemical and biological features of Burkholderia cepacia complex lipopolysaccharides. Innate Immun 14:127–144. doi: 10.1177/1753425908093984. [DOI] [PubMed] [Google Scholar]
- 52.Rose H, Baldwin A, Dowson CG, Mahenthiralingam E. 2009. Biocide susceptibility of the Burkholderia cepacia complex. J Antimicrob Chemother 63:502–510. doi: 10.1093/jac/dkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schutte BC, McCray PB Jr. 2002. Beta-defensins in lung host defense. Annu Rev Physiol 64:709–748. doi: 10.1146/annurev.physiol.64.081501.134340. [DOI] [PubMed] [Google Scholar]
- 54.Schnapp D, Harris A. 1998. Antibacterial peptides in bronchoalveolar lavage fluid. Am J Respir Cell Mol Biol 19:352–356. doi: 10.1165/ajrcmb.19.3.3384. [DOI] [PubMed] [Google Scholar]
- 55.Konstan MW, Chen PW, Sherman JM, Thomassen MJ, Wood RE, Boat TF. 1981. Human lung lysozyme: sources and properties. Am Rev Respir Dis 123:120–124. [DOI] [PubMed] [Google Scholar]
- 56.Cole AM, Dewan P, Ganz T. 1999. Innate antimicrobial activity of nasal secretions. Infect Immun 67:3267–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kennedy S, Beaudoin T, Yau YC, Caraher E, Zlosnik JE, Speert DP, LiPuma JJ, Tullis E, Waters V. 2016. Activity of tobramycin against cystic fibrosis isolates of Burkholderia cepacia complex grown as biofilms. Antimicrob Agents Chemother 60:348–355. doi: 10.1128/AAC.02068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva IN, Tavares AC, Ferreira AS, Moreira LM. 2013. Stress conditions triggering mucoid morphotype variation in Burkholderia species and effect on virulence in Galleria mellonella and biofilm formation in vitro. PLoS One 8:e82522. doi: 10.1371/journal.pone.0082522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Bakri AG, Gilbert P, Allison DG. 2005. Influence of gentamicin and tobramycin on binary biofilm formation by co-cultures of Burkholderia cepacia and Pseudomonas aeruginosa. J Basic Microbiol 45:392–396. doi: 10.1002/jobm.200510011. [DOI] [PubMed] [Google Scholar]
- 60.Splendiani A, Livingston AG, Nicolella C. 2006. Control of membrane-attached biofilms using surfactants. Biotechnol Bioeng 94:15–23. doi: 10.1002/bit.20752. [DOI] [PubMed] [Google Scholar]
- 61.Jiang D, Persinger R, Wu Q, Gross A, Chu HW. 2013. α1-Antitrypsin promotes SPLUNC1-mediated lung defense against Pseudomonas aeruginosa infection in mice. Respir Res 14:122. doi: 10.1186/1465-9921-14-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 63.Saiman L, Siegel J, Cystic Fibrosis Foundation Consensus Conference on Infection Control Participants. 2003. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Am J Infect Control 31:S1–S62. [PubMed] [Google Scholar]
- 64.Courtney JM, Dunbar KE, McDowell A, Moore JE, Warke TJ, Stevenson M, Elborn JS. 2004. Clinical outcome of Burkholderia cepacia complex infection in cystic fibrosis adults. J Cyst Fibros 3:93–98. doi: 10.1016/j.jcf.2004.01.005. [DOI] [PubMed] [Google Scholar]