Abstract
Eradication of established biofilm communities of pathogenic Gram-negative species is one of the pending challenges for the development of new antimicrobial agents. In particular, Pseudomonas aeruginosa is one of the main dreaded nosocomial species, with a tendency to form organized microbial communities that offer an enhanced resistance to conventional antibiotics. We describe here an engineered antimicrobial peptide (AMP) which combines bactericidal activity with a high bacterial cell agglutination and lipopolysaccharide (LPS) affinity. The RN3(5-17P22-36) peptide is a 30-mer derived from the eosinophil cationic protein (ECP), a host defense RNase secreted by eosinophils upon infection, with a wide spectrum of antipathogen activity. The protein displays high biofilm eradication activity that is not dependent on its RNase catalytic activity, as evaluated by using an active site-defective mutant. On the other hand, the peptide encompasses both the LPS-binding and aggregation-prone regions from the parental protein, which provide the appropriate structural features for the peptide's attachment to the bacterial exopolysaccharide layer and further improved removal of established biofilms. Moreover, the peptide's high cationicity and amphipathicity promote the cell membrane destabilization action. The results are also compared side by side with other reported AMPs effective against either planktonic and/or biofilm forms of Pseudomonas aeruginosa strain PAO1. The ECP and its derived peptide are unique in combining high bactericidal potency and cell agglutination activity, achieving effective biofilm eradication at a low micromolar range. We conclude that the designed RN3(5-17P22-36) peptide is a promising lead candidate against Gram-negative biofilms.
INTRODUCTION
Pseudomonas aeruginosa is a major nosocomial pathogen responsible for severe chronic and acute infections (1, 2). In particular, P. aeruginosa is one of the most frequent causes of ventilator-associated pneumonia and catheter-related bloodstream infections (3, 4). Patients with acquired immune deficiency syndrome (AIDS), burn wounds, or cystic fibrosis have a high risk of developing a serious infection, and this increased risk is related to a high death rate (5–7). P. aeruginosa can acquire resistance to certain antibiotics and, most importantly, is also able to form biofilms on inert surfaces such as medical devices of internal and external use. A biofilm, a microbial community attached to an abiotic surface, represents an additional challenge to antimicrobial therapies. Biofilm formation is induced by genotypic and phenotypic changes of the planktonic microorganisms which lead to an ensuing multilayered cell cluster structure coated by an external polysaccharide matrix composed of polysaccharides, proteins, and extracellular DNA (8). P. aeruginosa, once organized in a biofilm structure, becomes more resistant to antibiotic agents and immune system clearance and thus requires a more elaborate strategy for successful treatment of associated infections (9).
Antimicrobial proteins and peptides (AMPs) have been proposed as a new alternative to current treatments for biofilm-associated infections (10–13). AMPs are small, amphipathic, and frequently cationic molecules that present a rapid, high-potency, and broad-spectrum action against microorganisms (14–16). In our laboratory, we study human RNases as a new potential source of alternative antimicrobial agents (17, 18). Human RNase 3, also known as the eosinophil cationic protein (ECP), is a small, highly cationic protein (pI ∼11) that is stored in the secondary granules of eosinophils (19, 20). RNase 3 is secreted during the infection process, where the protein exerts high antimicrobial activity against a wide range of microorganisms, such as bacteria, yeast, viruses, and parasites (18, 21–24). Moreover, our laboratory has reported RNase 3's high affinity with negatively charged lipopolysaccharides (LPS) from Gram-negative bacteria outer membranes (25), a feature that mediates the protein's high bacterial agglutination ability (26). Indeed, RNase 3's interaction with the bacterial envelope promotes its self-aggregation and triggers bacterial cell death (27). Experimentally, we have determined that the N-terminal region of RNase 3 largely retains the antimicrobial, LPS-binding, and agglutinating properties of the whole protein (28). Exposure of a hydrophobic patch at the protein's N terminus induces the formation of amyloid-like protein aggregates (29) that can drive the cell agglutination process (27). The essential sequence requirements defining the protein's key active region were then outlined by a structure-based algorithm to identify new AMPs (30).
In the present work, we explored the potential of a new ECP-derived peptide, RN3(5-17P22-36), which combines a significant size reduction together with a high antimicrobial efficacy. In particular, the peptide, which encompasses the aggregation-prone region together with the LPS-binding domain, proved highly effective against both planktonic and established biofilms of P. aeruginosa. The peptide mechanism was evaluated by means of a variety of functional methodologies, and the direct action on the biofilm structure was visualized by confocal microscopy. The antimicrobial properties of the RNase 3-derived peptide were compared with properties of previously characterized antimicrobial peptides with reported activity against P. aeruginosa biofilms and LPS-binding or cell-agglutinating properties.
Our comparison demonstrated that the N-terminal peptide RN3(5-17P22-36) was the only peptide able to totally remove established biofilms through bacterial killing and agglutination, highlighting the huge potential of the RNase 3-derived peptide for P. aeruginosa biofilm eradication.
MATERIALS AND METHODS
Reagents and bacteral strains.
BODIPY TR cadaverine [BC; BODIPY is boron dipyrromethane (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene] was purchased from Invitrogen (Carlsbad, CA). Sytox Green was purchased from Invitrogen (Carlsbad, CA). LPS from P. aeruginosa, fluorescein isothiocyanate (FITC)-labeled lectin from Triticum vulgaris (wheat germ agglutinin [WGA]), crystal violet dye, FITC, and 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) were purchased from Sigma-Aldrich (St. Louis, MO). The Alexa Fluor 488 protein-labeling kit and the Live/Dead bacterial viability kit were purchased from Molecular Probes (Eugene, OR). The BacTiter-Glo assay kit was from Promega (Madison, WI). Escherichia coli BL21 was from Novagen (Madison, WI), and Staphylococcus aureus (ATCC 502A) was obtained from the Colección Española de Cultivos tipo (CECT), Universidad de Valencia, Valencia, Spain.
Pseudomonas aeruginosa PAO1 (31) was a kind gift from Isidre Gibert (Institut de Biotecnologia i Biomedicina, Universitat Autònoma de Barcelona, Cerdanyola del Vallès, Barcelona, Spain. P. aeruginosa clinical isolates were provided by the Microbiology Service of Valle Hebron Hospital, Spain. Fresh sheep blood was purchased from Oxoid (Basingstoke, United Kingdom). The MRC-5 human lung fibroblast cell line was supplied by the Cell Culture Service (Institut de Biotecnologia i Biomedicina, Universitat Autònoma de Barcelona, Cerdanyola del Vallès, Barcelona, Spain.
Protein and peptides.
Recombinant RNase 3 was expressed from a human synthetic gene cloned in pET11c. Protein expression in the E. coli BL21(DE3) strain, folding of the protein from inclusion bodies, and purification were carried out as previously described (32). The RNase 3-H15A mutant was constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutation was confirmed by DNA sequencing, and the purified proteins were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry and N-terminal sequencing.
All the peptides used in this work were acquired from Genecust (Dudelange, Luxembourg); the RNase 3 N-terminal-derived peptides [RN3(1-36) and RN3(5-17P22-36)] and the antimicrobial reference peptides human cathelicidin LL-37, cecropin A-melittin hybrid peptide (CA-M), the bacteria-agglutinating peptide derived from the parotid secretory protein (GL-13), and the β-boomerang peptide (WY-12) were all synthesized with C-terminal amidation.
Hemolysis.
Peptide hemolysis was determined as previously described (33). Briefly, fresh sheep red blood cells were washed three times with phosphate-buffered saline (PBS; 35 mM phosphate buffer, 0.15 M NaCl; pH 7.4) by centrifugation for 5 min at 3,000 × g and resuspended in PBS at 2 × 107 cells/ml. Red blood cells were incubated with increasing concentrations of peptides (from 0.01 to 400 μM) at 37°C for 4 h and centrifuged at 13,000 × g for 5 min. The supernatant was separated from the pellet, and its absorbance was measured at 570 nm. The negative control was red blood cells incubated in PBS. The 100% hemolysis point was defined as the absorbance obtained by incubating the blood with distilled water. The concentration required to achieve 50% hemolysis (HC50) was calculated by fitting the data to a sigmoidal function.
Cytotoxicity assay.
Cytotoxicity was measured for the MRC-5 human cell line in an 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium (MTT; Sigma-Aldrich) assay. MRC-5 cells were grown in 5% CO2 at 37°C with minimal essential medium supplemented with 10% fetal bovine serum (FBS). Cells were plated at 5 × 104 cells/well in a 96-well plate and incubated overnight. Next, the medium was removed and serial dilutions of proteins and peptides were added at concentrations ranging from 400 to 0.2 μM in 100 μl of medium without serum. After 4 h of incubation, the medium was replaced with fresh medium containing 10% MTT solution and the mixture was incubated for 3 h in 5% CO2 at 37°C. The medium was then removed. Formazan was dissolved by adding acidic isopropanol. The optical density (OD) was recorded by using an enzyme-linked immunosorbent assay reader (Infinite M200 microplate reader; Tecan) set at 550 nm and 630 nm as references. Reference absorbance at 630 nm was used to correct for nonspecific background values. Each experiment was repeated at least three times.
Minimum bactericidal concentration.
