Abstract
Clinical isolates producing hybrid CTX-M β-lactamases, presumably due to recombination between the blaCTX-M-15 and blaCTX-M-14 elements, have emerged in recent years. Among the hybrid enzymes, CTX-M-64 and CTX-M-14 display the most significant difference in catalytic activity. This study aims to investigate the mechanisms underlying such differential enzymatic activities in order to provide insight into the structure/function relationship of this class of enzymes. Sequence alignment analysis showed that the major differences between the amino acid composition of CTX-M-64 and CTX-M-14 lie at both the N and C termini of the enzymes. Single or multiple amino acid substitutions introduced into CTX-M-64 and CTX-M-14 were found to produce only minor effects on hydrolytic functions; such a finding is consistent with the notion that the discrepancy between the functional activities of the two enzymes is not the result of only a few amino acid changes but is attributable to interactions between a unique set of amino acid residues in each enzyme. This theory is supported by the results of the thermal stability assay, which confirmed that CTX-M-64 is significantly more stable than CTX-M-14. Our data confirmed that, in addition to the important residues located in the active site, residues distal to the active site also contribute to the catalytic activity of the enzyme through stabilizing its structural integrity.
INTRODUCTION
The utilization of extended-spectrum cephalosporins for the treatment of bacterial infections has been extremely effective since the 1980s. However, such effectiveness has gradually been eroded as a result of the dissemination of resistance elements that encode a variety of β-lactamases, especially the extended-spectrum β-lactamases (ESBLs). Having emerged as a result of the extensive usage of β-lactam antibiotics in humans and animals, the class A serine enzymes of the CTX-M-type ESBLs have become the most common type of plasmid-mediated ESBL enzymes produced by drug-resistant organisms. Sharing less than 40% amino acid sequence homology with the TEM- and SHV-type enzymes, the CTX-M enzymes can be subclassified into six groups (CTX-M-1, -2, -8, -9, -25, and the KLUC groups) (1, 2).
Unlike other class A β-lactamases, the evolution of genetic elements encoding CTX-M enzymes is known to be mediated by random mutations and recombination between different resistance genes, in particular those encoding the CTX-M-1 and CTX-M-9 enzymes (3). Several hybrid CTX-M β-lactamases (CTX-M-64, CTX-M-123, CTX-M-137, and CTX-M-132), presumably resulting from recombination between blaCTX-M-15 and blaCTX-M-14, which are the most common variants worldwide, have been reported in recent years (4–7). Among these hybrid enzymes, CTX-M-64, which contained the N- and C-terminal portion of CTX-M-15 and the middle fragment of CTX-M-14, exhibited even higher catalytic activity than its parental prototypes (4).
The CTX-M-type enzymes degrade a wide range of substrates, including penicillin and the first, second, and third generation of cephalosporins, with exceptionally high activity against cefotaxime. A number of amino acid residues that are functionally important for the extended-spectrum activity of the enzyme have been identified; among them, the structurally flexible B3 β-strand and Ω loop and residues Asn-104, Ser-237, Asp-240, and Arg-276, which are located in the vicinity of the active side, appear to be involved in the cefotaxime-hydrolyzing activity of CTX-M enzymes (8, 9). In previous studies on the comparative kinetic characterization of CTX-M-14, CTX-M-15, and their hybrids, it was suggested that residues distal to the active site contribute to variation in the catalytic activities of these CTX-M variants, yet exactly which residues are involved in the catalytic functions of CTX-M were not known. In this study, we used CTX-M-64 and CTX-M-14, which exhibited dramatically different catalytic activities, as candidate enzymes to investigate the molecular basis of differential mechanisms exhibited by these enzymes (4). The findings of this work will help identify the key functional components of the CTX-M class of enzymes.
MATERIALS AND METHODS
Materials.
Ampicillin, cefotaxime, ceftazidime, cefepime, cefuroxime, ceftriaxone, cefquinome, cephalothin, and thrombin were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Kanamycin and isopropyl-β-d-1-thiogalactopyranoside (IPTG) were purchased from IBI Inc. (Boca Raton, FL, USA). All media were purchased from BD Co. (Franklin Lakes, NJ, USA).
CTX-M constructs and site-directed mutagenesis.
Two versions of the blaCTX-M construct were utilized in this study, namely, the full-length gene of pET28-blaCTX-M-14 and pET28-blaCTX-M-64 and the truncated gene without signal peptide regions of pET28-blaH6-mCTX-M-14 and pET28-blaH6-mCTX-M-64, which were described in our previous study (4). Different residues between CTX-M-14 and CTX-M-64 were marked by using the MegAlign software of Lasergene 7 (DNAStar Inc., WI, USA). Amino acid substitutions were introduced into the CTX-M-14 and CTX-M-64 β-lactamases, with single, double, and multiple mutants generated sequentially. Escherichia coli strain DH5α was transformed with plasmids harboring the mutated blaCTX-M genes, which were generated using the GeneArt site-directed mutagenesis kit (Invitrogen Co., NY, USA). The transformants were screened on LB plates that were supplemented with 50 μg/ml of Kanamycin and confirmed by nucleotide sequencing. The constructs carrying selected mutations were transformed into E. coli strain BL21(DE3) for overexpression and purification.
MICs.
