Abstract
Spectinomycin is an aminocyclitol antibiotic used clinically to treat a variety of infections in animals. Here, we characterized drug resistance prevalence in clinical Streptococcus suis isolates and discovered a novel resistance mechanism in which the s5 mutation (Gly26Asp) results in high spectinomycin resistance. Additionally, a novel integrative and conjugative element encompassing a multidrug resistance spw_like-aadE-lnu(B)-lsa(E) cluster and a cadmium resistance operon were identified, suggesting a possible cause for the wide dissemination of spectinomycin resistance in S. suis.
TEXT
Streptococcus suis, the leading agent for human meningitis in several Asian countries, is a significant zoonotic pathogen worldwide. Recently, it has received growing attention from the global academic community not only for its increased incidence of infections in humans but also for its important implied role in drug resistance transmission (1). The aminocyclitol antibiotic spectinomycin, often combined with lincomycin, is widely used for the treatment of pathogen infections in farm animals, including swine (2). However, few studies have examined spectinomycin resistance in S. suis globally. In this study, 191 clinical S. suis isolates, collected from different provinces in China over the period from 2006 to 2012, were initially subjected to spectinomycin susceptibility analysis by Etest method. Based on the spectinomycin MIC breakpoints to cattle respiratory pathogens (3), 20 isolates (10.4%) were found to be resistant (MIC of ≥256 μg/ml) (see Table S1 in the supplemental material). To investigate resistance mechanisms within them, multi-PCR experiments were conducted to detect an array of adenyltransferase genes, including spc, aad9, spw, spd, and various aadA genes (4–8). In addition, the complete 16S RNA gene and the s5 gene were amplified and sequenced for mutation analysis in all resistant strains as well as certain susceptible strains (see Table S3 in the supplemental material). Unexpectedly, none of the widely reported resistance genes could be detected among these strains with the exception of the spw gene and its variant spw_like, which demonstrated approximately 94% amino acid identity corresponding to 15 site mutations (see Fig. S1 in the supplemental material). The spw gene and its variant spw_like were present in 13 isolates and 6 isolates, respectively (Table 1). In contrast, there were a variety of mutations within the 16S RNA and s5 genes in the resistant isolates in comparison with the susceptible strains, and, among these, two mutations were present in the 16S rRNA regions of all four operons corresponding to sites 1069(C1069T) and 1188(A1188G) in Escherichia coli 16S rRNA and one site within the 26 amino acids (aa) of the s5 protein (GGT → GAT, Gly26Asp). Interestingly, in the isolate with the s5 gene mutation, the spw or spw_like genes were also not detected, suggesting that spectinomycin resistance was likely caused by this mutation.
TABLE 1.
Characteristics of the spectinomycin-resistant strains primarily used in this study
| Strain | Yr of isolation | Source | Sample/origin | Serotype | MLST type | Resistance phenotypea | Resistance genotype |
|---|---|---|---|---|---|---|---|
| Cm4-1 | 2009 | Zhejiang, China | Lung/pig | 8 | ST617 | SPT, STR, LIN, CLI, KAN, ERY | spw_like aadE lnu(B) lsa(E) aphA erm(B) mef(A) msr(D) tet(M) tet(L) |
| A16h | 2009 | Zhejiang, China | Lung/pig | 28 | ST616 | SPT, STR, LIN, CLI, KAN, ERY, TET | spw_like aadE aacA-aphD lnu(B) lsa(E) aphA erm(B) mef(A) tet(M) tet(L) |
| Cm2-1 | 2009 | Zhejiang, China | Lung/pig | 4 | ST618 | SPT, STR, LIN, CLI, KAN, ERY, TET | spw_like aadE aacA-aphD lnu(B) lsa(E) aphA erm(B) mef(A) mrs(D) tet(O) tet(M) |
| D12 | 2006 | Sichuan, China | Lung/pig | 9 | ST619 | SPT, STR, LIN, CLI, ERY, TET | spw_like aadE erm(B) tet(O) tet(L) |
| G52-1 | 2009 | Zhejiang, China | Brain/pig | 26 | ST615 | SPT, STR, LIN, CLI, KAN, TET | spw_like aadE aacA-aphD lnu(B) lsa(E) aphA tet(M) tet(S) |
| NC28-6 | 2008 | Hubei, China | Lung/pig | 28 | ST75 | SPT, STR, LIN, CLI, ERY, TET | spw_like aadE lnu(B) lsa(E) erm(B) mef(A) tet(M) |
| W8h | 2009 | Hunan, China | Lung/pig | 28 | ST487 | SPT, LIN, CLI, ERY, TET | erm(B) tet(O) tet(L) rpsE(GGT77GAT Gly26Asp) |
SPT, spectinomycin; STR, streptomycin; LIN, lincomycin; CLI, clindamycin; KAN, kanamycin; ERY, erythromycin; TET, tetracycline.