Antimicrobial activity was calculated as the minimum bactericidal concentration (MBC), defined as the lowest protein/peptide concentration that completely eradicated microbial growth. The MBC of each protein/peptide was determined as described previously (27, 34). Bacteria were incubated at 37°C overnight in Luria-Bertani broth (LB) and diluted to approximately 5 × 105 CFU/ml. In each assay, protein/peptide serially was diluted from 50 to 0.2 μM in 10 mM sodium phosphate, 0.1 M NaCl, pH 7.5 buffer and added to 100-μl aliquots of the bacterial dilution and incubated for 4 h at 37°C. Subsequently, samples were plated onto petri dishes and incubated at 37°C overnight. MBC values were determined as a function of the total removal of CFU by the protein/peptide from two independent experiments performed in triplicate.
MIC.
The MIC for each protein/peptide was determined from two independent experiments performed in triplicate for each concentration. Bacterial cells were incubated at 37°C overnight in Mueller-Hinton (MH) broth and diluted to approximately 5 × 105 CFU/ml. The bacterial suspension was incubated in MH with peptide at various concentrations (0.1 to 200 μM) at 37°C in 96-well polypropylene plates, and optical densities were monitored overnight. The MIC is defined as the minimum concentration at which no bacterial growth was detected.
Minimum agglutination activity.
Bacterial cells were grown in LB at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 5 min, and resuspended in buffer containing 10 mM Tris-HCl, 0.1 M NaCl (pH 7.5). Aliquots of 100 μl of the bacterial suspension were treated with increasing protein/peptide concentrations (from 0.01 to 50 μM), and mixtures were incubated at room temperature for 1 h. The aggregation behavior was followed at 50× magnification by using a binocular stereomicroscope. The minimum agglutinating activity (MAC) is expressed as the minimum agglutinating concentration of the sample tested, as previously described (27). All assays were performed in triplicate.
LPS binding assay.
LPS binding was assessed by using the fluorescent probe BODIPY TR cadaverine as previously described (35). BC binds strongly to native LPS, specifically recognizing the lipid A portion. When a protein that interacts with LPS is added, BC is displaced from the complex, and its fluorescence is increased, decreasing its occupancy factor. LPS binding assays were carried out in 10 mM HEPES buffer at pH 7.5. The displacement assay was performed using a 96-well microtiter plate containing a stirred mixture of LPS (10 μg/ml) and BC (10 μM). Protein/peptides were serially diluted from 10 to 0.1 μM. Fluorescence measurements were performed on a Victor3 plate reader (PerkinElmer, Waltham, MA). The protein/peptide concentrations required to achieve half of the total and the total BC displacement were estimated from nonlinear regression analysis as previously described (25, 36).
Biofilm formation.
Biofilm formation assay was modified from the previously described protocol (37). Briefly, overnight cultures of P. aeruginosa were centrifuged at 5,000 × g for 5 min, washed, and adjusted to an OD600 of 0.1 in tryptic soy broth medium (TSB). Two hundred microliters of the bacterial suspension was dispensed in each well of a polystyrene 96-well microtiter plate (Greiner Bio-one, France). The plate was incubated at 37°C on an orbital shaker (30 rpm). The medium was exchanged with fresh prewarmed TSB after 4 h, in order to remove nonattached bacteria. After 24 h of incubation, the formation of biofilm was then evaluated by using the crystal violet staining technique (38). Briefly, the plates were washed three times with PBS, fixed at 60°C for 1 h, and stained for 15 min with 200 μl of 0.1% crystal violet. After the dye was discarded, plates were rinsed in PBS and allowed to dry for 30 min at 37°C. Bound crystal violet was dissolved in 250 μl of 95% ethanol for 15 min, and the OD570 of the extracted dye was measured using a Victor3 plate reader (Perkin-Elmer, Waltham, MA).
Biofilm eradication assay.
Biofilms were grown as described above, washed three times with PBS, and rinsed with 10 mM sodium phosphate buffer, pH 7.5. Wells were treated with increasing protein/peptide concentrations (from 0.01 to 200 μM) and incubated at 37°C for 4 h. Biofilm eradication was then evaluated by using the crystal violet staining technique (38) and by calculating the total number of remaining viable cells using the BacTiter-Glo assay. The ability of the antimicrobial peptides to disperse and reduce the biofilm mass of preformed biofilms was calculated. The peptide concentration required to achieve total preformed biofilm eradication after 4 h of incubation time was estimated. Nontreated cells incubated with PBS buffer under the same conditions were used as negative controls. A maximum biofilm eradication reference value was determined by exposure to 10% Triton X-100.
The biofilm eradication activity of the protein/peptide was also evaluated by counting the CFU. At 24 h, formed P. aeruginosa biofilms were washed three times with PBS and rinsed with 10 mM sodium phosphate buffer (pH 7.5). Wells were treated with increasing protein/peptide concentrations (400 to 0.01 μM) and incubated at 37°C for 4 h. For each well, the supernatant was also collected and seeded in LB agar petri dishes to evaluate the protein/peptide biofilm dispersion activity. Then, three washes were performed and the biofilm attached to each well was picked up with a sterile swab and diluted in an Eppendorf tube in 1 ml of PBS. Next, the whole volume was sonicated twice for 30 s to release the bacteria into the liquid, and the mixture was briefly vortexed before seeding a 50-μl aliquot on an LB-agar petri dish. CFU were counted after 16 h of incubation at 37°C. Three independent replicates were performed. The ED50 and the minimum biofilm eradication concentration (MBEC) were determined by fitting the data to a nonlinear function.
Bacterial cell viability luminescence assay.
Bacterial viability was assayed by using the BacTiter-Glo microbial cell viability kit. Briefly, protein/peptides were dissolved in buffer (10 mM sodium phosphate, 0.1 M NaCl; pH 7.5) serially diluted and tested in a 96-well microtiter plate containing P. aeruginosa planktonic cells (OD600, ∼ 0.2) or 24-h formed biofilms for 4 h at 37°C. For planktonic P. aeruginosa, the protein/peptide concentrations used ranged from 50 to 0.1 μM; in case of preformed biofilms, the concentrations used ranged from 500 to 0.1 μM. After incubation, 50 μl of BacTiter-Glo reagent (Promega) was added to each well according to the manufacturer's instructions and incubated at room temperature for 15 min. Luminescence was read on a Victor3 plate reader with a 3-s integration time. The correlation between fluorescent signal and viable cell counts was checked for the untreated P. aeruginosa biofilm controls (17,000 arbitrary units, corresponding to approximately 2 × 105 CFU), and all of the recorded fluorescent values were confirmed to fit into the assay linear relationship range provided by the manufacturer. The ED50 and total eradication concentrations were calculated by fitting the data to a dose-response curve.
Bacteria cytoplasmic membrane depolarization assay.
Membrane depolarization was determined using the method described earlier (25). Briefly, bacterial cells were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 7 min, washed with 5 mM HEPES (pH 7.2) containing 20 mM glucose, and resuspended in 5 mM HEPES-KOH, 20 mM glucose, and 100 mM KCl at pH 7.2 to an OD600 of 0.05. Aliquots of 200 μl were transferred to a microtiter plate for planktonic assays. For the biofilm assay, 24-h formed P. aeruginosa biofilms grown in a 96-well microtiter plate were washed three times with PBS. Afterwards P. aeruginosa biofilms were washed with 5 mM HEPES (pH 7.2) containing 20 mM glucose and rinsed in 5 mM HEPES-KOH, 20 mM glucose, and 100 mM KCl at pH 7.2. In both cases, DiSC3 (5) was added to a final concentration of 0.4 μM, and changes in the fluorescence were continuously recorded by using a Victor3 plate reader, after addition of peptides at 10 μM. A maximum reference value for total depolarization was calculated for cells exposed to 10% Triton X-100. Alternatively, changes in the fluorescence were continuously recorded after addition of serially diluted protein/peptides from 50 to 0.1 μM. The peptide concentrations required to achieve half of the total and the total membrane depolarization after a 40-min incubation time were estimated from nonlinear regression analysis as previously described (39).
Sytox Green fluorescence membrane permeabilization assay.
Membrane permeabilization was determined in a Sytox Green fluorescence membrane permeabilization assay. For the planktonic assay, bacterial cells were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 5 min, and resuspended in 1× PBS to an OD600 of 0.2. An aliquot of 200 μl was then transferred to a microtiter plate. For the biofilm assay, a 24-h formed P. aeruginosa biofilm in a 96-well microtiter plate was washed three times with PBS. Sytox Green was added to a final concentration of 1 μM, and the mixture was incubated for 15 min in the dark. Changes in the fluorescence signal were continuously recorded after the addition of serially diluted protein/peptides from 50 to 0.1 μM in a Victor3 plate reader. The peptide concentrations required to achieve half of the total and the total membrane permeabilization achieved after 40 min were estimated from nonlinear regression analysis as previously described (23). A maximum reference value was determined using 10% Triton X-100.
Confocal microscopy.
