Abstract
Tuberculosis is still a cause of major concern, partly due to the emergence of multidrug-resistant strains. New drugs are therefore needed. Vancomycin can target mycobacteria with cell envelope deficiency. In this study, we used a vancomycin susceptibility assay to detect drugs hampering lipid synthesis in Mycobacterium bovis BCG and in Mycobacterium tuberculosis. We tested three drugs already used to treat human obesity: tetrahydrolipstatin (THL), simvastatin, and fenofibrate. Only vancomycin and THL were able to synergize on M. bovis BCG and on M. tuberculosis, although mycobacteria could also be inhibited by simvastatin alone. Lipid analysis allowed us to identify several lipid modifications in M. tuberculosis H37Rv treated with those drugs. THL treatment mainly reduced the phthiocerol dimycocerosate (PDIM) content in the mycobacterial cell wall, providing an explanation for the synergy, since PDIM deficiency has been related to vancomycin susceptibility. Proteomic analysis suggested that bacteria treated with THL, in contrast to bacteria treated with simvastatin, tried to recover, inducing, among other reactions, lipid synthesis. The combination of THL and vancomycin should be considered a promising solution in developing new strategies to treat multidrug-resistant tuberculosis.
INTRODUCTION
Tuberculosis (TB), which is mainly caused by Mycobacterium tuberculosis, is still killing 1.5 million people annually in the world (1). Although the TB death rate has decreased by 47% since 1990 according to the Millennium Development Goals targets, reaching a 86% treatment success in 2013, the emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) infections (9.7% of MDR-TB cases) is highly worrisome. In some regions of Eastern Europe, the percentage of XDR-TB cases has even reached 25% (2). MDR-TB refers to TB with resistance to both isoniazid and rifampin, and XDR-TB refers to an MDR subgroup with additional resistance to any fluoroquinolone and at least one second-line injectable drug. In 2015, the overall cure rate for MDR infections, using restricted, expensive, prolonged, second-line treatment regimens decreased to 50% (1). There is growing concern to discover new drugs to fight MDR- and XDR-TB. As such, bedaquiline and delamanid have been recently approved for treating MDR-TB, and other drugs are currently in clinical trials (3, 4).
It is well established that M. tuberculosis antibiotic resistance is highly dependent on its unique waxy cell wall, consisting of a covalently linked skeleton composed from inside to outside of peptidoglycan, arabinogalactan, and mycolic acids (5). Unique, extractable lipids, such as acyltrehaloses, including sulfolipids (SLs), trehalose dimycolates (TDM), diacyltrehaloses (DAT), and penta- or polyacyltrehaloses (PAT), as well as phthiocerol dimycocerosates (PDIM), particularly those involved in virulence and antibiotic resistance, are embedded in an outer membrane. In addition, phosphatidylmyoinositol mannosides (PIM), as well as hypermannosylated PIM, lipomannans, and lipoarabinomannans, are anchored through their phosphatidylmyoinositol moieties to the inner and outer membranes of the cell wall and are involved in M. smegmatis antibiotic resistance (6).
Recently, we described an innovative approach to identify compounds targeting mycobacterial lipids biosynthesis that can synergize with glycopeptides to inhibit the growth of mycobacteria. Interestingly, we showed that a combination of vancomycin with a drug targeting long-chain lipid synthesis, such as cerulenin, acts synergistically against MDR and XDR clinical strains (7).
In this study, we used our vancomycin susceptibility assay to test commercially available drugs focusing on any that could potentially lower lipid biosynthesis in order to identify potential synergistic activity with vancomycin (7). This choice is justified by the importance of complex lipids composing the mycobacterial envelope, their roles in the cell wall impermeability, the susceptibility to antibiotics, and the virulence of M. tuberculosis (5, 7, 8). Three drugs commonly used to treat obesity and cholesterol disorders were tested: tetrahydrolipstatin (THL), simvastatin, and fenofibrate (7). THL is an esterase inhibitor well known for inhibiting pancreatic lipases (9), as well as several bacterial enzymes (10–17). Parker et al. reported that, in M. smegmatis, THL can target Rv3802, acting both as a thioesterase and as a phospholipase on phosphatidylinositol mannoside 2 (18) but also the ES-31 serine protease (19). Interestingly, Ravindran et al. pointed out nine lipases and one thioesterase (TesA) as potential THL targets (20). This last enzyme, TesA, is involved in the synthesis of phthiocerol dimycocerosate (PDIM) (21). PDIM are lipid compounds in the outer membrane of defined slow-growing mycobacterial species, including M. tuberculosis and M. bovis BCG, that are essential for cell envelope impermeability and antibiotic resistance, in particular for resistance to glycopeptides (5, 7). Simvastatin is a cholesterol-lowering drug that has recently been reported to enhance phagosomal maturation and autophagy, increasing the M. tuberculosis burden in macrophages and mice (22, 23). This drug has been reported to improve the efficiency of first-line antituberculous drugs in vivo, probably by lowering lipids in foamy macrophages (23). Finally, fenofibrate belongs to the fibrate lipid-lowering drug family. Garbe et al. reported some toxicity of fibrates on M. tuberculosis (24). Gemfibrozil has been indeed shown to inhibit M. tuberculosis growth with a potential action on a mycobacterial enoyl coenzyme A (24, 25). More recently, Kim et al. reported that fibrates could lower the bacterial burden and inflammation in an M. smegmatis macrophage invasion model by a PPARα-independent pathway (26).
