Abstract
A colistin-resistant Escherichia coli strain was recovered from a patient with a diabetic foot infection in Brazil. Whole-genome analysis revealed that the E. coli isolate belonged to the widespread sequence type (ST) 101 and harbored the mcr-1 gene on an IncX4 plasmid that was highly similar to mcr-1-bearing IncX4 plasmids that were recently identified in Enterobacteriaceae from food, animal, and human samples recovered on different continents. These results suggest that self-transmissible IncX4-type plasmids may represent promiscuous plasmids contributing to the intercontinental spread of the mcr-1 gene.
TEXT
The plasmid-mediated colistin resistance mechanism MCR-1 has become a great challenge to public health worldwide. In fact, since its initial identification in Enterobacteriaceae strains (mostly Escherichia coli) isolated from animals, food, and humans in China (1), MCR-1 has also been reported in other countries in Asia, Africa, Europe, and North America (2). In South America, E. coli harboring the mcr-1 gene has been present in food-producing animals since at least 2012 (3), being recently identified in human clinical samples from Argentina (4). We hereby report the first description (to our knowledge) of MCR-1 in a human E. coli isolate from Brazil.
In early 2016, a man in his late 60s with a medical history of type 2 diabetes mellitus, atrial fibrillation, obesity, dyslipidemia, and hypertension was admitted to a private hospital in northeastern Brazil with a 2-month history of a right calcaneal ulcer (informed consent was obtained from this patient). The patient underwent debridement with fasciotomy of necrosed tissue. Wound cultures taken during debridement were negative for aerobic bacteria, and empirical intravenous antibiotic therapy with ceftriaxone and clindamycin was initiated. Following 14 days of antibiotic therapy, the patient was discharged with clinical improvement. Nine days later, however, he returned to the hospital with worsened symptoms, and the combined therapy of ceftriaxone with clindamycin was restarted. As the patient's condition was deteriorating, the treatment was changed to piperacillin-tazobactam. Nevertheless, 10 days later the patient had developed recurrent episodes of fever associated with poor general condition. The antibiotic regimen was changed to meropenem, and the patient underwent another debridement, from which a fragment of soft tissue sent for culture yielded growth of colistin-resistant extended-spectrum β-lactamase (ESBL)-producing E. coli (ICBEC72H) and carbapenem-resistant Citrobacter freundii. Ciprofloxacin was added to the antibiotic regimen at that time, whereas meropenem was stopped after 3 days of combined therapy. The patient underwent lower-limb amputation due to foot necrosis; ciprofloxacin treatment was maintained for 14 days, and the patient was discharged after complete recovery.
The identification and antimicrobial susceptibility testing of the E. coli and C. freundii isolates were performed using the MicroScan system (Beckman Coulter). While the E. coli strain presented resistance to ampicillin (>16 μg/ml), ampicillin-sulbactam (>16/8 μg/ml), aztreonam (>8 μg/ml), cefepime (>8 μg/ml), cefotaxime (>16 μg/ml), cefuroxime (>16 μg/ml), cephalothin (>16 μg/ml), and colistin (>4 μg/ml), the C. freundii isolate showed resistance to aztreonam (>8 μg/ml), cefepime (>8 μg/ml), cefotaxime (>16 μg/ml), ceftazidime (8 μg/ml), ertapenem (>1 μg/ml), imipenem (4 μg/ml), meropenem (4 μg/ml), piperacillin-tazobactam (>64 μg/ml), tobramycin (8 μg/ml), and trimethoprim-sulfamethoxazole (>2/38 μg/ml) (5, 6). Both isolates remained susceptible to amikacin, ciprofloxacin, gentamicin, levofloxacin, and tigecycline. The presence of the blaKPC-2 gene in C. freundii was confirmed by PCR (7), whereas the total genomic DNA of E. coli ICBEC72H was used to construct a mate-paired library, which was sequenced using the MiSeq platform (Illumina, Inc.). Genome assembly was carried out using SPAdes v3.7.1 with the high-quality mate-pair option (8) and, after automatic annotation using Prokka (www.github.com/tseemann/prokka), the sequence was manually curated using the GenBank database and InterPro (www.ebi.ac.uk/interpro). Resistance genes were detected by BLASTn using the ResFinder 2.1 database (https://cge.cbs.dtu.dk/services/ResFinder), plasmid classification was carried out in silico by BLASTn using the PlasmidFinder 1.3 database (https://cge.cbs.dtu.dk/services/PlasmidFinder), and virulence genes were detected by BLASTn using the VirulenceFinder 1.5 database (https://cge.cbs.dtu.dk/services/VirulenceFinder).