Overnight cultures of P. aeruginosa were centrifuged at 5,000 × g for 5 min, washed with LB medium, and adjusted to an OD600 of 0.1. The bacterial suspension was added to a 35-cm2 plate and incubated at 37°C for 24 h. The formation of biofilm was then evaluated using the crystal violet staining technique (38). Bacterial cell viability within biofilms was also assessed using fluorescent markers. At 24 h, P. aeruginosa biofilms were washed three times with PBS and rinsed in 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.4, containing 1:1 Syto9/propidium iodide, DAPI, or FITC-WGA (both at 10 μg/μl) and incubated in the dark for 15 min. Afterwards, the stained biofilms were gently washed with PBS in order to remove the excess fluorescent dye. Syto9/propidium iodide staining allowed us to monitor the bacterial viability within the biofilm as the Syto9 dye is able to permeate the cell membrane and stain the nucleic acid of healthy cells; whereas, when the cell membrane is damaged, propidium iodide can diffuse into the cell and displace Syto9. On the other hand, DAPI/WGA staining allows us to monitor the antimicrobial action on the biofilm at both, exopolysaccharide (EPS) and cellular levels. The lectin WGA-FITC conjugated is able to interact with the β-N-acetylglucosamine bonds present on the EPS molecules, allowing the observation of the AMPs action on the EPS. At the same time, DAPI is able to stain the bacterial DNA, showing the antimicrobial effect on the bacteria that form the biofilm. Then biofilms were incubated with 10 μM protein/peptides for 4 h at 37°C. Changes were imaged using a laser scanning confocal microscope (Olympus FluoView 1000 equipped with a UPlansApo 60× lens in 1.4 oil immersion objective, United Kingdom).
For quantitative analysis of live/dead bacterial cells, biofilms were grown as above in a plate coverslip system. The grown biofilms were then washed three times with PBS. Then, each well was treated with 10 μM protein/peptide resuspended in 10 mM sodium phosphate buffer pH 7.5, 0.1 M NaCl and incubated at 37°C for 4 h. After incubation, the wells were washed three times with PBS to remove planktonic bacteria. Then, the biofilms were prestained using the SYTO9/propidium iodide 1:1 mixture provided by the Live/Dead staining kit (Molecular Probes, Eugene, OR, USA) and incubated in the dark at 37°C for 15 min. Three washes were performed to remove excess dye, and then the biofilm was imaged using a laser scanning confocal microscope (Olympus FluoView 1000, equipped with an UPlansApo 40× lens with a 1.4 numerical aperture oil immersion objective. Mortality was calculated as the mean area of 10 randomly selected fields and quantified using ImageJ software (40). Unspecific background was subtracted, and particles with a diameter larger than 0.8 μm were analyzed. Biofilm three-dimensional reconstruction and estimation of the biofilm depth were performed with the IMARIS Bitplane imaging software (Oxford Instruments, Zurich, Switzerland).
Statistical analysis.
Data obtained for each assay were tested by one-way analysis of variance (ANOVA) to determine statistical significance. If an effect was present, Scheffé and Bonferroni post hoc tests were used for individual comparisons, using SPSS software (IBM, USA).
RESULTS
Antimicrobial peptide design.
Human RNase 3 has previously been described as a potent antimicrobial agent, displaying a mechanism of action against Gram-negative bacteria that is mediated by cell aggregation in an LPS-dependent manner (26). In our laboratory, we demonstrated that this eosinophil-secreted RNase interacts with the negatively charged surfaces of Gram-negative bacteria and experiences a conformational change that triggers the amyloid-like self-aggregation of the protein, resulting in bacterial agglutination and cell death (27). The structural determinants for the antimicrobial action are encoded in the first two α-helices of the N-terminal region of the protein; the first α-helix (F5 to S17) is required for bacterial aggregation, and the second helix (T22 to R36) is required for the direct antimicrobial action through LPS interaction and membrane disruption (28, 29). Previous studies by rational structure minimization led us to identify the minimal entity of the protein N-terminal active regions, where the two first α-helices were bound by the chemical linker acid amino hexanoic (Ahx). The designed RNase 3-derived peptide fully retained the capacity to interact with both bacterial membranes and LPS, producing bacterial cell agglutination and leakage (33). In this work, we further refined the antimicrobial peptide by pruning both peptide ends, removing nonessential residues, and connecting the two active domains by a single proline residue (see Table S1 in the supplemental material). The RN3(5-17P22-36) antimicrobial properties of E. coli and Staphylococcus aureus were compared with the those for the parental protein and RN3(1-36) peptide, and we observed an enhancement of the antimicrobial and bacterial agglutinating activities (see Table S2 in the supplemental material) while retaining an equivalent affinity for LPS (see Table S3 in the supplemental material) and the ability to disrupt and aggregate dioleolylphosphatdylcholine/dioleoylphosphatidylglycercol (DOPC/DOPG) model membranes (see Fig. S1 and S2 in the supplemental material). No activity was detected under the same assayed conditions for pure neutral vesicles (data not shown), as previously observed for RNase 3 (41). The data also confirmed that the cell agglutination was specific for the tested Gram-negative species, as previously reported for the other assayed RNase peptides (17, 33). Most importantly, no detectable hemolysis was found at concentrations up to 25 μM, and 50% lysis was achieved at 217 ± 42.17 μM for the RNase 3 protein and 178,33 ± 81.72 μM for the RN3(5-17P22-36) peptide. Moreover, no significant cytotoxicity was observed for the MRC-5 human cell line when tested at values above 50- to 100-fold the peptide and protein bactericidal 50% effective dose, respectively, providing a suitable therapeutic index (see Table S4 in the supplemental material).
We proceeded to analyze the RN3(5-17P22-36) derived peptide's antimicrobial action against P. aeruginosa strain PAO1 and also three P. aeruginosa clinical isolates (see Table S5 in the supplemental material). The encouraging results led us to compare the RN3(5-17P22-36) mechanism of action with other reference peptides (see Table S1 in the supplemental material) with selected antimicrobial properties: either as potent P. aeruginosa inhibitors on both planktonic and biofilms or bacterial agglutinating agents. First, from the cathelicidin family, we selected the antimicrobial peptide LL-37, which presents a high interaction for the negatively charged bacterial membranes and LPS molecules and has reported antimicrobial activity against planktonic P. aeruginosa (42). Furthermore, LL-37 is able to both inhibit P. aeruginosa biofilm formation (43, 44) and remove preformed P. aeruginosa biofilms (43, 45, 46). Second, the antimicrobial synthetic fusion peptide cecropin A (1-7)-melittin (2–9), referred to here as CA-M, was chosen as it is a potent antimicrobial peptide against a wide variety of microorganisms. The hybrid peptide CA-M is a small, amphipathic, and cationic peptide which presents a high affinity to bacterial negatively charged membranes, exerting its antimicrobial action via pore formation (47, 48) and was recently described to be active against P. aeruginosa biofilms (45). The third choice was the β-boomerang WY-12 peptide, which adopts a particular β-stranded secondary structure that resembles a boomerang upon interaction with LPS (49). Last, we selected the human parotid secretory derived peptide GL-13, which is able to induce P. aeruginosa agglutination (50).
Antimicrobial action on planktonic P. aeruginosa.
To determine antimicrobial actions on planktonic cells, we first assessed peptide activity on P. aeruginosa cells by determining the MBC. All the AMPs under investigation were effective at a micromolar range, except for the GL-13 peptide, which showed no bactericidal activity even at the maximum tested concentration (500 μM). In particular, RNase 3 and the N-terminal peptide RN3(5-17P22-36) were the most active compounds, displaying MBC values of about 2.5 μM, followed by the well-known antimicrobial peptides LL-37 and CA-M, which required a 2-fold concentration to produce the same microbicidal activity. Last, the antimicrobial peptide WY-12 needed more than 10 μM to totally eliminate P. aeruginosa cells (Table 1).
TABLE 1.
Antimicrobial and agglutinating activities of RNase 3, RN3(5-17P22-36), and reference peptides on planktonic P. aeruginosaa
| Peptide | MBC (μM) | ED50 (μM) | MAC (μM) |
|---|---|---|---|
| RNase 3 | 2.50 ± 0.05 | 0.84 ± 0.03 | 2.00 ± 0.15 |
| RN3(5-17P22-36) | 2.50 ± 0.08 | 1.25 ± 0.10 | 1.50 ± 0.08 |
| GL-13 | >50 | ND | 0.70 ± 0.05 |
| LL-37 | 6.00 ± 0.05 | 2.28 ± 0.22 | >50 |
| WY-12 | 12.50 ± 0.12 | 7.72 ± 0.37 | >50 |
| CA-M | 6.00 ± 0.05 | 2.54 ± 1.08 | >50 |
The peptides GL-13 (50), LL-37 (46), WY-12 (49), and CA-M (48) were included as reference peptides. The MBC, the ED50 for bacterial cell viability reduction, and the MAC were calculated as described in Materials and Methods. ND, not detected at the maximum assayed concentration (50 μM). All assays were performed in triplicate. Reported values are means ± standard errors of the means.
We next monitored microbial cell viability by using the BacTiter-Glo luminescent kit. Viable P. aeruginosa cells that were metabolically active were measured based on ATP quantification, using a coupled luciferin/oxyluciferin in the presence of luciferase, where luminescence is proportional to ATP and hence to the number of viable cells in the culture. Results (Table 1) highlighted RNase 3 as the most active compound, with ED50 values below 1 μM, followed by RN3(5-17P22-36) and then LL-37, CA-M, and WY-12, which needed, respectively, increasing concentrations to reduce cell viability to 50%. No cell viability reduction was detected for the peptide GL-13, even at the maximum concentration assayed (50 μM).