Here, using three different antibacterial assays, we show demonstrate the antimycobacterial activity of simvastatin and THL and show that THL synergizes with vancomycin. Lipid and protein analysis indicated a distinct mode of action, with THL inhibiting the cell wall synthesis as expected.
MATERIALS AND METHODS
Bacteria and cultures.
M. bovis BCG GL2 and M. tuberculosis H37Rv were used. Mycobacteria were grown in 7H9 medium containing 0.05% Tween 80 supplemented with 10% albumin-dextrose complex (Difco Laboratories) or on 7H11 Middlebrook agar supplemented with oleic acid-albumin-dextrose complex (Difco Laboratories).
Drug susceptibility tests.
Drug susceptibility was investigated using three methods: the macrodilution method, the standardized agar proportion method, and the BacT/Alert MP method (Mycobacteria Process) according to the bioMérieux protocol and previous reports (27, 28). For M. bovis BCG, in the context of a first assay, we used the macrodilution method to determine more rapidly and using a smaller volume the MIC and the fractional inhibitory concentration index (FICI). For M. tuberculosis, we also used the standardized agar proportion method. The BacT/Alert MP method allowed us to perform a fast preliminary test on M. bovis BCG and to evaluate the potential synergy of drug combination on M. tuberculosis. For all methods, cultures that were less than 4 weeks old were used to prepare a homogeneous suspension inoculum.
The macrodilution method, used for M. bovis BCG, was performed in 7H9 medium without Tween supplemented with albumin-dextrose complex in a polycarbonate tube to reach a final volume of 1.2 ml, thus mimicking the BacT/Alert method at a 1:10 scale. A 600-μl inoculum, at an optical density at 600 nm (OD600) of 0.2, diluted 1 in 24, was added to 600-μl serial drug dilutions (one in two). The FICI of the drug in combination with vancomycin was calculated as follows: FICI = FICa + FICb = MICab/MICa + MICba/MICb. In agreement with the checkerboard method, synergy is reached when the FICI is ≤0.5, indifference could be observed when the FICI is between 0.5 and 2, and an antagonistic effect is noted when the FICI is >2 (29).
For the standard agar proportion method (27), we used mycobacterial suspensions (1 McFarland turbidity) to perform serial dilutions from 10−1 to 10−4 in the presence or absence of various concentrations of THL, simvastatin, fenofibrate, and vancomycin alone or in combination. The MIC was defined as the lowest drug concentration that inhibited more than 99% of the bacterial population. Vancomycin, THL, fenofibrate, and simvastatin were purchased from Sigma Aldrich. THL was also commercially obtained from Sandoz and fenofibrate from Eurogenerics.
BacT/Alert MP system (Mycobacteria Process) samples using BacT/Alert MP bottles (11 ml) supplemented with restoring fluid were inoculated with drugs and mycobacterial suspensions as previously described (7). Growth index (GI) values were recorded every 10 min. A 1/100 proportional growth control condition consisted of a 100-fold-diluted bacterial inoculum injected in a drug-free control vial. The concentration of the drug(s) simultaneously giving a flagged positive bottle as the 1/100 control vial was considered the MIC. The combined drug effect was investigated using concentrations at sub-MICs and previously published as the “x/y” methodology (30–33). Briefly, when the 1:100 control vial was flagged and the daily ΔGI of the 1:100 control vial reached at least 30, the GI was read for at least one additional day to calculate the ΔGI from the previous day. In the case of a two-drug combination, a “Δx/Δy” quotient of <0.5 indicates synergy, with “Δx” being the ΔGI value obtained for the vial with the combination of drugs and “Δy” being the lowest ΔGI value obtained with any of the single drugs used within the combinations tested (7).
Lipid analysis.
Lipids from M. tuberculosis H37Rv (a 350-ml 7H9 culture without Tween 80 at an OD600 of 0.5) treated with THL (50 μg/ml) or treated with the same volume of dimethyl sulfoxide (DMSO) as a control were extracted first with CHCl3/CH3OH (1:2 [vol/vol]) for 24 h at room temperature and then twice with CHCl3/CH3OH (2:1 [vol/vol]) for 2 days. Crude extracts were washed twice with distilled water and evaporated to dryness according to standard protocols as described previously (34).
The lipid extracts were comparatively analyzed by high-performance thin-layer chromatography (HPTLC) on a HPTLC Silica Gel 60 (Merck), using various solvent systems, mainly petroleum ether/diethyl ether (9:1 [vol/vol]) for phthiocerol dimycocerosates (PDIM) and CHCl3/CH3OH/H2O (30:8:1, 65:25:4, and 60:35:8 [vol/vol/vol]) for more polar lipids (trehalose mycolates and phospholipids). Visualizations were performed by immersion of the plates in primuline solution or by spraying the plates with 10% phosphomolybdic acid and charring for PDIM or 0.2% anthrone solution (wt/vol) in concentrated H2SO4 and charring for glycolipids. The various lipid spots were quantified by either absorption measurements at the specific wavelength with a TLC Scanner 3 using wincats software and/or colorimetric measurements using ChemiDoc XRS+ with Image Lab software (Bio-Rad).
SWATH acquisition liquid chromatography-tandem mass spectrometry (MS/MS) analysis.
Quantitative proteomic analyses were performed on proteome extracts of the different M. tuberculosis samples. Cells from three biological replicates were harvested by centrifugation (16,000 × g, 10 min, 4°C) when the OD600 reached 0.5. Proteins were extracted by sonication (three times for 10 s, amplitude 40%, using an IKA U50 sonicator) in 6 M guanidinium chloride solution. Extracted proteins were reduced, alkylated, and precipitated with acetone. Proteolytic peptides were obtained by overnight enzymatic digestion using trypsin at a ratio of 1:50 (wt/wt).