Antibiotic susceptibility levels of the organisms containing pET28-blaCTX-M-14, pET28-blaCTX-M-64, and other mutant constructs were determined by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (10). The strains were inoculated onto Mueller-Hinton agar (MHA) plates, grown at 37°C overnight, transferred to Mueller-Hinton broth (MHB) media that were supplemented with serial concentrations of selected β-lactams and 1 mM IPTG the next day, and then incubated at 37°C for 16 to 20 h. MIC values were determined by recording the lowest concentration of β-lactams that inhibited the growth of the E. coli BL21. The experiment was repeated at least three times for each antibiotic tested. E. coli strain ATCC 25922 was used as a quality control.
Expression and purification of CTX-M derivatives.
Overnight cultures were inoculated into 0.5 liters of LB to achieve an optical density at 600 nm (OD600) of 0.6, induced with 1 mM IPTG, and incubated at 16°C overnight. The cells were recovered, resuspended in lysis buffer (10 mM Tris-HCl, pH 7.6, 0.3 M sucrose, 1% NP-40, 0.5% Triton X-100, and 0.5% Tween 20), and broken with a French press (Thermo Scientific Inc., MA, USA). The soluble fractions were passed through a Ni-nitrilotriacetic acid (NTA) column and eluted with 20 mM Tris-HCl, pH 7.9, 500 mM NaCl, and 250 mM imidazole. The eluted proteins were concentrated using the Amicon Ultra-15 (nominal molecular weight limit [NMWL] = 10,000) centrifugal filter device, and the buffer was exchanged with 50 mM phosphate buffer, pH 7.4, during the process. The purified enzyme was incubated with thrombin to remove the His6 tag at room temperature for 1 h. The thrombin-treated proteins were further subjected to gel filtration chromatography by using a Sephacryl S-200 column. The protein fractions were then pooled and further concentrated using the Amicon Ultra-15 centrifugal filter device. The purity of the proteins was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The concentrations of enzymes were determined using the NanoDrop Lite (Thermo Scientific Inc., MA, USA). The mCTX-M enzyme yields were all approximately 14 mg/500 ml.
In vitro expression analysis of CTX-M enzymes.
The expression levels of H6-mCTX-M-14, H6-mCTX-M-64, and their derivatives in E. coli were determined by probing the total lysate with the anti-His6 tag antibody. Briefly, E. coli cultures carrying pET28-blaH6-mCTX-M were subjected to enzyme induction as mentioned above. The cells were recovered and resuspended in B-PER buffer (Thermo Scientific, MA, USA). The total lysate was subjected to SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with an anti-H6 antibody to determine the expression levels of E. coli strains carrying determinants of different CTX-M enzymes and their mutants. The membrane was also probed by anti-GADPH antibody to ensure equal loading.
Enzyme kinetics of CTX-M and its derivatives.
The kinetic parameters were determined by incubating the enzyme with different concentrations of β-lactams at 25°C in 500 μl of assay buffer (50 mM phosphate buffer, pH 7.0). The rate of the hydrolysis of substrates was measured by monitoring the changes in absorption, due to the opening of the β-lactam ring, at specific wavelengths by means of a spectrometer (Perkin Elmer Lambda Bio 20) for 5 min. The initial velocities versus substrate concentrations were measured at least thrice and fitted to the Michaelis-Menten equation using GraphPad Prism 5 (San Diego, CA, USA), which was also used to calculate the standard errors of Km and kcat. Bovine serum albumin (BSA) (100 μg/ml) was added to the diluted enzymes and the reaction mixture to prevent enzyme degradation. The molar extinction coefficients for tested substrates were obtained from previous studies (4, 11). For ceftazidime, which was a poor substrate, the Ki values of mCTX-M enzymes were determined by a competitive inhibition assay using 100 μM nitrocefin as the reporter substrate (6, 12).
Circular dichroism analysis of CTX-M derivatives.
The thermal stability assay was performed by using 0.5 μM enzymes in milli-Q water in a Jasco J-810 spectropolarimeter (Easton, MD, USA) at 25°C, 40°C, and 60°C. Quartz cells with a 10-mm path length were used for all experiments. Circular dichroism (CD) spectra were obtained and recorded every 0.1 nm between λ190 and λ250 with a scan rate of 50 nm/min and a response time of 2 s. All experiments were performed in triplicate; the CD signals were expressed as molar ellipticity.
CTX-M protein structural modeling and analysis.
The structure of CTX-M-14 (PDB ID 1YLT) was obtained from the Protein Data Bank (http://www.rcsb.org/pdb/); the structure of CTX-M-64 was generated on the basis of a currently available amino acid sequence (GenBank accession number ACU87987.1) and by using the comparative protein modeling SWISS-MODEL server (available at http://swissmodel.expasy.org), with 1YLT as the template. The global model quality estimation (GMQE) scores, the values used to estimate the accuracy of a model built with the template, were between 0.93 and 0.97 for the models built in this study. The protein structures and built models were analyzed by PyMOL.