We previously described the importance of the bacterial agglutination of RNase 3 for the antimicrobial activity of the protein (27). Therefore, we assayed the protein's capability to agglutinate P. aeruginosa cells in a MAC assay (Table 1). As described previously for other Gram-negative species, RNase 3 was able to agglutinate P. aeruginosa at a micromolar range. Interestingly, the N-terminal RNase 3-derived peptide RN3(5-17P22-36) displayed an even lower MAC value, producing bacterial agglutination at 1.5 μM. In addition, the peptide GL-13 was able to agglutinate P. aeruginosa in the submicromolar range, as previously described (50), but was devoid of any antimicrobial activity. Last, LL-37, CA-M, and WY-12 peptides were not able to produce any bacterial agglutination.
LPS binding affinity was monitored in a fluorometric assay which measured the competitive displacement of a cadaverine fluorescent probe (BC) by the protein and peptides. The 50% LPS-binding affinity (ED50), defined as the AMP concentration needed to achieve displacement of half of the BC, and the total LPS-binding affinity, expressed as a percentage, were determined (Table 2). Interaction with negatively charged surfaces of bacteria is the first step for cationic peptides to reach the bacterial membranes and exert their antimicrobial action. All tested peptides were able to displace the fluorescent BC probe in a micromolar range, confirming LPS interaction. It is worth noticing that RN3(5-17P22-36), together with LL-37 and WY-12, achieved total displacement of the BC molecule under the assayed conditions.
TABLE 2.
LPS-binding properties of RNase 3, RN3(5-17P22-36), GL-13, LL-37, WY-12, and CA-M peptidesa
| Peptide | LPS binding |
|
|---|---|---|
| ED50 (μM) | % of maximum | |
| RNase 3 | 1.79 ± 0.32 | 99.85 ± 9.51 |
| RN3(5-17P22-36) | 2.11 ± 0.12 | 92.16 ± 2.06 |
| GL-13 | 1.63 ± 0.11 | 44.37 ± 3.84 |
| LL-37 | 2.65 ± 0.11 | 100.00 ± 3.13 |
| WY-12 | 2.90 ± 0.02 | 99.08 ± 0.50 |
| CA-M | 5.69 ± 0.89 | 69.16 ± 6.23 |
The ED50 for displacement of the dye and the percentage of maximum displacement were calculated as described in Materials and Methods. A value of 100% indicates total displacement, whereas 0% indicates no displacement of the dye, i.e., no binding. Values are means ± standard errors of the means.
To further investigate the antimicrobial mechanism of action of the RNase 3-derived peptide, we evaluated its ability to depolarize P. aeruginosa membranes. Depolarization activity was analyzed in a 3,3′-dipropylthiadicarbocyanine iodide (DiSC3) assay (5) (Table 3). DiSC3 is a fluorescent probe that is sensitive to membrane potential. DiSC3 fluorescence is quenched upon interaction with intact cell membranes. When the membrane potential is lost, the probe is released to the medium, resulting in an increase of fluorescence that can be recorded as a function of peptide concentration. Thus, we measured the ED50 in terms of depolarization action and the total depolarization activity at the final incubation time (Table 3). All the AMPs tested were able to depolarize P. aeruginosa cytoplasmic membranes when present in the micromolar range, except for the peptide GL-13, which was not able to produce any depolarization at the concentrations tested. Moreover, RNase 3 and its N-terminal-derived peptide RN3(5-17P22-36) were able to achieve close to total depolarization of the P. aeruginosa population, showing even higher values than the antimicrobial peptide CA-M, which is frequently used as a positive control due to its high depolarization activity (47, 48).
TABLE 3.
Membrane permeabilization and membrane depolarization activities of RNase 3, RN3(5-17P22-36), GL-13, LL-37, WY-12, and CA-M peptides on planktonic P. aeruginosa cellsa
| Peptide | Depolarization |
Permeabilization |
||
|---|---|---|---|---|
| ED50 (μM) | % of maximum | ED50 (μM) | % of maximum | |
| RNase 3 | 1.54 ± 0.06 | 80.03 ± 2.67 | 0.92 ± 0.17 | 45.67 ± 2.29 |
| RN3(5-17P22-36) | 1.26 ± 0.03 | 78.19 ± 6.33 | 1.67 ± 0.90 | 63.63 ± 1.42* |
| GL-13 | ND | ND | ND | ND |
| LL-37 | 1.31 ± 0.01 | 28.24 ± 2.91 | 2.20 ± 0.23 | 38.80 ± 1.13 |
| WY-12 | 4.23 ± 0.21 | 44.02 ± 4.01 | 7.18 ± 1.15 | 39.43 ± 1.19 |
| CA-M | 3.01 ± 0.31 | 71.36 ± 7.60 | 6.11 ± 0.58 | 34.69 ± 2.18 |
Depolarization and permeabilization activities were calculated as described in Materials and Methods. The ED50 values for total activity and the mean percentage of maximum activity are shown (percentages were calculated by using 10% Triton X-100 to obtain the maximum reference value). ND, not detected at the maximum assayed concentration (50 μM). Values are means ± standard errors of the means.*, P < 0.05; significant difference based on ANOVA with the Scheffé and Bonferroni post hoc tests.
Further insight into the membrane-permeabilizing effect of the selected AMPs against P. aeruginosa was evaluated in a Sytox Green assay. Local membrane disturbances allow the cell uptake of the fluorescent Sytox Green dye, which subsequently stains the cell's nucleic acids; a fluorescence increase is determined to monitor cell membrane permeabilization. Therefore, we measured the ED50 in terms of membrane permeabilization and the total permeabilization activity, expressed as a percentage (Table 3). All the studied AMPs were able to permeabilize P. aeruginosa cytoplasmic membranes when present at a micromolar range, with the exception of GL-13, which was unable to cause any cell membrane permeabilization under the concentrations tested. RNase 3 was the most active compound, with an ED50 below 1 μM, while the RN3(5-17P22-36) and LL-37 peptides needed approximately double this concentration to achieve the ED50. On the other hand, WY-12 and CA-M required 6- to 7-fold-greater concentrations than RNase 3 to produce their respective ED50s. Regarding the total permeabilizing effect, we can highlight that RNase 3 and the RN3(5-17P22-36) peptide showed significantly higher activities.
Antimicrobial action on P. aeruginosa biofilms.
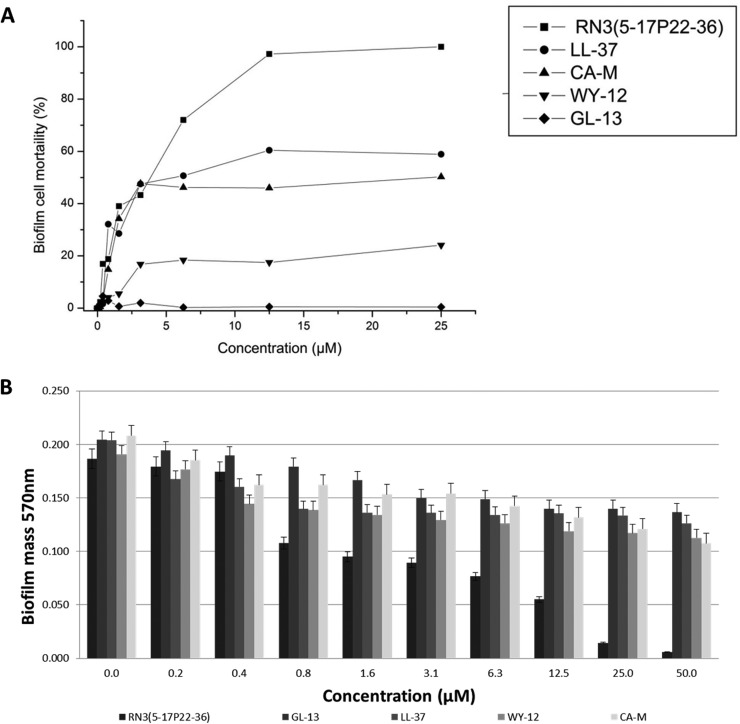
After the encouraging results achieved against planktonic P. aeruginosa cells, we decided to further characterize the antimicrobial action of the RNase 3-derived peptide against P. aeruginosa biofilms. As a first approach, we monitored the biofilm cell viability by using the BacTiter-Glo luminescent assay. The antimicrobial action of the studied AMPs was determined on already-established P. aeruginosa biolfilms (Fig. 1 and Table 4). All the AMPs tested were able to exert their bactericidal action in a micromolar range, with the exception of GL-13, which was unable to cause any cell mortality even at the maximum tested concentration (50 μM). However, for all the active peptides, the concentrations needed to produce an equivalent effect on biofilms were between 2 and 10 times higher than for planktonic cultures. Interestingly, we determined that the only peptide that could reduce the bacterial population on the established biofilm in its totality was the RN3(5-17P22-36), showing comparable results to the parental protein, which achieved total cell death at concentrations well below 50 μM. At the same time, we applied the crystal violet technique to assess the peptide's ability to disperse preestablished P. aeruginosa biofilms. The biofilm eradication activity was determined. Interestingly, the derived N-terminal peptide RN3(5-17P22-36) presented a biofilm eradication activity that was similar to that of its parental protein. On the other hand, the reference peptides LL-37, CA-M, WY-12, and GL-13 showed moderate effects, being able to disperse from 30 to 50% of the total biofilm at 50 μM (Table 4). Moreover, the biofilm eradication activities of all the tested reference peptides were well above the maximum concentration tested (50 μM), except for RN3(5-17P22-36), which was able to totally remove the biofilm cell community layer well below this threshold. We then evaluated the protein and RN3 peptide biofilm dispersion and eradication actions by monitoring the remaining CFU in both the biofilm layer and supernatant fractions as a function of the protein/peptide concentration. Results indicated that biofilm dispersion precedes bacterial mortality, with a calculated ED50 of <1 μM and MBEC of ≤5 μM for biofilm layer removal (see Fig. S3 in the supplemental material).