MS experiments were conducted according to a label-free strategy on UHPLC-HRMS/MS platform (Eksigent 2D Ultra and AB Sciex TripleTOF 5600) in SWATH data independent acquisition mode. Portions (2 μg) of peptides were separated on a C18 column (Acclaim PepMap100, 3 μm, 150 μm by 25 cm, Dionex) with a linear acetonitrile gradient (5 to 35% [vol/vol], 450 nl·min−1, 30 min) in water containing 0.1% (vol/vol) formic acid. Each MS survey scan (400 to 1,500 m/z, 50-ms accumulation time) was followed by analysis using 34 SWATH acquisition overlapping windows (25 Da) covering the precursor m/z range. For each window, ions were fragmented using a rolling-collision energy, and fragment ion spectra were accumulated for 95 ms in high-sensitivity mode.
SWATH spectra were identified by comparison to a reference spectral library obtained with traditional data-dependent acquisition (DDA) experiments on proteins extracted from the different M. tuberculosis samples. The sample preparation and separation procedures were identical to the one previously described. DDA spectra were acquired using the following parameters: MS scan (400 to 1,500 m/z, 500 ms accumulation time) in high resolution mode (>35,000), followed by 50 MS/MS scans (100 to 1,800 m/z, a 50-ms accumulation time, and an intensity threshold at 200 cps). The DDA MS data were processed with AB Sciex ProteinPilot 4.5 software. Spectrum identification was performed by searching against the M. tuberculosis H37Rv UniProt entries with parameters, including carbamidomethyl cysteine, oxidized methionine, all biological modifications, amino acid substitutions, and missed cleavage sites. Proteins identified at a false-discovery rate below 1% were used as the SWATH reference spectral library.
SWATH wiff files were processed by using AB Sciex PeakView 2.1 software and the SWATH acquisition MicroApp. Up to six peptides with at least 99% confidence were selected with six transitions per peptide. XIC extraction windows was set to 25 min, and the XIC width was set to 75 ppm. The XIC peak area was extracted and exported using AB Sciex MarkerView 1.2 software for normalization and statistical analysis. The TubercuList protein functional classification (http://genolist.pasteur.fr/TubercuList/) was used. Only proteins with a significant change in abundance (P <0.05) were selected and analyzed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) version 9.1 (http://string.embl.de/). STRING uses a score to define interaction confidence; all interactions with a confidence score of >0.9 (highest confidence) were collected (35).
RESULTS
Preliminary test on M. bovis BCG.
We tested tetrahydrolipstatin (THL), simvastatin, and fenofibrate for their ability to weaken the mycobacterial cell wall and consequently to potentiate the vancomycin inhibitory effect (7). The first experiment was performed using M. bovis BCG according to the BacT/Alert MP method and a fixed 5-μg/ml vancomycin concentration, which is far below the vancomycin MIC (>500 μg/ml), as previously reported (7). We observed that only THL remarkably increased the susceptibility of the bacteria to vancomycin (Fig. 1). In order to investigate whether this growth inhibition resulted from a synergy or from the effect of THL alone, we determined the MIC of THL and its FICI in combination with vancomycin. As shown in Table 1, the MIC for THL alone, as determined by the macrodilution method, was 5 μg/ml. The MIC dropped to 0.625 to 1.25 μg/ml in the presence of vancomycin (10 μg/ml), revealing that this combination acts synergistically (FICI = 0.185 to 0.31). In contrast, simvastatin alone displayed an MIC of 100 μg/ml and could act with vancomycin but not in synergy (FICI = 1). Interestingly, no synergistic effect of THL was observed in the presence of amoxicillin-clavulanic acid, suggesting a specific mechanism responsible for the combined effect of THL with vancomycin (data not show).
FIG 1.
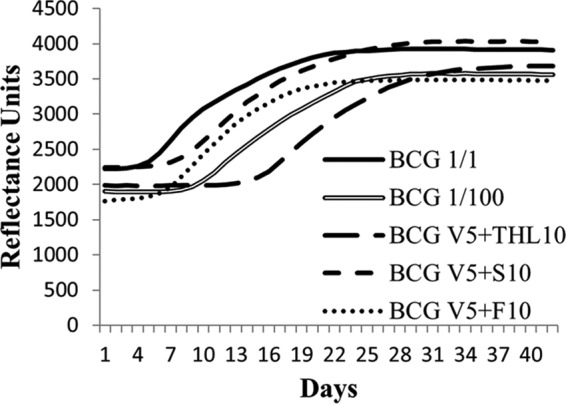
Drug susceptibility assay on M. bovis BCG. A fluorometric reflectance experiment was performed to record the normal and delayed growth of M. bovis BCG with or without drugs. Growth curves were compared to a 1/100 inoculum dilution to evaluate the MIC. Representative growth curves of the initial inoculum (BCG 1/1), of the dilution (BCG 1/100), and of the initial inoculum with a fixed 5-μg/ml amount of vancomycin (V5) combined with 10 μg/ml THL (THL10), 10 μg/ml simvastatin (S10), or 10 μg/ml fenofibrate (F10) are shown.
TABLE 1.