RESULTS AND DISCUSSION
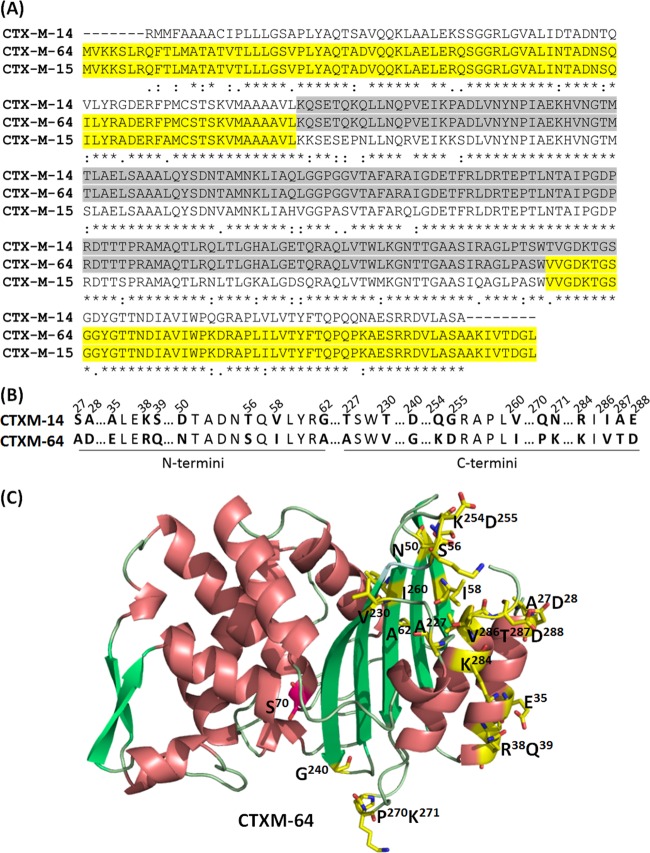
CTX-M-64, a hybrid enzyme comprising the N and C termini of CTX-M-15 and the middle part of CTX-M-14, exhibited higher hydrolytic activities against cephalosporins than other hybrid enzymes as well as the parental enzymes CTX-M-14 and CTX-M-15, thereby resulting in higher MIC values in organisms producing this enzyme (11). Compared to CTX-M-14, CTX-M-64 was found to harbor a number of amino acid substitutions at both the C and N termini (Fig. 1A). As shown in Fig. 1B, which depicts the sites of these amino acid variations, most of the changes were found to be located at α-helices and β-sheets that are most likely involved in maintaining the stability of protein structure through forming hydrogen bonds to each other as well as among residues at the lower edge of the enzymes, with the exception of residue G240, which is located at the vicinity of the active site. In order to identify residues, which play an important role in the enhancement of the catalytic activity of CTX-M-64, the enzyme was subjected to sequential amino acid substitutions to the corresponding residues in CTX-M-14. Two types of constructs were used in this study to ensure the accuracy of the data. The strain containing pET28-blaCTX-Ms, which encoded the full length of CTX-M plus the signal peptide, was used to determine MICs of the proteins, whereas the organism containing pET28-blaH6-mCTX-Ms, which encoded a His-tagged CTX-M without a signal peptide, was used to determine the kinetic constants of the enzyme. The expression levels of all pET28-blaCTX-Ms constructs were confirmed by probing the total lysate with the anti-His6 tag antibody, the results of which depicted a stable and similar level of expression for all of the constructs, indicating that the difference in MIC exhibited by different constructs was due to the alteration of amino acid profiles and not the expression level of different constructs (data not shown). To test the effects of amino acid substitutions in mCTX-M, CD analysis was performed on the proteins concerned, with results showing that all of the mutant proteins exhibited similar CD spectra. This finding suggests that the mutations tested did not lead to a significant alteration of the overall protein structure (data not shown).
FIG 1.
Amino acid residues subjected to mutagenesis studies on CTX-M-14 and CTX-M-64. (A) Sequence alignment of CTX-M14, CTX-M15, and CTX-M64. (B) The amino acid changes in CTX-M-14 and CTX-M-64 were shown, respectively. (C) Structure of CTX-M-64 depicting the specific location of the residues, which are different from those of CTX-M-14 (yellow); S70, the active site (magenta). The amino acid residues were numbered according to Ambler's system (18).
Residues in CTX-M-64 contributed to its high catalytic activity.
G240, one of the residues that differ from the corresponding site in CTX-M-14, was the only amino acid near the center of the active site in CTX-M-64. It was reported that D240G resulted in a higher MIC toward ceftazidime but a variable MIC toward cefotaxime (13, 14) and that the coexistence of V77, P167, and G240 could result in an increased resistance to cefotaxime and ceftazidime in the background of CTX-M-10 (15). In order to test whether G240 was related to the hydrolytic activity of CTX-M-64, different amino acid substitutions were introduced into this site resulting in the G240D, G240A, G240E, G240L, and G240R changes. Several enzymes harboring these amino acid substitutions displayed 2-fold reductions in MICs of cephalothin, cefuroxime, and ceftriaxone and 4∼128-fold reductions in MICs of cefotaxime and ceftazidime (Table 1). Among these mutant enzymes, CTX-M-64(G240R) was found to exhibit the most dramatic effects. Consistent with the changes in MICs, the mutant enzymes all exhibited decreased hydrolytic activity on various β-lactam agents; in particular, the G240R substitution resulted in significantly decreased catalytic activity toward cephalothin, cefuroxime, ceftriaxone, and cefotaxime, with 36-, 19-, 25-, and 73-fold reductions, respectively (Table 2). In other class A ESBLs, such as TEM and SHV, the positively charged residue Arginine was able to elevate the catalytic activity against ceftazidime through the formation of an electrostatic bond with the carboxylic acid group in the oxyimino substituents of ceftazidime (16). However, the G240R substitution was found to exhibit a negative effect on the catalytic activity of CTX-M-64. Other substitutions also consistently resulted in the reduction of MIC and the kcat/Km values, suggesting that a bulky side chain in this position is not functionally acceptable, which may narrow down the active site pocket.