FIG 1.
(A) Biofilm cell mortality caused by RN3(5-17P22-36), GL-13, LL-37, WY-12, and CA-M peptides on established P. aeruginosa biofilms, assessed by using the BactTiter-Glo luminescence assay. (B) Biofilm eradication by peptides on established P. aeruginosa biofilms, assayed by using the crystal violet technique as described in Materials and Methods.
TABLE 4.
Biofilm eradication and killing activities of RNase 3, RN3(5-17P22-36), GL-13, LL-37, WY-12, and CA-M peptides on preestablished P. aeruginosa biofilmsa
| Peptide | Biofilm eradication (% of maximum) | Biofilm cell killing (% of maximum) |
|---|---|---|
| RNase 3 | 63.29 ± 4.08 | 98.50 ± 1.50* |
| RN3(5-17P22-36) | 100.00 ± 5.00* | 96.40 ± 2.41* |
| GL-13 | 33.28 ± 1.07 | ND |
| LL-37 | 35.54 ± 1.13 | 64.05 ± 3.33 |
| WY-12 | 38.36 ± 2.36 | 39.67 ± 8.93 |
| CA-M | 53.03 ± 1.46 | 54.32 ± 5.51 |
The mean percentage for maximum activity was calculated for biofilm eradication and cell killing activities by using a bacteria viability cell luminescent assay as described in Materials and Methods, with values for 10% Triton X-100 treatment used as the maximum reference values. ND, not detected at the maximum assayed concentration (50 μM). Values are means ± standard errors of the means. *, P < 0.05; significant differences were determined by ANOVA with Scheffé and Bonferroni post hoc tests.
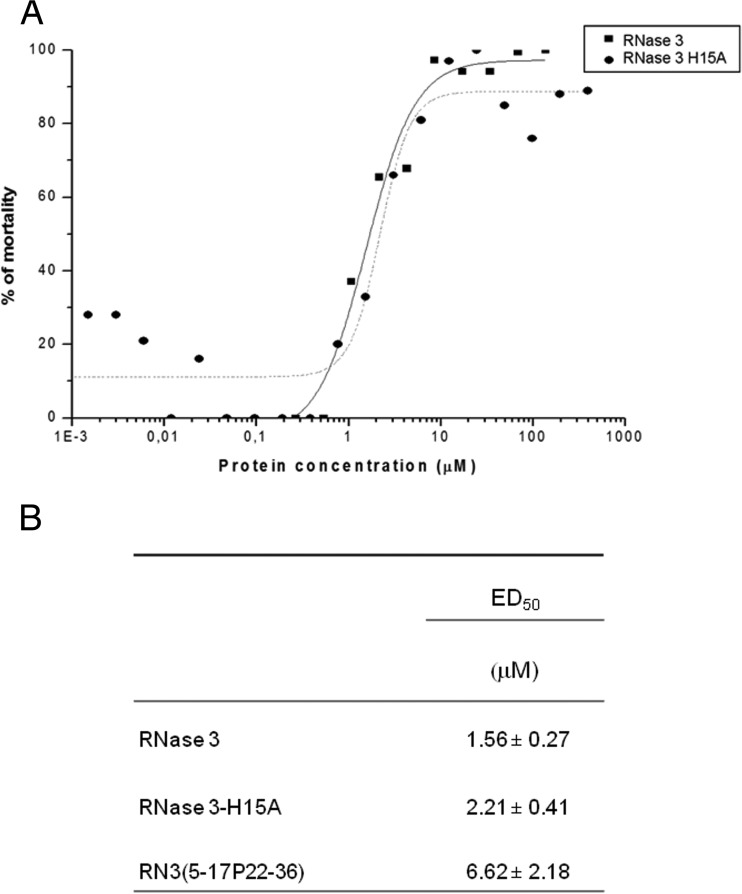
Interestingly, the biofilm eradication activity was found to not depend on the RNase catalytic activity, as illustrated by the comparative cell viability profiles of both the wild-type and the defective RNase 3 active site mutant (Fig. 2).
FIG 2.
Effects on preestablished P. aeruginosa biofilms by wild-type RNase 3, an active site mutant (RNase 3 H15A), and the RN3 peptide. (A) Comparison of mortality percentage profiles for the wild type and the H15A mutant. Bacterial cell mortality was assessed by using the Bac-Titer Glo assay as described in Materials and Methods. (B) Calculated ED50 values for biofilm eradication, determined as described in Materials and Methods. We used 10% Triton X-100 to determine the maximum reference value. Values indicated are mean results ± standard errors of the means.
The antimicrobial mechanism of action of the RNase 3-derived peptide on P. aeruginosa biofilms was further characterized in the depolarization and permeabilization assays. As for the P. aeruginosa planktonic culture, the fluorescent DisC3 assay was also used here in order to monitor the depolarization activities of the studied protein and peptides against established biofilms. Therefore, we measured ED50 (in terms of depolarization) and the total depolarization activity, expressed as a percentage (Table 5). Results indicated that the biofilm structure hinders the depolarization activity of the studied AMPs; the depolarization activities of all the peptides tested were reduced 2- to 3-fold compared to the effect in the planktonic P. aeruginosa culture. Similar results were obtained in the Sytox Green permeabilization assay, where the total permeabilization effect was hindered for all the AMPs tested, and between 2- and 10-fold higher concentrations were required to achieve half of the biofilm cell permeabilizing effect, in comparison to the planktonic cells. Nevertheless, significantly higher mean percentage values at the maximum concentration tested were achieved for the RN3(5-17P22-36) peptide.
TABLE 5.
Biofilm cell membrane depolarization and permeabilization activities of RNase 3, RN3(5-17P22-36), GL-13, LL-37, WY-12, and CA-M peptides on established P. aeruginosa biofilmsa
| Peptide | Depolarization |
Permeabilization |
||
|---|---|---|---|---|
| ED50 (μM) ± SEM | Mean % ± SEM | ED50 (μM) ± SEM | Mean % ± SEM | |
| RNase 3 | 9.45 ± 0.97 | 46.11 ± 3.21 | 8.59 ± 1.70 | 39.4 ± 2.88 |
| RN3(5-17P22-36) | 3.69 ± 0.37 | 73.51 ± 1.94* | 6.60 ± 2.01 | 64.26 ± 5.44* |
| GL-13 | ND | ND | ND | ND |
| LL-37 | 8.83 ± 0.54 | 31.85 ± 1.15 | 12.35 ± 3.13 | 24.21 ± 3.71 |
| WY-12 | 18.11 ± 0.89 | 32.58 ± 1.87 | 18.79 ± 8.23 | 18.91 ± 2.23 |
| CA-M | 15.79 ± 0.77 | 39.09 ± 2.46 | 10.64 ± 0.11 | 23.01 ± 1.13 |
Data shown are the ED50s and the percentages of the maximums at the final incubation time for membrane depolarization and permeabilization, calculated as described in Materials and Methods. Results with 10% Triton X-100 were considered the maximum reference values. ND, not detected at the highest assayed concentration (50 μM). Values are means ± standard errors of the means. *, P < 0.05; significant differences were determined by ANOVA with Scheffé and Bonferroni post hoc tests.