Drug susceptibility results obtained in the macrodilution series and analyzed by the checkerboard method for M. bovis BCG
| Treatment | MIC (μg/ml)/FIC (μg/ml)/FICIa |
||
|---|---|---|---|
| Vancomycin | THL | Simvastatin | |
| Alone | 250/–/– | 5/–/– | 100/–/– |
| + 1 μg/ml THL | 15/0.06/0.185–0.31 | –/–/– | –/–/– |
| + 25 μg/ml simvastatin | 125/0.5/1 | –/–/– | –/–/– |
| + 10 μg/ml vancomycin | –/–/– | 0.625–1.25/0.125–0.25/0.185–0.31 | 50/0.5/1 |
–, not applicable.
Synergistic inhibition confirmation on M. tuberculosis.
The BacT/Alert assay on M. tuberculosis H37Rv showed an effect of simvastatin at 100 μg/ml and THL at 50 μg/ml when combined with vancomycin at 5 μg/ml, in contrast to fenofibrate showing no effect at concentrations up to 100 μg/ml (Fig. 2). We further investigated with the BacT/Alert system the combination of THL (25 μg/ml) with vancomycin (5 μg/ml) and obtained an Δx/Δy value of 0.03, showing a synergy for those drugs. To calculate the FICI by the checkerboard method, we performed macrodilution experiments for both THL and simvastatin with vancomycin (Table 2). The THL MIC was 50 μg/ml when tested alone and fell to 3.1 μg/ml when combined with vancomycin (10 μg/ml). Simvastatin alone showed an MIC of 100 μg/ml that decreased (50 μg/ml) in combination with vancomycin (10 μg/ml). Based on these data, the FICI calculation showed synergism for THL with vancomycin (FICI = 0.31) and a nonsynergistic effect for simvastatin with vancomycin (FICI = 1.5). Finally, we used the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) agar proportion method to confirm these data (Table 2). Using this method, the THL MIC was 5 μg/ml and fell to 0.625 μg/ml in combination with vancomycin at 2 μg/ml. Confirming the BacT/Alert results, simvastatin showed an MIC of 100 μg/ml, which decreased to 50 μg/ml in the presence of 2 μg/ml vancomycin. The checkerboard allowed us to calculate a FICI of 0.375 for THL with vancomycin and a FICI of 1 for simvastatin with vancomycin. Fenofibrate was also retested using the agar proportion method, which confirmed its inability to inhibit the growth of H37Rv (up to 200 μg/ml), including in combination with 2 μg/ml vancomycin.
FIG 2.
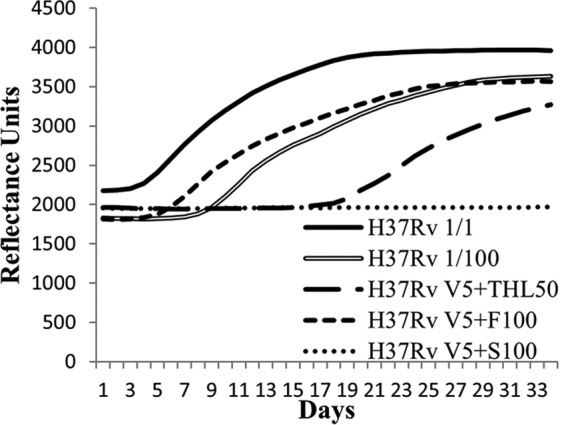
THL synergizes with vancomycin. Representative growth curves of the initial inoculum (H37Rv 1/1), the dilution (H37Rv 1/100), and the initial inoculum with a fixed 5-μg/ml amount of vancomycin (V5) added to 50 μg/ml THL (THL50), 100 μg/ml fenofibrate (F100), or 100 μg/ml simvastatin (S100) are shown.
TABLE 2.
Drug susceptibility results obtained with a macrodilution series and the CLSI agar proportion method and then analyzed by the checkerboard method with M. tuberculosis
| Treatment | Methoda | MIC (μg/ml)/FIC (μg/ml)/FICIb |
|||
|---|---|---|---|---|---|
| Vancomycin | THL | Simvastatin | Fenofibrate | ||
| Alone | MS | 100/–/– | 50/–/– | 100/–/– | –/–/– |
| CLSI | 40/–/– | 5/–/– | 100/–/– | >200/–/– | |
| + 1 μg/ml THL | MS | 25/0.25/0.31 | –/–/– | –/–/– | –/–/– |
| CLSI | 10/0.25/0.375 | –/–/– | –/–/– | –/–/– | |
| + 25 μg/ml simvastatin | MS | 100/1/1.5 | –/–/– | –/–/– | –/–/– |
| CLSI | 20/0.5/1 | –/–/– | –/–/– | –/–/– | |
| + 50 μg/ml fenofibrate | MS | –/–/– | –/–/– | –/–/– | –/–/– |
| CLSI | 40/1/2 | –/–/– | –/–/– | –/–/– | |
| + 10 μg/ml vancomycin | MS | –/–/– | 3.1/0.06/0.31 | 50/0.5/1.5 | –/–/– |
| CLSI | –/–/– | –/–/– | –/–/– | –/–/– | |
| + 2 μg/ml vancomycin | MS | –/–/– | –/–/– | –/–/– | –/–/– |
| CLSI | –/–/– | 0.625/0.125/0.375 | 50/0.5/1 | >200/1/2 | |
MS, macrodilution series; CLSI, agar proportion method.
–, not applicable.
Membrane lipid alteration by THL and simvastatin on M. tuberculosis.