TABLE 1.
MICs of CTX-M-64 mutants toward different β-lactamsa
| Enzyme/mutant | MIC (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|
| AMP | CEP | CFX | CRO | CTX | CAZ | |
| CTX-M-64 WT | ≥2,048 | 1,024 | 2,048 | 1,024 | 2,048 | 32 |
| Vector control | 2 | 2 | 0.5 | <0.25 | <0.25 | 0.03 |
| I58V | 2,048 | 512 | 1,024 | 512 | 512 | 4 |
| G240D | ≥2,048 | 512 | 1,024 | 1,024 | 512 | 2 |
| G240L | ≥2,048 | 512 | 2,048 | 1,024 | 256 | 2 |
| G240A | ≥2,048 | 512 | 1,024 | 1,024 | 512 | 4 |
| G240E | ≥2,048 | 512 | 1,024 | 512 | 256 | 2 |
| G240R | ≥2,048 | 512 | 1,024 | 512 | 128 | 0.25 |
| G240D, I58V | ≥2,048 | 512 | 2,048 | 1,024 | 512 | 2 |
| G240D, K254Q, D255G | ≥2,048 | 512 | 2,048 | 1,024 | 512 | 2 |
| G240D, I58V, K254Q, D255G | ≥2,048 | 1,024 | 2,048 | 1,024 | 512 | 1 |
| I58V, K254Q, D 255G | ≥2,048 | 512 | 1,024 | 512 | 512 | 16 |
| G240D, P270Q, K271N | ≥2,048 | 1,024 | 2,048 | 1,024 | 1,024 | 2 |
| I58V, L33A | 1,024 | 512 | 1,024 | 1,024 | 512 | 4 |
| I58V, V52A | 512 | 512 | 512 | 512 | 256 | 4 |
| I58V, L287A | 512 | 512 | 512 | 512 | 256 | 4 |
| CTX-M-14 WT | 2,048 | 256 | 1,024 | 512 | 128 | 1 |
AMP, ampicillin; CEP, cephalothin; CFX, cefuroxime; CRO, ceftriaxone; CTX, cefotaxime; CAZ, ceftazidime.
TABLE 2.
Kinetic parameters of CTX-M-64 mutant enzymes toward seven β-lactams
| CTX-M-64 | Parameter | AMP | NCFa | CEP | CFX | CRO | CTX | CAZ |
|---|---|---|---|---|---|---|---|---|
| WT | Km (μM) | 17 ± 2 | 10 ± 2 | 25 ± 3 | 31 ± 4 | 10 ± 1 | 24 ± 4 | 1,516 ± 166 |
| kcat (s−1) | 107 ± 3 | 345 ± 34 | 91 ± 3 | 235 ± 12 | 195 ± 7 | 174 ± 9 | NDb | |
| kcat/Km (μM−1 s−1) | 6.3 | 34.5 | 3.6 | 7.6 | 19.5 | 7.3 | ND | |
| I58V | Km (μM) | 37 ± 3 | 5 ± 0.4 | 14 ± 4 | 34 ± 4 | 13 ± 0.7 | 116 ± 16 | >8,000 |
| kcat (s−1) | 54 ± 1 | 57 ± 2 | 13 ± 2 | 17 ± 1 | 33 ± 0.6 | 10 ± 0.8 | ND | |
| kcat/Km (μM−1 s−1) | 1.5 | 11 | 0.9 | 0.5 | 2.5 | 0.1 | ND | |
| G240D | Km (μM) | 36 ± 9 | 14 ± 1 | 43 ± 10 | 20 ± 2 | 7 ± 1 | 29 ± 4 | 2,130 ± 172 |
| kcat (s−1) | 114 ± 13 | 301 ± 15 | 174 ± 12 | 111 ± 5 | 58 ± 3 | 66 ± 3 | ND | |
| kcat/Km (μM−1 s−1) | 3.2 | 22 | 4 | 5.6 | 8.3 | 2.3 | ND | |
| G240A | Km (μM) | 31 ± 2 | 5 ± 0.8 | 33 ± 2 | 21 ± 0.5 | 9 ± 0.6 | 32 ± 3 | 1,966 ± 95 |
| kcat (s−1) | 119 ± 3 | 99 ± 6 | 112 ± 8 | 132 ± 1 | 38 ± 0.7 | 132 ± 6 | ND | |
| kcat/Km (μM−1 s−1) | 3.8 | 20 | 4.9 | 6.3 | 4.2 | 4.1 | ND | |
| G240E | Km (μM) | 39 ± 1 | 9 ± 0.6 | 61 ± 5 | 12 ± 1 | 13 ± 1 | 40 ± 3 | 2,023 ± 131 |
| kcat (s−1) | 156 ± 3 | 165 ± 6 | 197 ± 10 | 60 ± 2 | 47 ± 1 | 60 ± 2 | ND | |
| kcat/Km (μM−1 s−1) | 4 | 18 | 3.2 | 5 | 3.6 | 1.5 | ND | |
| G240R | Km (μM) | 44 ± 7 | 4 ± 0.5 | 20 ± 2 | 21 ± 4 | 32 ± 3 | 75 ± 8 | >8,000 |
| kcat (s−1) | 133 ± 9 | 78 ± 4 | 2.3 ± 0.07 | 8 ± 0.4 | 27 ± 1 | 11 ± 0.5 | ND | |
| kcat/Km (μM−1 s−1) | 3 | 20 | 0.1 | 0.4 | 0.8 | 0.1 | ND | |
| G240L | Km (μM) | 29 ± 3 | 0.4 ± 0.04 | 15 ± 2 | 20 ± 2 | 32 ± 4 | 45 ± 9 | 2,000 ± 100 |