To verify these potential mechanisms, we cloned the complete spw_like genes and mutated alleles of the 16S RNA and s5 genes into the shuttle vector pAT18 and then electrotransformed them into the spectinomycin-susceptible SC19 strain (9). The spectinomycin MICs of all transformants were determined by the Etest method. The results showed that the MIC values of the transformants harboring the plasmids containing the mutated s5 or spw_like genes increased more than 85-fold (from 12 μg/ml to ≥1,024 μg/ml) compared with those of the transformant containing the empty plasmid (12 μg/ml), while no changes in the MIC values could be observed in the SC19 cells transformed with the mutated 16S rRNA. To the best of our knowledge, this is the first report that the mutation from glycine to aspartic acid in site 26 of the s5 protein could lead to spectinomycin resistance in bacteria.
The spw gene was discovered to be widespread in clinical Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, and Erysipelothrix rhusiopathiae isolates from swine in China by residing in the aadE-spw-lsa(E)-lnu(B) cluster (10–12). In this study, of the 13 isolates carrying the spw gene, we detected this complete multiresistance cluster in three isolates.
The dissemination mechanism and genetic context of the spw_like genes in S. suis were previously entirely unknown. Given that no plasmids were isolated from the six strains containing the spw_like gene (data not shown), mating assays were performed to evaluate dissemination and genetic context. The mating assays were performed as previously described (13), using a ciprofloxacin-resistant derivative of S. suis A7 and a rifampin- and fusidic acid-resistant derivative of E. faecalis JH2-2 as the recipients and the six spw_like-positive S. suis strains as donors. The results indicated that the spw_like gene within S. suis strain NC28-6 was capable of transferring into S. suis A7, although at very low frequency (3.7 × 10−9 per donor). In all transconjugants obtained, the MIC values not only for spectinomycin but also for lincomycin and streptomycin increased significantly (data not shown), together pointing to the possibility that the spw_like gene was residing in a mobile genetic element associated with other resistance determinants. Subsequently, to determine the nucleotide sequence of this mobile element harboring the spw_like gene, the transconjugant was subjected to whole-genome resequencing utilizing the Illumina HiSeq 2000 sequencing platform and employing methods as previously reported (14).
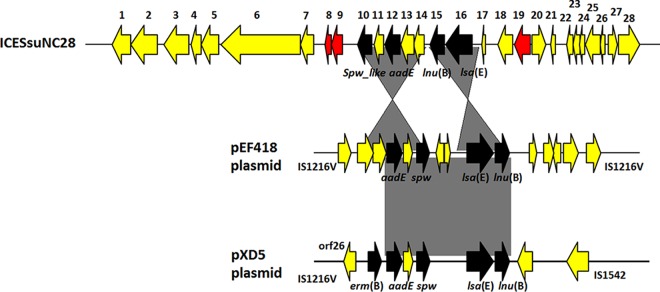
The results indicate that this element, designated ICESsuNC28 here, was integrated into the 3′ end of the rpsI gene in the S. suis A7 genome, generating approximately 83 imperfect direct-repeat flanking sequences, which was consistent with the donor strain S. suis NC28-6. Moreover, in all of the tested transconjugants, ICESsuNC28 was found to be integrated into this common site. Remarkably, thus far no integrative conjugative elements or transposons have been reported to integrate into the bacterial chromosome at this site, and integration was presumably mediated by a unique integrase distinct from other frequently reported or well-recognized integrases in other elements. The complete sequence of ICESsuNC28 was 29,661 bp in size and had an average G+C content of 34.3%, which was far below that of the S. suis chromosome (41.2%). ICESsuNC28 encompasses 28 open reading frames (ORFs), of which 25 are transcribed in the same direction (Fig. 1). In the ICESsuNC28 central region, orf8 and orf9 were identical to the cadmium resistance operon in ICESde3396 from Streptococcus dysgalactiae subsp. equisimilis, which was found to be responsible for a cadmium resistance phenotype (15). Downstream of the cadmium resistance operon, there was a multiresistance spw_like-aadE-lnu(B)-lsa(E) cluster, which is in a different arrangement from those of the reported spw-containing clusters in plasmids pEF418 and pXD5 with the aadE-spw-lsa(E)-lnu(B) configuration common to all members of the genus reported thus far (Fig. 1). Notably, in S. suis, lincosamide resistance is commonly associated with the erm(B) gene (1). No lnu family genes with specific resistance to the lincosamide group exclusively have previously been reported in S. suis except for a truncated lnu(E), although six different types of lnu family genes are currently well recognized (16). In addition to the cadmium resistance operon, another predicted cation transporter, encoded by orf 19, that is likely responsible for cadmium, cobalt, and zinc resistance was also observed in this element (see Table S2 in the supplemental material).