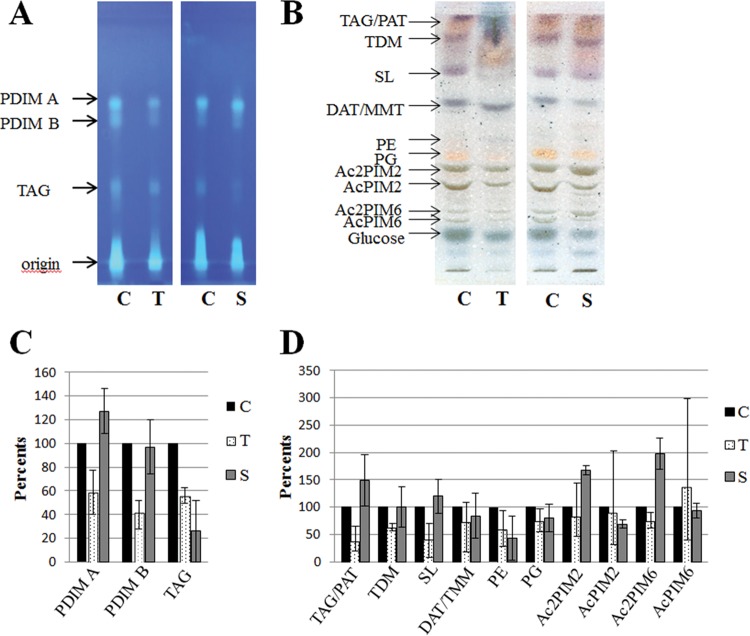
Total lipid extracts from treated M. tuberculosis H37Rv were compared to those from untreated samples to assess the effects of these drugs on cell wall lipids. As shown in Fig. 3, 24 h after the addition of 50 μg/ml THL, the amount of phthiocerol dimycocerosate (PDIM A) and phthiodiolone dimycocerosate (PDIM B) outer membrane lipids clearly decreased, as well as the amount of triacylglycerol (TAG) TDM and SL-1. Some compounds, exhibiting a mobility close to those of TDM and the major sulfated compounds (SL-1), were observed only in THL-treated extracts. The mycolic acids were unchanged (data not shown).
FIG 3.
Lipid modifications detected in M. tuberculosis treated with THL (lanes T) or simvastatin (lanes S) compared to the DMSO control (lanes C). (A) HPTLC analysis of PDIM A, PDIM B, and TAG separated with petroleum ether-diethyl ether (9:1 [vol/vol]) and visualized by primuline. (B) HPTLC analysis of lipids separated with CHCl3-CH3OH-H2O (60:35:8 [vol/vol/vol]), visualized by spraying with a 0.2% anthrone solution (wt/vol) in concentrated H2SO4, followed by heating. (C) Lipid quantification of PDIM A, PDIM B, and TAG based on colorimetric and fluorometric analysis of TLC. (D) Lipid quantification of representative lipids based on colorimetric and fluorometric analysis of TLC. In each graph, means ± the standard error bars of three independent experiments, converted to a percentage, with untreated samples taken as 100%, are shown. For THL-treated or simvastatin-treated samples versus untreated samples, three independent experiments were analyzed by TLC and quantified.
In contrast to THL, treatment with 100 μg/ml simvastatin did not affect the amount of PDIM A and PDIM B in the cell wall. We also observed increased amounts of phosphatidylmyoinositol dimannosides (Ac2PIM2 and Ac2PIM6) and reduced amounts of phosphatidylethanolamine (PE), AcPIM2, and TAG. The mycolic acids were equally unchanged (data not shown).
Proteomic response to THL and simvastatin in M. tuberculosis.
To improve our comprehension of the drug actions, we compared the proteome of untreated M. tuberculosis with the proteome of M. tuberculosis treated for 24 h with a 2-fold-reduced sub-MIC of either THL (25 μg/ml) or simvastatin (50 μg/ml) and no vancomycin in order to study bacteria reaction under conditions avoiding cell killing. Differential proteomic data were obtained using the SWATH (sequential window acquisition of all theoretical spectra) approach. SWATH-MS is a recently developed, label-free, quantitative proteomics-based MS method that combines data-independent acquisition (DIA) and targeted data analysis. In this way, robust quantitative data for thousands of proteins in a single measurement were obtained in MS2 mode (36). As shown in Table 3, the THL treatment induced more protein changes (ca. 7.5% of the protein content) in M. tuberculosis than the simvastatin treatment (ca. 1.8% change). THL treatment mainly involved protein upregulation (97%), in contrast to simvastatin inducing mainly protein downregulation (78%). Based on this simple analysis, it is clear that the bacterial response is completely different after a THL or simvastatin treatment. However, few proteins were commonly regulated (0.3%), such as four proteins downregulated from the intermediary metabolism (e.g., AldC and ProB) and two upregulated ribosomal proteins (RplW and RpmJ). THL treatment induced many protein upregulations, mainly involved in intermediary metabolism and respiration (34%), information pathways (16%) and lipid metabolism (14%) (Table 3; see also Fig. S1 and S2 in the supplemental material). Proteins involved in transcription (RpoA, RpoC, Rho, NusG, Mfd, …), transduction (RpmJ, RpsM, RplT, RpsR1, RpmD, …), the metabolic tricarboxylic acid cycle (MDH, GltA2, CitA, Can, Icd2, and Kgd), and glutamate metabolism (GltB, GlnA1, and Gdh) and numerous enzymes involved in FAS I (Fas) and II (AcpM, KasA, FabG1, Pks13, …) lipid synthesis were upregulated in THL-treated M. tuberculosis (see Fig. S2 and Table S3 in the supplemental material). Simvastatin downregulated proteins mainly involved in intermediary metabolism and respiration (33%) and lipid metabolism (12%) (Table 3; see also Fig. S1, Fig. S2, and Table S3 in the supplemental material).
TABLE 3.