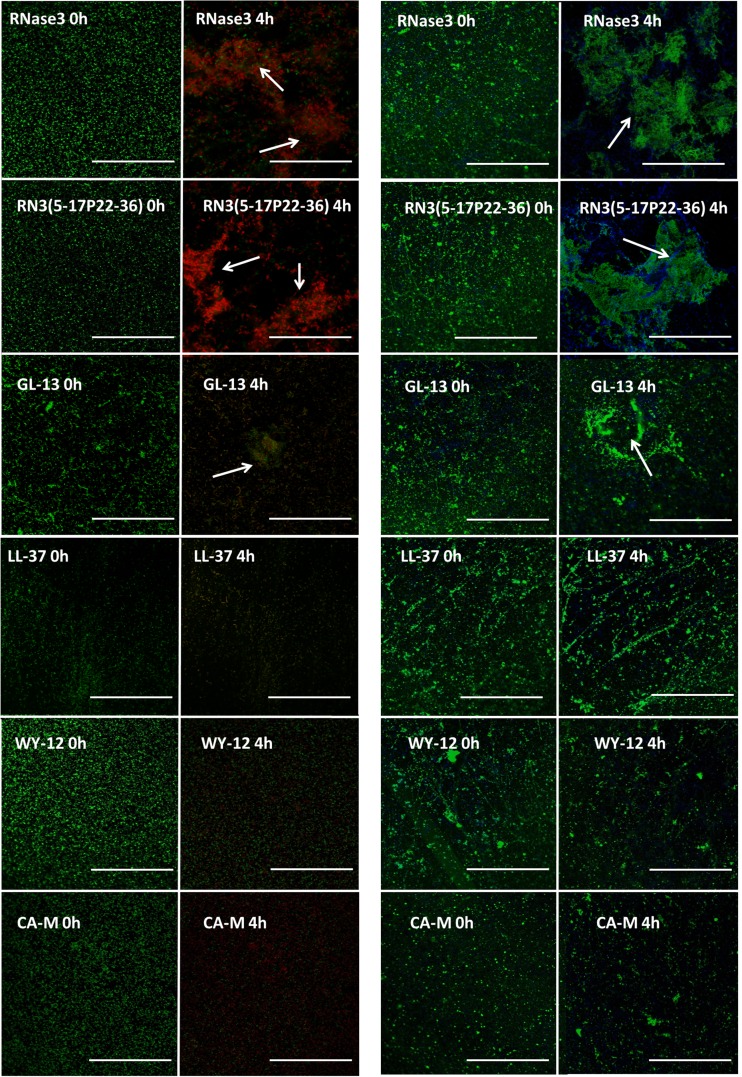
Finally, confocal microscopy studies were performed in order to visualize the antibiofilm action of the tested peptides. As a first approach, we used the Live/Dead staining technique, which allowed us to monitor the cell viability on P. aeruginosa biofilms, where live cells stain green by SYTO9 and dead cells are stained red by propidium iodide. The images captured after 4 h of incubation showed that all the AMPs tested were able to induce bacterial killing on the established biofilms (Fig. 3). Interestingly, RNase 3 and its N-terminal-derived peptide were able to induce prominent biofilm cell aggregation. This property was also shared by the GL-13 peptide, although the aggregates were smaller and less frequent. Established biofilms were then stained with WGA-FITC (WGA from Triticum vulgaris, which is able to recognize the β-N-acetylglucosamine bonds of the exopolysaccharide (EPS) molecules and with the nucleic acid DAPI dye, which labels the bacterial cells. This technique allowed us to visualize how the antimicrobial actions of the tested AMPs produced evident damage to the biofilm extracellular matrix structure. In addition, we confirmed that the biofilm-dispersed areas were devoid of EPS. Moreover, the results suggested that the EPS molecules could be involved in the biofilm agglutination process, as visualized aggregates were composed of both EPS and bacteria. Quantitative analysis of RNase 3 and RN3(5-17P22-36) peptide action on preestablished biofilms by using confocal microscopy following Live/Dead staining was applied to estimate the percentage of cell mortality on the biofilm total population, together with the respective reductions of the biofilm layer depth (see Fig. S4 in the supplemental material). Both the protein and its derived peptide were able to almost totally reduce the viability of the entire biofilm at 10 μM, damaging the biofilm architecture and decreasing its depth by more than 50%.
FIG 3.
Effects against established P. aeruginosa biofilms, determined via confocal microscopy. Live/Dead stain (green and red cells, respectively) and DAPI-FITC and WGA stains (blue and green, respectively) were used. White arrows show bacterial agglutination on the biofilm at a protein/peptide concentration of 10 μM. Bars, 20 μm.
DISCUSSION
Infections caused by P. aeruginosa pose a serious clinical challenge. Unfortunately, despite the advances in antimicrobial therapy, P. aeruginosa in its planktonic form causes acute infections, with mortalities as high as nearly 60% in immunocompromised patients (51). This opportunistic pathogen, once it is established within the host, is able to form biofilms, a sine qua non feature for the development of a variety of chronic infections. Indeed, chronic infections by this bacterium are the most common cause of fatal lung disease in patients with cystic fibrosis (7). In addition, the challenge is increased by the appearance of P. aeruginosa clinical strains resistant to the common prototypical antibiotic molecules (52, 53). Therefore, there is an urgent need to develop new effective antimicrobial strategies against both planktonic and biofilm forms of this dreadful pathogen.
Human antimicrobial RNases, and in particular the human RNase 3, display a rapid and potent antimicrobial effect against a wide variety of microorganisms (18). The antimicrobial activity of the secreted human RNases is mainly located at the N-terminal region (17). Accordingly, the RNase antimicrobial scaffold was engineered to produce new antimicrobial peptides (23, 33, 54). The present study investigated the antimicrobial potency of a new optimized version of the RNase 3 N-terminal peptide against both planktonic and biofilm forms of P. aeruginosa. The mechanism of action of the RNase 3-derived peptide was compared with previously characterized peptides with reported activity against planktonic and/or biofilm forms of P. aeruginosa (see Table S1 in the supplemental material).
Our findings revealed that human RNase 3 displays a strong antimicrobial activity against the Gram-negative pathogen P. aeruginosa. Interestingly, the N-terminal-derived peptide RN3(5-17P22-36) is able to achieve the same bactericidal effects as its parental protein at similar, low concentrations (Table 1), and is also effective against other tested P. aeruginosa clinical isolates (see Table S5 in the supplemental material). In tune with findings from previous functional assays on RNase 3 antimicrobial properties (55, 56), we also confirmed here that the protein antibiofilm activity is not dependent on its enzymatic activity (Fig. 2). The potential contribution of the protein catalytic activity was blocked by assaying the activity of the RNase 3 H15A variant, in which one of the two catalytic His residues is replaced. Previous work in our laboratory already confirmed that the H15A mutant was an appropriate construct to test the effect of total removal of the RNase enzymatic activity without alteration of the protein's three-dimensional structure (PDB ID 4OWZ) (24). The present results corroborate that the active site-defective mutant achieves an antibiofilm activity equivalent to that of the wild-type protein (Fig. 2).
Interestingly, the RNase 3 antimicrobial action is mediated through bacterial agglutination of Gram-negative bacteria (27). Here, we show that RNase 3 is able to agglutinate P. aeruginosa at concentrations as low as the those required for its antimicrobial action (Table 1). Additionally, the N-terminal-derived peptide RN3(5-17P22-36) retains the biophysical properties of the whole protein, promoting the same agglutinating activity at even lower concentrations. It is noteworthy that the antimicrobial activity of RN3(5-17P22-36) is higher than those recorded for the tested reference peptides. Moreover, the agglutinating activities of both RNase 3 and RN3(5-17P22-36) against P. aeruginosa are similar to the activity displayed by the potent agglutinating peptide GL-13. However, the GL-13 peptide does not display detectable antimicrobial activity.
Another key feature that mediates the peptide's activity against Gram-negative bacteria and facilitates its antimicrobial action is the ability to interact with the negatively charged surfaces of bacteria, in particular, the outer membrane LPS. Although LPS could pose a hydrophilic barrier to hydrophobic antimicrobial compounds, several AMPs are able to take advantage of their cationicity and use the LPS molecule as a first point of interaction in exerting their bactericidal activities. Previously, we showed that RNase 3 presents a high affinity for interacting with the Gram-negative LPS (25) and that the structural determinants for this LPS recognition are encoded at the N-terminal part of the protein (28). The selected reference AMPs tested in this work were also previously reported to display a high affinity toward LPS (42, 49, 57, 58). Our findings are in tune with the previous published data; not only human RNase 3 and its N-terminal derivative peptide but also the reference peptides are able to interact with P. aeruginosa LPS at low concentrations (Table 2). After the interaction with the negatively charged molecules of the bacterial cell envelopes, antimicrobial peptides exert their bactericidal action by producing loss of membrane potential and permeabilizing the cytoplasmic membrane, thus inducing cell lysis and death. Our results (Table 3) showed that both RNase 3 and RN3(5-17P22-36) display even a higher membrane-depolarizing effect than CA-M, which was previously reported to display a high depolarization activity against a wide range of Gram-positive and Gram-negative bacteria (47). In addition, the antimicrobial peptides LL-37 and WY-12 present lower maximum depolarization against P. aeruginosa, indicating that other factors contribute to the peptide action at the cytoplasmic membrane. Recent insights into AMP interactions with LPS indicate that binding to the bacterium's outer membrane does not guarantee the peptide subsequent membrane-permeabilizing action. On the contrary, LPS may induce peptide aggregation at the cell wall, acting as an inactivating trap and impeding access to the cytoplasmic membrane (59). Indeed, a recent biophysical approach toward the underlying AMP structural requirements for biofilm eradication pointed to a delicate balance between peptide-bacterium adhesion and self-oligomerization (12). Interestingly, the N-terminal-derived peptide RN3(5-17P22-36) can overcome the outer membrane barrier and exerts a greater permeabilization effect against P. aeruginosa planktonic cells.