| kcat (s−1) | 108 ± 43 | 13 ± 0.2 | 66 ± 3 | 77 ± 3 | 155 ± 7 | 114 ± 11 | ND | |
| kcat/Km (μM−1 s−1) | 3.7 | 32.5 | 4.4 | 3.9 | 4.8 | 2.5 | ND | |
| G240D/I58V/KD 254/255QG | Km (μM) | 39 ± 7 | 2 ± 0.3 | 37 ± 13 | 23 ± 3 | 9 ± 3 | 42 ± 4 | >8,000 |
| kcat (s−1) | 133 ± 10 | 36 ± 2 | 132 ± 23 | 106 ± 5 | 56 ± 4 | 85 ± 4 | ND | |
| kcat/Km (μM−1 s−1) | 3.4 | 18 | 3.6 | 4.6 | 6.2 | 2 | ND | |
| CTX-M-14 WT | Km (μM) | 72 ± 11 | 11 ± 2 | 64 ± 6 | 22 ± 3 | 17 ± 3 | 34 ± 5 | >8,000 |
| kcat (s−1) | 25 ± 2 | 114 ± 12 | 357 ± 18 | 56 ± 3 | 42 ± 4 | 37 ± 2 | ND | |
| kcat/Km (μM−1 s−1) | 0.35 | 10 | 5.6 | 2.5 | 2.5 | 1.1 | ND | |
| CTX-M-14 (D240G) | Km (μM) | 17 ± 2 | 2 ± 0.4 | 68 ± 21 | 28 ± 2 | 14 ± 2 | 30 ± 3 | >8,000 |
| kcat (s−1) | 25 ± 0.8 | 41 ± 3 | 265 ± 57 | 50 ± 2 | 68 ± 4 | 54 ± 3 | ND | |
| kcat/Km (μM−1 s−1) | 1.5 | 21 | 3.9 | 1.8 | 4.9 | 1.8 | ND | |
| C9c mutations | Km (μM) | 25 ± 2 | 14 ± 2 | 23 ± 3 | 54 ± 3 | 16 ± 3 | 28 ± 5 | >8,000 |
| kcat (s−1) | 89 ± 3 | 285 ± 32 | 78 ± 3 | 232 ± 11 | 187 ± 9 | 134 ± 7 | ND | |
| kcat/Km (μM−1 s−1) | 3.6 | 20.3 | 3.4 | 4.3 | 11.68 | 4.8 | ND | |
| NC22c mutations | Km (μM) | 18 ± 1 | 9 ± 2 | 28 ± 2 | 34 ± 3 | 8 ± 1 | 26 ± 4 | 1,986 ± 131 |
| kcat (s−1) | 112 ± 3 | 325 ± 31 | 102 ± 5 | 245 ± 15 | 165 ± 5 | 154 ± 8 | ND | |
| kcat/Km (μM−1 s−1) | 6.2 | 36.1 | 3.6 | 7.2 | 20.6 | 5.9 | ND |
NCF, nitrocefin.
ND, the kinetic constants could not be determined due to low activity of the enzymes toward ceftazidime.
Please see the exact mutations in Table 3.
The effects of the amino acid substitutions located at both the N and C termini of CTX-M-64 were further tested. These amino acid changes were found to result in only a slight reduction in the catalytic activity of CTX-M-64, except for I58V, which exhibited 4- and 8-fold reductions in the rate of catalytic degradation of cefotaxime and ceftazidime, respectively (Table 2). Mutation I58V resulted in attenuation of kcat/Km against cephalothin, cefuroxime, ceftriaxone, and cefotaxime by 4-, 15-, 8-, and 73-fold, respectively, while the MICs toward these antibiotics only reduced slightly. Double mutations that result in the G240D/I58V changes were found to cause a similar degree of reduction in the catalytic activity of CTX-M-64 as that caused by the G240D substitution (Table 1). Other combinations of amino acid substitutions, including G240D/I58V and G240D/I58V/K254Q/D255G, also resulted in only minor change in catalytic activities (Table 2). These data suggest that amino acids located at the N and C termini of CTX-M-64 contribute slightly to the catalytic activity of CTX-M-64, with G240 and I58 being the more important residues, but single or double mutations are apparently not sufficient to bring about significant functional changes
Residues in CTX-M-14 contributed to lower catalytic activity.