Numbers of protein changes (up- and downregulation) 24 h after THL or simvastatin treatmenta
| Functional classification | No. of proteins detected |
|||
|---|---|---|---|---|
| THL |
Simvastatin |
|||
| Up | Down | Up | Down | |
| Intermediary metabolism and respiration | 99 | 6 | 4 | 19 |
| Lipid metabolism | 40 | 0 | 2 | 7 |
| Information pathways | 48 | 1 | 4 | 3 |
| Virulence, detoxification, adaptation | 12 | 0 | 1 | 2 |
| Cell wall and processes | 26 | 0 | 0 | 4 |
| Regulatory proteins | 13 | 0 | 0 | 4 |
| Conserved hypotheticals | 55 | 2 | 5 | 19 |
That is, proteins with a corresponding P value of <0.05.
DISCUSSION
THL, simvastatin, and fenofibrate were tested for their ability to potentiate the antimycobacterial activity of glycopeptides by weakening the mycobacterial cell wall (7). Our first approach, using the BacT/Alert system on M. bovis BCG, identified an interesting inhibitory effect of THL with vancomycin. A synergy could be identified by the checkerboard method from the macrodilution assay. This synergy was also observed on M. tuberculosis by both the BacT/Alert method and the checkerboard method using the macrodilution and the CLSI agar proportion results. This synergy suggested that THL destabilized the outer membrane of the cell envelope and facilitated the action of vancomycin. The impact of THL on the outer membrane integrity was verified by lipid analysis. As expected for a drug synergizing with vancomycin against M. tuberculosis and M. bovis BCG growth, we observed a reduction of PDIM A and PDIM B in THL-treated M. tuberculosis. The interaction of THL with TesA might explain, at least partly, this result (20). In agreement with Belardinelli et al., we also observed that the outer membrane SL-1 lipids were strongly decreased (37). Belardinelli et al. recently showed that THL inhibits PAT biosynthesis and, even more, SL-1 biosynthesis, suggesting a potential action on Chp 1 (37). The amount of mycolic acids was unchanged by THL treatment. The effect of THL on M. tuberculosis is therefore different than on M. kansasii, as Kremer et al., describing for the first time the antimycobacterial activity of this drug in M. kansasii, underlined its partial inhibitory effect on mycolic acids and meromycolyl-diacylglycerol synthesis (38). It is worth noting that we also observed an accumulation of degradation products of the cell wall mycolyl-arabinogalactan at 48 and 72 h after treatment. These products consisted mainly in dimycolyl-diarabinoglycerol, monomycolyl-arabinoglycerol and monomycolyl-diarabinoglycerol (data not shown). We also observed a reduction of TAG. Altogether, our results suggest that THL induced a loss of cell wall integrity by targeting several cell envelope lipid pathways, potentially including Chp 1, involved in SL-1 synthesis as previously reported (30), and TesA, involved in PDIM synthesis, as suggested by Ravindran et al. (20).
Simvastatin, although not able to synergize with vancomycin, inhibited already alone (100 μg/ml) the growth of both M. bovis BCG and M. tuberculosis. This result, observed in three antimycobacterial drug susceptibility assays, is in contrast to a previous publication in which no inhibitory effect for the simvastatin alone was detected (23). Our hypothesis is that this could be due either to the fact that the earlier study used a different macrodilution method or that the quality of the simvastatin was not identical (23). Lipid analysis of simvastatin-treated M. tuberculosis confirmed that in contrast to THL-treated cells, PDIM levels were not reduced, a finding in agreement with the absence of synergy with vancomycin. The amounts of PE and TAG were reduced, as were the amounts of AcPIM2, but the levels of Ac2PIM2 and Ac2PIM6 were increased. The action of THL and simvastatin on the mycobacterial cell wall was thus clearly different. For the fenofibrate, we could not detect any effect, either alone or in combination with vancomycin, on M. bovis BCG or M. tuberculosis.
It is interesting to understand how the bacteria respond to an antibiotic/drug stress, since this could provide a clue to the drug tolerance or resistance developed by bacteria. We observed here that M. tuberculosis treated with either THL or simvastatin has lower intracellular TAG contents than do untreated cells. The TAG reservoir could have served as a fatty acyl/carbon source for the recovery metabolism but could also have been decreased due to the action of these lipid-lowering drugs. Proteomic analyses suggested that M. tuberculosis under THL stress is trying to recover, expressing more proteins, especially those involved in intermediary metabolism, respiration, and information pathways, probably in order to gain energy. The protein upregulation observed in the FAS I and FAS II lipid synthesis pathways in THL-treated M. tuberculosis suggests that the bacterium is remodeling its cell wall. These modifications in lipid metabolism were consistent with three gene cluster signature profiles (GC82, GC89, and GC120) of six proposed by Boshoff et al. for inhibitors of cell wall biosynthesis (isoniazid, ethionamide, and cerulenin), which is in agreement with our lipid results and previous reports as described above (39). Our proteomic data with THL were compared to the microarray analysis of Waddell et al. providing a gene expression profile for THL-treated M. tuberculosis (40). In agreement with our results, Waddell et al. generally observed gene upregulation with THL treatment.
In contrast, M. tuberculosis treated with simvastatin did not seem to try to recover, since very few change in protein expression were detected. Interestingly, some downregulated proteins are listed in three gene clusters—GC29, GC34, and GC76—described by Boshoff et al. as being downregulated by (i) 5-chloropyrazinamide, rifampin, and H2O2 (GC29), (ii) rifampin, pyrazinamide, nicotinamide, and benzamide (GC34), and (iii) rifampin, pyrazinamide, nicotinamide, H2O2, and UV (GC76), respectively (39). It would be therefore interesting to carry out further studies to investigate whether simvastatin could damage DNA.