Whole-genome sequencing revealed that E. coli ICBEC72H belonged to the widespread sequence type (ST) 101, which was found previously in Australia, Asia, Europe, and North America, where it was found to harbor blaNDM-1 and, less commonly, blaCTX-M (9–11). The presence of the iroN (siderophore), mcmA (microcin), mchB (microcin), mchC (microcin), mchF (microcin), lpfA (fimbriae), and iss (increased serum survival) virulence genes found in ICBEC72H can be associated with the low-virulence B1 phylogenetic group. Further pulsed-field gel electrophoresis (PFGE) characterization, using XbaI restriction, showed that E. coli ICBEC72H was clonally unrelated to previously identified mcr-1-positive E. coli strains isolated from Brazilian livestock (3).
E. coli ICBEC72H carried the blaCTX-M-8 and mcr-1 genes on two different plasmids, which were successfully transferred to E. coli strain EC600 by conjugation (broth mating method). Transconjugants were selected on MacConkey agar plates containing streptomycin (200 μg/ml) and colistin (2 μg/ml). Conjugative transfer of the plasmid mediating colistin resistance from E. coli strain ICBEC72H to recipient E. coli strain EC600 was accompanied by cotransfer of the compatible plasmid conferring third-generation cephalosporin resistance. Plasmid pICBEC72Hctx (92.2 kb in length) was classified as belonging to the IncI1 incompatibility group and carried only blaCTX-M-8, whereas the plasmid carrying mcr-1 (pICBEC72Hmcr) was 33.67 kb in length and belonged to the IncX4 incompatibility group.
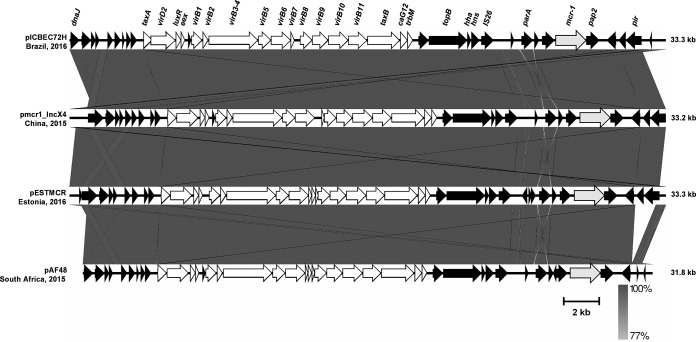
Multiple MAFFT alignments between pICBEC72Hmcr and the other three IncX4 plasmids bearing mcr-1 that are available in GenBank showed very high levels of architectural conservation among these plasmids, with pICBEC72Hmcr presenting 99.9% nucleotide identity with pmcr1_IncX4 (GenBank accession no. KU761327.1), which was found in a ST25 Klebsiella pneumoniae strain isolated from a human clinical sample in China (12), and pESTMCR (GenBank accession no. KU743383.1), which was found in an E. coli strain isolated from pig slurry in Estonia. Plasmid pICBEC72Hmcr presented 96.3% identity with pAF48 (GenBank accession no. KX032520.1), which was found in an E. coli strain isolated from a human clinical sample in South Africa (13), because the latter plasmid presents an ∼1,200-bp deletion that includes a partial deletion of the replication initiation protein PI and a few indels, mostly in noncoding sequences (Fig. 1). The insertion sequence ISApl1, which was initially found to be associated with mcr-1 in pHNSHP45 (1), was not present in the IncX4 plasmid (Fig. 1) (12, 13).
FIG 1.
Backbone of pICBEC72Hmcr IncX4 plasmid (GenBank accession no. CP015977) carrying the mcr-1 gene in a human E. coli strain isolated in Brazil. The pICBEC72H plasmid was compared with plasmid pmcr1_IncX4 (GenBank accession no. KU761327.1) from a MCR-1-positive ST25 Klebsiella pneumoniae strain isolated from a human clinical sample in China (12), pESTMCR (GenBank accession no. KU743383.1) from a MCR-1-positive E. coli strain isolated from pig slurry in Estonia, and pAF48 (GenBank accession no. KX032520.1) from a MCR-1-positive ST624 E. coli strain isolated from a human clinical sample in South Africa (13). Comparative analysis was performed using Easyfig version 2.2.2 for genome comparisons. White arrows, genes that are part of transfer modules.