So far, our results highlight that human RNase 3 and its engineered N-terminal peptide are highly active against the planktonic form of P. aeruginosa. However, when bacteria are organized in a biofilm community, they acquire a great enhancement of their resistance toward antimicrobial agents. Biofilms constitute a complex structure where bacteria are coated by a dense layer of extracellular matrix. This matrix is mainly composed of polysaccharides and is generally referred as the EPS. Gram-negative EPS is mostly composed of extracellular matrix polysaccharides, such as LPS, proteoglycans, and extracellular DNA; most of these molecules are largely negatively charged, inhibiting the diffusion of antimicrobials via charge repulsion and steric hindrance (60).Therefore, it has been described that classic antibiotic compounds require between 10- and 1,000-fold-higher concentrations against biofilms to be as effective as they are against planktonic cells (61–63). Our findings demonstrate that human RNase 3 exhibits a high antibiofilm activity and is able to drastically reduce the viability of the cell population and disperse most of the established biofilm, with better results than the reference tested peptides (Table 4). In addition, our experimental data highlight that membrane depolarization and permeabilization also contribute to the RNase 3 antimicrobial mechanism of action on established biofilms (Table 5). The complex structure of P. aeruginosa biofilms hinders the antimicrobial action of most antimicrobial agents, as observed for the antimicrobial peptides LL-37, CA-M, and WY-12, which exhibit high antimicrobial activity against planktonic P. aeruginosa but are unable to totally eradicate established biofilms at the maximum tested concentrations and also show lower depolarization and permeabilization activities. Interestingly, the N-terminal-derived peptide RN3(5-17P22-36) not only retains the antimicrobial action of its parental protein but, in addition, can better overcome the complexity of the biofilm EPS barrier and reach the encased bacterial membranes. Most importantly, the RN3(5-17P22-36) peptide achieves total eradication of the established biofilm, mediated by membrane disruption and bacterial killing, and is able to eliminate both the cells forming the biofilm community and those released into the supernatant fraction (see Fig. S3 in the supplemental material). It is worth noting that the GL-13 peptide, which is unable to permeabilize the cytoplasmic membrane and cannot reduce the bacterial population of P. aeruginosa biofilms, does indeed show significant biofilm eradication activity. This surprising result suggests that bacterial agglutination may occur inside the biofilm, and therefore biofilm dispersion could be driven by the mechanical forces of bacterial agglutination. Notwithstanding this result, GL-13 has recently been further engineered to acquire antimicrobial activity by introducing Lys residues at three internal positions (58, 64, 65). In any case, the antibiofilm activity achieved with the new engineered GL-13 is still lower than that of the RNase 3 peptide, which combines potent antimicrobial and LPS-binding activities together with high cell agglutination activity. By using confocal microscopy, we were able to visualize the antimicrobial activity of the tested AMPs against established P. aeruginosa biofilms. Staining with the lectin WGA, which labels the EPS structure, showed how the AMPs tested were able to induce structural damage and eradication on the established P. aeruginosa biofilms. The confocal results indicated that the human RNase 3, its derived peptide, and GL-13 are the only ones able to induce bacterial agglutination inside the biofilm. It is known that RNase 3 mediates its antimicrobial action through bacterial agglutination. When the protein interacts with the negatively charged cell wall or cytoplasmic membranes, conformational changes take place that lead to protein self-aggregation and the ensuing bacterial agglutination. These results therefore suggest that an equivalent process could occur on biofilm surfaces where RNase 3 could interact with the heavily charged molecules of the extracellular matrix of the biofilm, triggering cell agglutination and leading to cell death. In addition, the N-terminal-derived peptide RN3(5-17P22-36), due to its size reduction and biophysical properties, could better overcome the EPS structure, causing greater havoc among the bacterial population. Thus, the combination of the mechanical dispersion promoted by bacterial agglutination and the antimicrobial action driven by membrane disruption could lead to the total eradication of an established biofilm (see Fig. S3 in the supplemental material). Recent studies have also identified peptides active against biofilms formed by P. aeruginosa and other Gram-negative multidrug-resistant species by using a library of truncated peptides derived from LL-37 (10, 46) and also the synthesis of d-enantiomer forms (43); however, these studies reported higher concentrations were required for removal of established P. aeruginosa biofilms. A third study recently highlighted the antimicrobial effects of the peptides LL-37 and CA-M on their own and in combination with traditional antibiotic compounds against P. aeruginosa biofilms (45). Very recently, biofilm eradication was also reported for a modified form of the lipopeptide battacin, the attached fatty acid that is the main determinant for the achieved efficiency (11). Therefore, despite the promising results, the antimicrobial performance of the assayed cationic peptides against mature biofilms did not achieve full biofilm eradication on its own. In order to overcome this problem, we present here an engineered RNase 3 N-terminal-derived peptide that combines an enhanced antimicrobial activity paired with high LPS-binding activity and the capability to agglutinate bacterial community cells, leading to total biofilm eradication at comparatively lower concentrations.
In conclusion, our results show for the first time that human RNase 3 presents high antibiofilm activity against P. aeruginosa. The antibiofilm activity of the protein is mediated by a combination of bacterial agglutination and direct cell killing. In addition, we demonstrated that the N terminal of the human RNase 3 scaffold is a lead candidate for development of new antibiofilm agents. Indeed, the engineered N-terminal-derived peptide not only retains the agglutinating and antimicrobial properties of the parental protein but also is able to eradicate mature P. aeruginosa biofilms. The results highlight the potential of human host defense proteins as useful templates to engineer alternative antibiotics. Further work is envisaged to optimize the RNase 3-derived peptide and eventually select the best antibiofilm candidates for clinical applications.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Ministerio de Economía y Competitividad (BFU2012-38965; SAF2015-66007-P) and cofinanced by FEDER funds and by the Generalitat de Catalunya (2014 SGR 728).
We gratefully acknowledge I. Gibert at the Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona (UAB), Cerdanyola del Vallés, Spain, for providing us with the Pseudomonas aeruginosa PAO1 strain and P. Martínez and P. Huedo at the Institut de Biotecnologia i de Biomedicina, UAB, Cerdanyola del Vallés, Spain, for help in optimizing the biofilm formation assays. We also thank Estela Noguera, M. Llorens, Lluis Santamaria, and Diego Velázquez for their support and technical help. Confocal studies were performed at the Servei de Microscopia of the UAB. Spectrofluorescence assays were performed at the Laboratori d'Anàlisi i Fotodocumentació, UAB. We are also thankful to M. Victòria Nogués for her critical reading of the manuscript.
We declare we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00830-16.
REFERENCES
- 1.Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. 2014. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 3:32. doi: 10.1186/2047-2994-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dettman JR, Rodrigue N, Kassen R. 2015. Genome-wide patterns of recombination in the opportunistic human pathogen Pseudomonas aeruginosa. Genome Biol Evol 7:18–34. doi: 10.1093/gbe/evu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections. Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 4.Kerr KG, Snelling AM. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect 73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Manfredi R, Nanetti A, Ferri M, Chiodo F. 2000. Pseudomonas spp. complications in patients with HIV disease: an eight-year clinical and microbiological survey. Eur J Epidemiol 16:111–118. doi: 10.1023/A:1007626410724. [DOI] [PubMed] [Google Scholar]
- 6.Bouza E, Burillo A, Munoz P. 2002. Catheter-related infections: diagnosis and intravascular treatment. Clin Microbiol Infect 8:265–274. doi: 10.1046/j.1469-0691.2002.00385.x. [DOI] [PubMed] [Google Scholar]
- 7.Valderrey AD, Pozuelo MJ, Jimenez PA, Macia MD, Oliver A, Rotger R. 2010. Chronic colonization by Pseudomonas aeruginosa of patients with obstructive lung diseases: cystic fibrosis, bronchiectasis, and chronic obstructive pulmonary disease. Diagn Microbiol Infect Dis 68:20–27. doi: 10.1016/j.diagmicrobio.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laverty G, Gorman SP, Gilmore BF. 2014. Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 3:596–632. doi: 10.3390/pathogens3030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X, Sambanthamoorthy K, Palys T, Paranavitana C. 2013. The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant Acinetobacter baumannii. Peptides 49:131–137. doi: 10.1016/j.peptides.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 11.De Zoysa GH, Cameron AJ, Hegde VV, Raghothama S, Sarojini V. 2015. Antimicrobial peptides with potential for biofilm eradication: synthesis and structure activity relationship studies of battacin peptides. J Med Chem 58:625–639. doi: 10.1021/jm501084q. [DOI] [PubMed] [Google Scholar]
- 12.Segev-Zarko LA, Saar-Dover R, Brumfeld V, Mangoni ML, Shai Y. 2015. Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem J 468:259–270. doi: 10.1042/BJ20141251. [DOI] [PubMed] [Google Scholar]
- 13.Laverty G, Gorman SP, Gilmore BF. 2015. Biofilm eradication kinetics of the ultrashort lipopeptide C12-OOWW-NH2 utilizing a modified MBEC assay™. Chem Biol Drug Des 85:645–652. doi: 10.1111/cbdd.12441. [DOI] [PubMed] [Google Scholar]
- 14.Yeung ATY, Gellatly SL, Hancock REW. 2011. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci 68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anunthawan T, de la Fuente-Nunez C, Hancock RE, Klaynongsruang S. 2015. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim Biophys Acta 1848:1352–1358. doi: 10.1016/j.bbamem.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Beloin C, Renard S, Ghigo JM, Lebeaux D. 2014. Novel approaches to combat bacterial biofilms. Curr Opin Pharmacol 18:61–68. doi: 10.1016/j.coph.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Torrent M, Pulido D, Valle J, Nogues MV, Andreu D, Boix E. 2013. Ribonucleases as a host-defence family: evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem J 456:99–108. doi: 10.1042/BJ20130123. [DOI] [PubMed] [Google Scholar]