In conclusion, we identified a synergy between THL and vancomycin to inhibit M. bovis BCG and M. tuberculosis growth. Lipids analysis confirmed that THL destabilized the outer membrane of the cell envelope by reducing the amount of PDIM, facilitating the vancomycin action. The concordance of the results obtained by the macrodilution method, the CLSI agar proportion experiments, and the BacT/Alert MP assays, suggests that the BacT/Alert MP system is suitable for antituberculosis drug screening. Simvastatin, by itself, also seems to be a promising antimycobacterial drug, since mycobacteria appeared to be unable to act in order to recover.
Further studies, especially in vivo assays, should be performed with these drugs in order to assess their potential use against TB. New formulations should be developed to allow their delivery into the lungs.
Supplementary Material
ACKNOWLEDGMENTS
Céline Rens was supported by the Amis des Instituts Pasteur à Bruxelles.
We thank Christine Farin, Zumal Gunal, Brigitte Lecocq, and Sara Dulanto for their technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00872-16.
REFERENCES
- 1.World Health Organization. 2015. WHO report: global tuberculosis control. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Acosta CD, Dadu A, Ramsay A, Dara M. 2014. Drug-resistant tuberculosis in Eastern Europe: challenges and ways forward. Public Health Action 4:S3–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olaru ID, von Groote-Bidlingmaier F, Heyckendorf J, Yew WW, Lange C, Chang KC. 2015. Novel drugs against tuberculosis: a clinician's perspective. Eur Respir J 45:1119–1131. doi: 10.1183/09031936.00162314. [DOI] [PubMed] [Google Scholar]
- 4.Mdluli K, Kaneko T, Upton A. 2015. The tuberculosis drug discovery and development pipeline and emerging drug targets. Cold Spring Harb Perspect Med 5:a021154. doi: 10.1101/cshperspect.a021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neyrolles O, Guilhot C. 2011. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91:187–195. doi: 10.1016/j.tube.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Parish T, Liu J, Nikaido H, Stoker NG. 1997. A Mycobacterium smegmatis mutant with a defective inositol monophosphate phosphatase gene homolog has altered cell envelope permeability. J Bacteriol 179:7827–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soetaert K, Rens C, Wang XM, De Bruyn J, Lanéelle MA, Laval F, Lemassu A, Daffé M, Bifani P, Fontaine V, Lefèvre P. 2015. Increased vancomycin susceptibility in mycobacteria: a new approach to identify synergistic activity against multidrug-resistant mycobacteria. Antimicrob Agents Chemother 59:5057–5060. doi: 10.1128/AAC.04856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffé M, Crick D, Jackson M. 2014. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectrum 2:MGM2-0021-2013. doi: 10.1128/microbiolspec.MGM2-0021-2013. [DOI] [PubMed] [Google Scholar]
- 9.Hadváry P, Sidler W, Meister W, Vetter W, Wolfer H. 1991. The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J Biol Chem 266:2021–2027. [PubMed] [Google Scholar]
- 10.Haalck L, Spener F. 1997. On the inhibition of microbial lipases by tetrahydrolipstatin. Methods Enzymol 286:252–263. doi: 10.1016/S0076-6879(97)86014-4. [DOI] [PubMed] [Google Scholar]
- 11.Saxena AK, Roy KK, Singh S, Vishnoi SP, Kumar A, Kashyap VK, Kremer L, Srivastava R, Srivastava BS. 2013. Identification and characterisation of small-molecule inhibitors of Rv3097c-encoded lipase (LipY) of Mycobacterium tuberculosis that selectively inhibit growth of bacilli in hypoxia. Int J Antimicrob Agents 42:27–35. doi: 10.1016/j.ijantimicag.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Dhouib R, Ducret A, Hubert P, Carrière F, Dukan S, Canaan S. 2011. Watching intracellular lipolysis in mycobacteria using time lapse fluorescence microscopy. Biochim Biophys Acta 1811:234–241. doi: 10.1016/j.bbalip.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Singh G, Arya S, Narang D, Jadeja D, Singh G, Gupta UD, Singh K, Kaur J. 2014. Characterization of an acid inducible lipase Rv3203 from Mycobacterium tuberculosis H37Rv. Mol Biol Rep 41:285–296. doi: 10.1007/s11033-013-2861-3. [DOI] [PubMed] [Google Scholar]
- 14.Côtes K, Dhouib R, Douchet I, Chahinian H, de Caro A, Carrière F, Canaan S. 2007. Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem J 408:417–427. doi: 10.1042/BJ20070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low KL, Rao PS, Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. 2009. Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guerin. J Bacteriol 191:5037–5043. doi: 10.1128/JB.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crellin PK, Vivian JP, Scoble J, Chow FM, West NP, Brammananth R, Proellocks NI, Shahine A, Le Nours J, Wilce MC, Britton WJ, Coppel RL, Rossjohn J, Beddoe T. 2010. Tetrahydrolipstatin inhibition, functional analyses, and three-dimensional structure of a lipase essential for mycobacterial viability. J Biol Chem 285:30050–30060. doi: 10.1074/jbc.M110.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeliger JC, Holsclaw CM, Schelle MW, Botyanszki Z, Gilmore SA, Tully SE, Niederweis M, Cravatt BF, Leary JA, Bertozzi CR. 2012. Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. J Biol Chem 287:7990–8000. doi: 10.1074/jbc.M111.315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker SK, Barkley RM, Rino JG, Vasil ML. 2009. Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin. PLoS One 4:e4281. doi: 10.1371/journal.pone.0004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majumdar A, Wankhade G, Kamble PD, Harinath BC. 2011. Effect of HIV protease inhibitors and orlistat on mycobacterial ES-31 serine protease, a potential drug target in Mycobacterium tuberculosis. Indian J Tuberc 58:4–10. [PubMed] [Google Scholar]