To further show that integrated coordination of the functions of multiple residues at the N and C termini of CTX-M-64 is required to confer a significant enhancement of activity, we performed sequential substitutions of CTX-M-64 residues to CTX-M-14 residues to investigate the degree of contribution of these residues in enhancing the activity of CTX-M-14. Single, double, or triple mutations resulting in the production of the mutant CTX-M-14 enzymes were first created. Interestingly, the ceftazidime MICs of strains expressing these mutant enzymes were reduced by 2- to 16-fold, whereas the MICs of other β-lactams only decreased or increased slightly (Table 3). Surprisingly, the CTX-M-14(D240G) enzyme was found to exhibit less effect on the MICs of the cephalosporins tested except for ceftazidime, where a 2-fold reduction was observed compared to the wild-type (WT) CTX-M-14. This result is not consistent with the fact that G240 is the most optimal site at this position for CTX-M-64 or with the reported data on CTX-M-27, the exact construct of CTX-M-14(D240G), that showed an increased MIC for ceftazidime (from 1∼8 μg/ml) (Table 3) (17). The discrepancy of these data cannot be addressed at this point since we have confirmed that the data reported in this study were very reproducible. We then assessed the combined effects of substitutions on CTX-M-14 hydrolytic activity by adopting an additive substitution strategy in which amino acid substitutions were introduced cumulatively as shown in Table 3. Multiple mutations were introduced together if the target sites were close enough to be mutated simultaneously. At the N terminus, one to nine cumulative amino acid substitutions did not cause a significant increase in the activity of CTX-M-14 toward ceftazidime, yet the MIC of cefotaxime was increased by up to 4-fold. Similarly, substitutions of up to 5 residues at the C terminus of CTX-M-14 resulted in a reduction of the ceftazidime MIC by 2-fold, yet the addition of 2 more C-terminal residues caused a 2-fold increase in the ceftazidime MIC (Table 3). All of the N-terminal substitutions, together with three C-terminal substitutions, did not lead to additional improvement in the hydrolytic activity of CTX-M-14, yet the addition of one more substitution, D240G (NC13 mutations), resulted in an increase in the MIC of ceftazidime by 4-fold (Table 3). It should be noted that combining the N- and C-terminal substitutions (NC19 mutations) in CTX-M-14 may mediated E. coli resistance to a level similar to CTX-M-64 but still slightly lower than the original CTX-M-64 (Table 3). These data suggest that the higher observed resistance of CTX-M-64 in E. coli is due to the combinatorial effects of various different residues located at both the C and N termini.
TABLE 3.
MICs of CTX-M-14 mutants toward different β-lactams
| Enzyme | MIC (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|
| AMP | CEP | CFX | CRO | CTX | CAZ | |
| Vector control | 2 | 2 | 0.5 | <0.25 | <0.25 | 0.03 |
| CTX-M-14 WT | 2,048 | 256 | 1,024 | 512 | 128 | 1 |
| V58I | 1,024 | 128 | 512 | 512 | 128 | 0.25 |
| D240G | 512 | 128 | 1,024 | 512 | 256 | 0.5 |
| S27A, A28D, A35E, K38R, S39Q | 2,048 | 128 | 1,024 | 512 | 512 | 1 |
| T56S, V58I, G62A | 2,048 | 128 | 1,024 | 1,024 | 512 | 1 |
| N9 mutationsa | 2,048 | 64 | 1,024 | 512 | 512 | 1 |
| C7 mutationsb | 1,024 | 256 | 512 | 512 | 256 | 0.5 |
| C9 mutationsc | 2,048 | 1,024 | 1,024 | 1,024 | 256 | 2 |
| NC12 mutationsd | 1,024 | 128 | 512 | 512 | 128 | 1 |
| NC13 mutationse | 1,024 | 256 | 1,024 | 1,024 | 256 | 4 |
| NC15 mutationsf | 2,048 | 512 | 1,024 | 1,024 | 512 | 1 |
| NC19 mutationsg | 2,048 | 512 | 1,024 | 1,024 | 512 | 2 |
| NC22 mutationsh | 2,048 | 1,024 | 2,048 | 1,024 | 1,024 | 8 |
| CTX-M-64 WT | ≥2,048 | 1,024 | 2,048 | 1,024 | 2,048 | 32 |
N9 mutations: SAAKSDTVG27/28/35/38/39/50/56/58/62ADERQNSIA.
C7 mutations: DQNRIAE240/270/271/284/286/287/288GPKKVTD.
C9 mutations: QGVQNRIAE254/255/260/270/271/284/286/287/288KDIPKKVTD.
NC12 mutations: SAAKSDTVGDTT27/28/35/38/39/50/56/58/62/227/230ADERQNSIAAV.
NC13 mutations: SAAKSDTVGDTTD27/28/35/38/39/50/56/58/62/227/230/240ADERQNSIAAVG.
NC15 mutations: SAAKSDTVGDTTDQGV27/28/35/38/39/50/56/58/62/227/230/240/254/255/260ADERQNSIAAVGKDI.
NC19 mutations: SAAKSDTVGDTTDQGVQN27/28/35/38/39/50/56/58/62/227/230/240/254/255/260/270/271ADERQNSIAAVGKDIPK.