IncX plasmids are self-transmissible plasmids that include at least five subtypes (IncX1 to IncX5) and previously were thought to be of low prevalence (14, 15). Instead, IncX plasmids have been implicated in the spread of many resistance genes, including ESBL- and carbapenemase-encoding genes (15–18). Recently, IncX4 plasmids carrying the mcr-1 gene were identified in Salmonella enterica (serovars Typhimurium, Parathyphi B, Java, Anatum, and Schwartzengrund) from human infections, ready-to-cook guinea fowl pie, and poultry meat in England, France, and Netherlands (19–21), respectively, and in E. coli isolated from imported chicken meat in Denmark (22), swine in Germany (23), livestock in the Netherlands (21), and human samples in China (12) and South Africa (13). What is surprising is the fact that the mcr-1-bearing IncX4 plasmids obtained from different bacterial species, belonging to different STs, isolated in different clinical contexts, and found on different continents are highly similar in the plasmid backbone sequences. This strongly suggests that self-transmissible IncX4-type plasmids may represent promiscuous plasmids contributing to the intercontinental spread of the mcr-1 gene.
Accession number(s).
Sequences have been deposited in GenBank, and the accession numbers can be found via BioSample record number PRJNA322664.
ACKNOWLEDGMENTS
FAPESP and CNPq research grants are gratefully acknowledged. N.L. and J.A.M. are research grant fellows of CNPq and FAPESP, respectively.
We thank Cefar Diagnóstica Ltda. (Brazil) for kindly supplying antibiotic discs for susceptibility testing.
Funding Statement
This work was funded by research grant 2013/12107-4 from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grants 485438/2012-7 and 457421/2014-2.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Skov RL, Monnet DL. 2016. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 21(9):pii=30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Gonçalves DD, Dropa M, Matté MH, Monte DF, Landgraf M, Francisco GR, Bueno MF, de Oliveira Garcia D, Knöbl T, Moreno AM, Lincopan N. 2016. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 21(17):pii=30214. doi: 10.2807/1560-7917.ES.2016.21.17.30214. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, MCR Group, Corso A. 2016. mcr-1-mediated colistin resistance in human infections caused by Escherichia coli: first description in Latin America. Antimicrob Agents Chemother doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters, v. 6. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf.
- 6.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasilinetc I, Prjibelski AD, Gurevich A, Korobeynikov A, Pevzner PA. 2015. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics 31:3262–3268. doi: 10.1093/bioinformatics/btv337. [DOI] [PubMed] [Google Scholar]
- 9.Yoo JS, Kim HM, Koo HS, Yang JW, Yoo JI, Kim HS, Park HK, Lee YS. 2013. Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J Antimicrob Chemother 68:2170–2172. doi: 10.1093/jac/dkt126. [DOI] [PubMed] [Google Scholar]
- 10.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 11.Mora A, Blanco M, López C, Mamani R, Blanco JE, Alonso MP, García-Garrote F, Dahbi G, Herrera A, Fernández A, Fernández B, Agulla A, Bou G, Blanco J. 2011. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents 37:16–21. doi: 10.1016/j.ijantimicag.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates, South Africa. Antimicrob Agents Chemother doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Lo WU, Chow KH, Law PY, Ng KY, Cheung YY, Lai EL, Ho PL. 2014. Highly conjugative IncX4 plasmids carrying blaCTX-M in Escherichia coli from humans and food animals. J Med Microbiol 63:835–840. doi: 10.1099/jmm.0.074021-0. [DOI] [PubMed] [Google Scholar]
- 16.Stokes MO, Abuoun M, Umur S, Wu G, Partridge SR, Mevius DJ, Coldham NG, Fielder MD. 2013. Complete sequence of pSAM7, an IncX4 plasmid carrying a novel blaCTX-M-14b transposition unit isolated from Escherichia coli and Enterobacter cloacae from cattle. Antimicrob Agents Chemother 57:4590–4594. doi: 10.1128/AAC.01157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho PL, Cheung YY, Lo WU, Li Z, Chow KH, Lin CH, Chan JF, Cheng VC. 2013. Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying blaKPC-2 in a Klebsiella pneumoniae. Curr Microbiol 67:493–498. doi: 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 20.Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:144–145. doi: 10.1016/S1473-3099(15)00538-1. [DOI] [PubMed] [Google Scholar]
- 21.Veldman K, van Essen-Zandbergen A, Rapallini M, Wit B, Heymans R, van Pelt W, Mevius D. 2016. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother doi: 10.1093/jac/dkw181. [DOI] [PubMed] [Google Scholar]
- 22.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20(49):pii=30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 23.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]