- 18.Boix E, Salazar VA, Torrent M, Pulido D, Nogues MV, Moussaoui M. 2012. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol Chem 393:801–815. doi: 10.1515/hsz-2012-0160. [DOI] [PubMed] [Google Scholar]
- 19.Venge P, Bystrom J, Carlson M, Hakansson L, Karawacjzyk M, Peterson C, Seveus L, Trulson A. 1999. Eosinophil cationic protein (ECP): molecular and biological properties and the use of ECP as a marker of eosinophil activation in disease. Clin Exp Allergy 29:1172–1186. doi: 10.1046/j.1365-2222.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- 20.Acharya KR, Ackerman SJ. 2014. Eosinophil granule proteins: form and function. J Biol Chem 289:17406–17415. doi: 10.1074/jbc.R113.546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bystrom J, Amin K, Bishop-Bailey D. 2011. Analysing the eosinophil cationic protein—a clue to the function of the eosinophil granulocyte. Respir Res 12:10. doi: 10.1186/1465-9921-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik A, Batra JK. 2012. Antimicrobial activity of human eosinophil granule proteins: involvement in host defence against pathogens. Crit Rev Microbiol 38:168–181. doi: 10.3109/1040841X.2011.645519. [DOI] [PubMed] [Google Scholar]
- 23.Pulido D, Torrent M, Andreu D, Nogues MV, Boix E. 2013. Two human host defense ribonucleases against mycobacteria, the eosinophil cationic protein (RNase 3) and RNase 7. Antimicrob Agents Chemother 57:3797–3805. doi: 10.1128/AAC.00428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salazar VA, Arranz-Trullen J, Navarro S, Blanco JA, Sánchez D, Moussaoui M, Boix E. 8 June 2016. Exploring the mechanisms of action of human secretory RNase 3 and RNase 7 against Candida albicans. Microbiologyopen doi: 10.1002/mbo3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torrent M, Navarro S, Moussaoui M, Nogues MV, Boix E. 2008. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 47:3544–3555. doi: 10.1021/bi702065b. [DOI] [PubMed] [Google Scholar]
- 26.Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. 2012. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob Agents Chemother 56:2378–2385. doi: 10.1128/AAC.06107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torrent M, Pulido D, Nogués MV, Boix E. 2012. Exploring new biological functions of amyloids: bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog 8:e1003005. doi: 10.1371/journal.ppat.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torrent M, de la Torre BG, Nogues VM, Andreu D, Boix E. 2009. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochem J 421:425–434. doi: 10.1042/BJ20082330. [DOI] [PubMed] [Google Scholar]
- 29.Torrent M, Odorizzi F, Nogues MV, Boix E. 2010. Eosinophil cationic protein aggregation: identification of an N-terminus amyloid prone region. Biomacromolecules 11:1983–1990. doi: 10.1021/bm100334u. [DOI] [PubMed] [Google Scholar]
- 30.Torrent M, Di Tommaso P, Pulido D, Nogués M, Notredame C, Boix E, Andreu D. 2012. AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics 28:130–131. doi: 10.1093/bioinformatics/btr604. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boix E, Nikolovski Z, Moiseyev GP, Rosenberg HF, Cuchillo CM, Nogues MV. 1999. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a subsite arrangement that favors exonuclease-type activity. J Biol Chem 274:15605–15614. doi: 10.1074/jbc.274.22.15605. [DOI] [PubMed] [Google Scholar]
- 33.Torrent M, Pulido D, De la Torre BG, Garcia-Mayoral MF, Nogues MV, Bruix M, Andreu D, Boix E. 2011. Refining the eosinophil cationic protein antibacterial pharmacophore by rational structure minimization. J Med Chem 54:5237–5244. doi: 10.1021/jm200701g. [DOI] [PubMed] [Google Scholar]
- 34.Salazar VA, Rubin J, Moussaoui M, Pulido D, Nogues MV, Venge P, Boix E. 2014. Protein post-translational modification in host defense: the antimicrobial mechanism of action of human eosinophil cationic protein native forms. FEBS J 281:5432–5446. doi: 10.1111/febs.13082. [DOI] [PubMed] [Google Scholar]
- 35.Pulido D, Moussaoui M, Nogues MV, Torrent M, Boix E. 2013. Towards the rational design of antimicrobial proteins: single point mutations can switch on bactericidal and agglutinating activities on the RNase A superfamily lineage. FEBS J 280:5841–5852. doi: 10.1111/febs.12506. [DOI] [PubMed] [Google Scholar]
- 36.Wood SJ, Miller KA, David SA. 2004. Anti-endotoxin agents. 1. Development of a fluorescent probe displacement method optimized for the rapid identification of lipopolysaccharide-binding agents. Comb Chem High Throughput Screen 7:239–249. doi: 10.2174/1386207043328832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huedo P, Yero D, Martinez-Servat S, Estibariz I, Planell R, Martinez P, Ruyra A, Roher N, Roca I, Vila J, Daura X, Gibert I. 2014. Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J Bacteriol 196:2431–2442. doi: 10.1128/JB.01540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 39.Torrent M, Badia M, Moussaoui M, Sanchez D, Nogues MV, Boix E. 2010. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J 277:1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrent M, Cuyas E, Carreras E, Navarro S, Lopez O, de la Maza A, Nogues MV, Reshetnyak YK, Boix E. 2007. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry 46:720–733. doi: 10.1021/bi061190e. [DOI] [PubMed] [Google Scholar]
- 42.Gough M, Hancock RE, Kelly NM. 1996. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect Immun 64:4922–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean SN, Bishop BM, van Hoek ML. 2011. Susceptibility of Pseudomonas aeruginosa biofilm to alpha-helical peptides: D-enantiomer of LL-37. Front Microbiol 2:128. doi: 10.3389/fmicb.2011.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Fuente-Nunez C, Korolik V, Bains M, Nguyen U, Breidenstein EB, Horsman S, Lewenza S, Burrows L, Hancock RE. 2012. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob Agents Chemother 56:2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dosler S, Karaaslan E. 2014. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 62:32–37. doi: 10.1016/j.peptides.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 46.Nagant C, Pitts B, Nazmi K, Vandenbranden M, Bolscher JG, Stewart PS, Dehaye JP. 2012. Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments. Antimicrob Agents Chemother 56:5698–5708. doi: 10.1128/AAC.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milani A, Benedusi M, Aquila M, Rispoli G. 2009. Pore forming properties of cecropin-melittin hybrid peptide in a natural membrane. Molecules 14:5179–5188. doi: 10.3390/molecules14125179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saugar JM, Rodriguez-Hernandez MJ, de la Torre BG, Pachon-Ibanez ME, Fernandez-Reyes M, Andreu D, Pachon J, Rivas L. 2006. Activity of cecropin A-melittin hybrid peptides against colistin-resistant clinical strains of Acinetobacter baumannii: molecular basis for the differential mechanisms of action. Antimicrob Agents Chemother 50:1251–1256. doi: 10.1128/AAC.50.4.1251-1256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhunia A, Mohanram H, Domadia PN, Torres J, Bhattacharjya S. 2009. Designed beta-boomerang antiendotoxic and antimicrobial peptides: structures and activities in lipopolysaccharide. J Biol Chem 284:21991–22004. doi: 10.1074/jbc.M109.013573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorr SU, Sotsky JB, Shelar AP, Demuth DR. 2008. Design of bacteria-agglutinating peptides derived from parotid secretory protein, a member of the bactericidal/permeability increasing-like protein family. Peptides 29:2118–2127. doi: 10.1016/j.peptides.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Bassetti M, Righi E, Viscoli C. 2008. Pseudomonas aeruginosa serious infections: mono or combination antimicrobial therapy? Curr Med Chem 15:517–522. doi: 10.2174/092986708783503186. [DOI] [PubMed] [Google Scholar]
- 52.Hawkey PM. 2008. The growing burden of antimicrobial resistance. J Antimicrob Chemother 62(Suppl 1):i1–i9. doi: 10.1093/jac/dkn241. [DOI] [PubMed] [Google Scholar]
- 53.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 54.Pulido D, Arranz-Trullen J, Prats-Ejarque G, Velazquez D, Torrent M, Moussaoui M, Boix E. 2016. Insights into the antimicrobial mechanism of action of human RNase 6: structural determinants for bacterial cell agglutination and membrane permeation. Int J Mol Sci 17:552. doi: 10.3390/ijms17040552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberg HF. 1995. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J Biol Chem 270:7876–7881. [DOI] [PubMed] [Google Scholar]
- 56.Carreras E, Boix E, Rosenberg HF, Cuchillo CM, Nogues MV. 2003. Both aromatic and cationic residues contribute to the membrane-lytic and bactericidal activity of eosinophil cationic protein. Biochemistry 42:6636–6644. doi: 10.1021/bi0273011. [DOI] [PubMed] [Google Scholar]
- 57.Piers KL, Hancock REW. 1994. The interaction of a recombinant cecropin/melittin hybrid peptide with the outer-membrane of Pseudomonas aeruginosa. Mol Microbiol 12:951–958. doi: 10.1111/j.1365-2958.1994.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 58.Abdolhosseini M, Sotsky JB, Shelar AP, Joyce PB, Gorr SU. 2012. Human parotid secretory protein is a lipopolysaccharide-binding protein: identification of an anti-inflammatory peptide domain. Mol Cell Biochem 359:1–8. doi: 10.1007/s11010-011-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohanram H, Bhattacharjya S. 2014. Resurrecting inactive antimicrobial peptides from the lipopolysaccharide trap. Antimicrob Agents Chemother 58:1987–1996. doi: 10.1128/AAC.02321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beckloff N, Laube D, Castro T, Furgang D, Park S, Perlin D, Clements D, Tang H, Scott RW, Tew GN, Diamond G. 2007. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob Agents Chemother 51:4125–4132. doi: 10.1128/AAC.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 62.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 63.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 64.Hirt H, Gorr SU. 2013. Antimicrobial peptide GL13K is effective in reducing biofilms of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4903–4910. doi: 10.1128/AAC.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Hirt H, Li Y, Gorr SU, Aparicio C. 2014. Antimicrobial GL13K peptide coatings killed and ruptured the wall of Streptococcus gordonii and prevented formation and growth of biofilms. PLoS One 9:e111579. doi: 10.1371/journal.pone.0111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.