NC22 mutations: SAAKSDTVGDTTDQGVQNRIAE27/28/35/38/39/50/56/58/62/227/230/240/254/255/260/270/271/284/286/287/288ADERQNSIAAVGKDIPKKVTD.
Potential mechanisms of the contribution of N- and C-terminal residues to CTX-M activity.
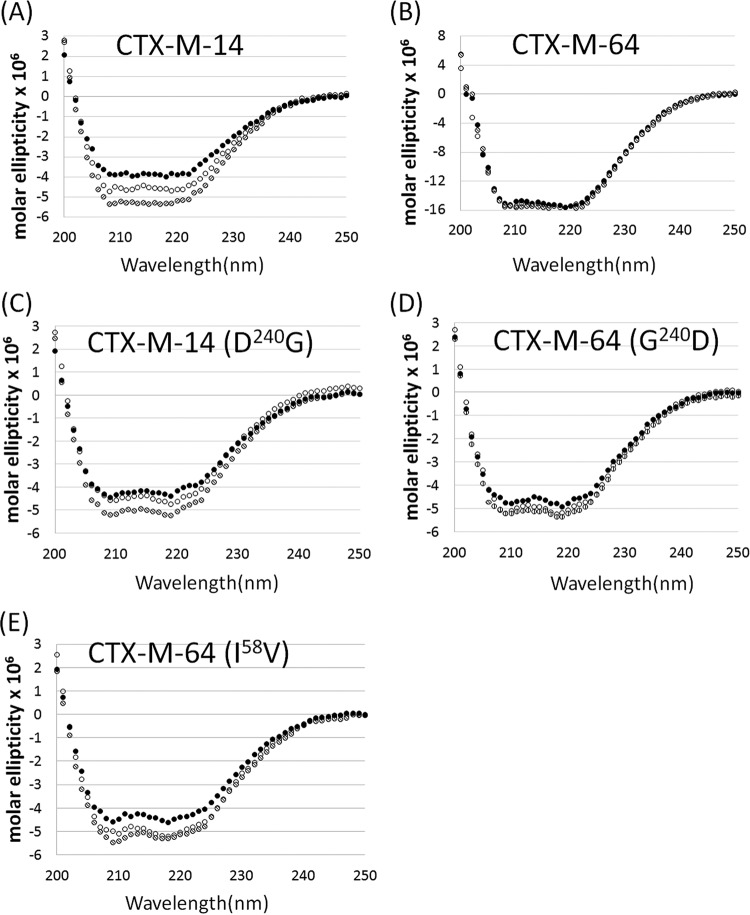
To further investigate how the N- and C-terminal substitutions that were mainly located distal to the active site of CTX-M-64 can confer a higher hydrolytic activity compared to CTX-M-14, a thermal stability assay was performed to determine whether the distal amino acid substitutions can enhance protein stability. The thermal stability assay using far-UV CD showed that CTX-M-64 exhibited highly similar spectra at 25°C, 40°C, and 60°C, but CTX-M-14 exhibited obvious loss in helicity throughout 200 to 230 nm at 40°C and 60°C compared to this state at 25°C (Fig. 2A and B), suggesting that CTX-M-64 may be more structurally stable than CTX-M-14. Such stability was presumably maintained by the interaction of a significant number of amino acid residues located at different regions of the protein.
FIG 2.
Determination of the secondary structure of CTX-M-14 (A), CTX-M-64 (B), and their derivatives (C, D, and E) at 25°C (○), 40°C (●), and 60°C (△) by circular dichroism.
The degree of contribution of the two most significant residues, G240 and I58, to the stability of CTX-M-64 was also investigated. As expected, the stability of the single mutant CTX-M-64(G240D) decreased slightly compared to CTX-M-64, whereas that of CTX-M-14(D240G) was slightly improved, suggesting that G240 may be contributing to the stability of CTX-M-64 (Fig. 2C and D). I58 was located in the B2 β-strand of CTX-M-64, suggesting that the residue plays a role in the maintenance of the stability of the protein structure but is not directly involved in the recognition of the substrate. This residue may be able to form strong hydrophobic interactions with amino acids V29, L48, and V286 located in the H1 α-helices, B1 β-strand, and H11 α-helices, respectively, in order to maintain the tertiary structures in this region (Fig. 3A). Structural analysis of CTX-M-14 and the in silico model of CTX-M-64 showed that V58 of CTX-M-14 formed less stable hydrophobic interactions with other residues, such as V29, L48, and V286, presumably due to the shorter side chain of V58 compared to I58 (Fig. 3B). The result of the thermal stability assay was consistent with our structural analysis, in that the I58V substitution resulted in a loss of helicity at both 40°C and 60°C (Fig. 2E). To further check whether the hydrophobic interaction in this area is important to protein stability, three potential interacting residues, V29, L48, and V286, were subjected to amino acid substitution analysis, in which six types of amino acid substitutions were generated, including V29A, L48A, V286A, I58V/V29A, I58V/L48A, and I58V/V286A. Our data showed that, upon replacement by Ala at these three sites, a mild reduction in MIC was observable, suggesting the possible contribution of the hydrophobic interactions in this region to protein stability (Table 1).
FIG 3.