- 20.Ravindran MS, Rao SP, Cheng X, Shukla A, Cazenave-Gassiot A, Yao SQ, Wenk MR. 2014. Targeting lipid esterases in mycobacteria grown under different physiological conditions using activity-based profiling with tetrahydrolipstatin (THL). Mol Cell Proteomics 13:435–448. doi: 10.1074/mcp.M113.029942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao A, Ranganathan A. 2004. Interaction studies on proteins encoded by the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Mol Genet Genomics 272:571–579. doi: 10.1007/s00438-004-1088-3. [DOI] [PubMed] [Google Scholar]
- 22.Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, Suzuki H, Marais AD, Brombacher F. 2014. Statin therapy reduces the Mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J Infect Dis 209:754–763. doi: 10.1093/infdis/jit550. [DOI] [PubMed] [Google Scholar]
- 23.Skerry C, Pinn ML, Bruiners N, Pine R, Gennaro ML, Karakousis PC. 2014. Simvastatin increases the in vivo activity of the first-line tuberculosis regimen. J Antimicrob Chemother 69:2453–2457. doi: 10.1093/jac/dku166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garbe TR. 2004. Coinduction of methyltransferase Rv0560c by naphthoquinones and fibric acids suggests attenuation of isoprenoid quinone action in Mycobacterium tuberculosis. Can J Microbiol 50:771–778. doi: 10.1139/w04-067. [DOI] [PubMed] [Google Scholar]
- 25.Reich-Slotky R, Kabbash C, Della-Latta P, Blanchard J, Feinmark S, Freeman S, Kaplan G, Shuman H, Silverstein S. 2009. Gemfibrozil inhibits Legionella pneumophila and Mycobacterium tuberculosis enoyl coenzyme a reductases and blocks intracellular growth of these bacteria in macrophages. J Bacteriol 191:5262–5271. doi: 10.1128/JB.00175-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SJ, Hong M, Song KD, Lee HK, Ryoo S, Heo TH. 2014. Normalization of the levels of inflammatory molecules in Mycobacterium smegmatis-infected U937 cells by fibrate pretreatment. Biol Res 47:42. doi: 10.1186/0717-6287-47-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed] [Google Scholar]
- 28.Lorian V. 2005. Antibiotics in laboratory medicine, 5th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Hsieh MH, Yu CM, Yu VL, Chow JW. 1993. Synergy assessed by checkerboard: a critical analysis. Diagn Microbiol Infect Dis 16:343–349. doi: 10.1016/0732-8893(93)90087-N. [DOI] [PubMed] [Google Scholar]
- 30.Mathys V, Wintjens R, Lefevre P, Bertout J, Singhal A, Kiass M, Kurepina N, Wang XM, Mathema B, Baulard A, Kreiswirth BN, Bifani P. 2009. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 53:2100–2109. doi: 10.1128/AAC.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werngren J, Klintz L, Hoffner SE. 2006. Evaluation of a novel kit for use with the BacT/ALERT 3D system for drug susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 44:2130–2132. doi: 10.1128/JCM.02218-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David S. 2001. Synergic activity of d-cycloserine and β-chloro-d-alanine against Mycobacterium tuberculosis. J Antimicrob Chemother 47:203–206. doi: 10.1093/jac/47.2.203. [DOI] [PubMed] [Google Scholar]
- 33.Singh P, Wesley C, Jadaun GP, Malonia SK, Das R, Upadhyay P, Faujdar J, Sharma P, Gupta P, Mishra AK, Singh K, Chauhan DS, Sharma VD, Gupta UD, Venkatesan K, Katoch VM. 2007. Comparative evaluation of Löwenstein-Jensen proportion method, BacT/ALERT 3D system, and enzymatic pyrazinamidase assay for pyrazinamide susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 45:76–80. doi: 10.1128/JCM.00951-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roxane Simeone Gaelle Huet Patricia Constant Wladimir Malaga Anne Lemassu Francoise Laval Mamadou Daffe Christophe Guilhot Christian Chalut. 2013. Functional characterization of three o-methyltransferases involved in the biosynthesis of phenolglycolipids in Mycobacterium tuberculosis. PLoS One 8:e58954. doi: 10.1371/journal.pone.0058954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11:O111.016717-1–17. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belardinelli JM, Larrouy-Maumus G, Jones V, Sorio de Carvalho LP, McNeil MR, Jackson M. 2014. Biosynthesis and translocation of unsulfated acyltrehaloses in Mycobacterium tuberculosis. J Biol Chem 289:27952–27965. doi: 10.1074/jbc.M114.581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremer L, de Chastellier C, Dobson G, Gibson KJ, Bifani P, Balor S, Gorvel JP, Locht C, Minnikin DE, Besra GS. 2005. Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol Microbiol 57:1113–1126. doi: 10.1111/j.1365-2958.2005.04717.x. [DOI] [PubMed] [Google Scholar]
- 39.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem 279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 40.Waddell SJ, Stabler RA, Laing K, Kremer L, Reynolds RC, Besra GS. 2004. The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberculosis 84:263–274. doi: 10.1016/j.tube.2003.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.