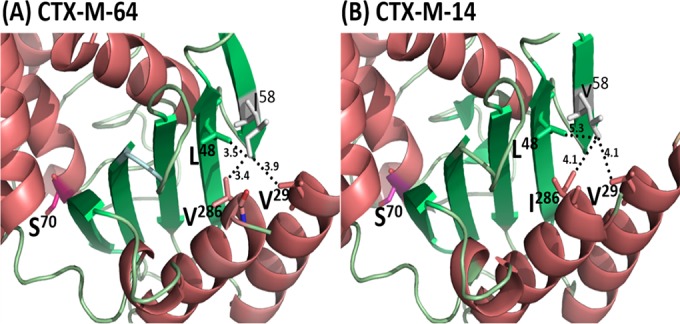
Interaction between V58/I58 and other residues in the adjacent helixes and sheets of CTX-M-64 and CTX-M-14. (A) V58 in CTX-M-64 interacting with residues V29, L48, and V286. The distances between V58 and V29, L48, and V286 were 3.9 Å, 3.5 Å, and 3.4 Å, respectively. (B) I58 in CTX-M-14 interacting with residues V29, L48, and I286. The distances between I58 and V29, L48, and I286 were 4.1 Å, 5.3 Å, and 4.1 Å, respectively.
In conclusion, this study characterized the contribution of a single residue located at both the N and C termini of CTX-M-14 and CTX-M-64 to the higher activity of CTX-M-64 compared to CTX-M-14. Our data indicated that the enhanced extended-spectrum activity of CTX-M-64 seems to be due to the combinational effect of the residues residing at both the N and C termini (through recombination process), rather than the contribution of a few amino acid substitutions (accumulation of single mutations). The data from this study demonstrates the complexity of β-lactamases. The hydrolytic activity of this class of enzyme is not only determined by active-site substrate recognition and catalysis but also by the maintenance of the integrity and the stability of the protein.
ACKNOWLEDGMENT
This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200).
We declare no conflict of interest.
REFERENCES
- 1.Bonnet R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Canton R, Gonzalez-Alba JM, Galan JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He D, Chiou J, Zeng Z, Liu L, Chen X, Zeng L, Chan EW, Liu JH, Chen S. 2015. Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid beta-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob Agents Chemother 59:5976–5983. doi: 10.1128/AAC.04920-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, He D, Lv L, Liu W, Chen X, Zeng Z, Partridge SR, Liu JH. 2015. blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrob Agents Chemother 59:4464–4470. doi: 10.1128/AAC.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagano Y, Nagano N, Wachino J, Ishikawa K, Arakawa Y. 2009. Novel chimeric beta-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 beta-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob Agents Chemother 53:69–74. doi: 10.1128/AAC.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian GB, Huang YM, Fang ZL, Qing Y, Zhang XF, Huang X. 2014. CTX-M-137, a hybrid of CTX-M-14-like and CTX-M-15-like beta-lactamases identified in an Escherichia coli clinical isolate. J Antimicrob Chemother 69:2081–2085. doi: 10.1093/jac/dku126. [DOI] [PubMed] [Google Scholar]
- 8.Gazouli M, Tzelepi E, Sidorenko SV, Tzouvelekis LS. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A beta-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother 42:1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibuka A, Taguchi A, Ishiguro M, Fushinobu S, Ishii Y, Kamitori S, Okuyama K, Yamaguchi K, Konno M, Matsuzawa H. 1999. Crystal structure of the E166A mutant of extended-spectrum beta-lactamase Toho-1 at 1.8 Å resolution. J Mol Biol 285:2079–2087. doi: 10.1006/jmbi.1998.2432. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Laraki N, Franceschini N, Rossolini GM, Santucci P, Meunier C, de Pauw E, Amicosante G, Frere JM, Galleni M. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-beta-lactamase IMP-1 produced by Escherichia coli. Antimicrob Agents Chemother 43:902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii Y, Galleni M, Ma L, Frere JM, Yamaguchi K. 2007. Biochemical characterisation of the CTX-M-14 beta-lactamase. Int J Antimicrob Agents 29:159–164. doi: 10.1016/j.ijantimicag.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Delmas J, Sirot J, Shoichet B, Bonnet R. 2005. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J Mol Biol 348:349–362. doi: 10.1016/j.jmb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Delmas J, Chen Y, Prati F, Robin F, Shoichet BK, Bonnet R. 2008. Structure and dynamics of CTX-M enzymes reveal insights into substrate accommodation by extended-spectrum beta-lactamases. J Mol Biol 375:192–201. doi: 10.1016/j.jmb.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Novais A, Canton R, Coque TM, Moya A, Baquero F, Galan JC. 2008. Mutational events in cefotaximase extended-spectrum beta-lactamases of the CTX-M-1 cluster involved in ceftazidime resistance. Antimicrob Agents Chemother 52:2377–2382. doi: 10.1128/AAC.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantu C III, Huang W, Palzkill T. 1996. Selection and characterization of amino acid substitutions at residues 237-240 of TEM-1 beta-lactamase with altered substrate specificity for aztreonam and ceftazidime. J Biol Chem 271:22538–22545. doi: 10.1074/jbc.271.37.22538. [DOI] [PubMed] [Google Scholar]
- 17.Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, De Champs C, Sirot J. 2003. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother 52:29–35. doi: 10.1093/jac/dkg256. [DOI] [PubMed] [Google Scholar]
- 18.Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem J 276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]