FIG 1.
Genetic organization of the novel mobile genetic element ICESsuNC28 and alignment between ICESsuNC28 and two other plasmids with the highest BLASTN scores. The direction of the arrow indicates the direction of transcription. The resistance determinants presented in black and red indicate the metal resistance genes.
In conclusion, in this study, we characterized the spectinomycin resistance that is primarily caused by spw and spw_like determinants in clinical S. suis isolates and discovered a novel resistance mechanism by which a mutation in the 30S ribosomal protein s5 (GGT → GAT, Gly26Asp) could confer high spectinomycin resistance in S. suis. Additionally, a novel mobile genetic element capable of transferring within S. suis by conjugation, harboring the spw_like-aadE-lnu(B)-lsa(E) multidrug resistance cluster and cadmium resistance operon, was identified, indicating the ongoing dissemination and evolution of multiresistance determinants between various pathogens via the swine reservoir.
Accession number.
The complete sequence of ICESsuNC28 has been deposited in GenBank under the accession number KU215704.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program (973) of China (2012CB518805) and the Chinese Major Special Science and Technology Project (2012ZX10004214-005).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01157-16.
REFERENCES
- 1.Palmieri C, Varaldo PE, Facinelli B. 2011. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front Microbiol 2:235. doi: 10.3389/fmicb.2011.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarz S, Werckenthin C, Alesik E, Wieler LH, Wallmann J. 2007. Susceptibility of bacterial pathogens against lincomycin/spectinomycin (1/2), penicillin G/neomycin (1/1), and penicillin G/dihydrostreptomycin (1/1) as determined in the BfT-GermVet monitoring program 2004-2006. Berl Munch Tierarztl Wochenschr 120:363–371. [PubMed] [Google Scholar]
- 3.Wisselink HJ, Veldman KT, Van den Eede C, Salmon SA, Mevius DJ. 2006. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet Microbiol 113:73–82. doi: 10.1016/j.vetmic.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 4.Kehrenberg C, Catry B, Haesebrouck F, de Kruif A, Schwarz S. 2005. Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob Agents Chemother 49:3046–3049. doi: 10.1128/AAC.49.7.3046-3049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CY, Chang LL, Chang YH, Lee TM, Li YH, Chang SF. 2000. Two new gene cassettes, dfr17 (for trimethoprim resistance) and aadA4 (for spectinomycin/streptomycin resistance), inserted in an Escherichia coli class 1 integron. J Antimicrob Chemother 46:87–89. doi: 10.1093/jac/46.1.87. [DOI] [PubMed] [Google Scholar]
- 6.LeBlanc DJ, Lee LN, Inamine JM. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother 35:1804–1810. doi: 10.1128/AAC.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wendlandt S, Li B, Lozano C, Ma Z, Torres C, Schwarz S. 2013. Identification of the novel spectinomycin resistance gene spw in methicillin-resistant and methicillin-susceptible Staphylococcus aureus of human and animal origin. J Antimicrob Chemother 68:1679–1680. doi: 10.1093/jac/dkt081. [DOI] [PubMed] [Google Scholar]
- 8.Wendlandt S, Fessler AT, Kadlec K, van Duijkeren E, Schwarz S. 2014. Identification of the novel spectinomycin resistance gene spd in a different plasmid background among methicillin-resistant Staphylococcus aureus CC398 and methicillin-susceptible S. aureus ST433. J Antimicrob Chemother 69:2000–2003. doi: 10.1093/jac/dku067. [DOI] [PubMed] [Google Scholar]
- 9.Si Y, Yuan F, Chang H, Liu X, Li H, Cai K, Xu Z, Huang Q, Bei W, Chen H. 2009. Contribution of glutamine synthetase to the virulence of Streptococcus suis serotype 2. Vet Microbiol 139:80–88. doi: 10.1016/j.vetmic.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Li XS, Dong WC, Wang XM, Hu GZ, Wang YB, Cai BY, Wu CM, Wang Y, Du XD. 2014. Presence and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in enterococci of human and swine origin. J Antimicrob Chemother 69:1424–1426. doi: 10.1093/jac/dkt502. [DOI] [PubMed] [Google Scholar]
- 11.Zhang A, Xu C, Wang H, Lei C, Liu B, Guan Z, Yang C, Yang Y, Peng L. 2015. Presence and new genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in Erysipelothrix rhusiopathiae of swine origin. Vet Microbiol 177:162–167. doi: 10.1016/j.vetmic.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Si H, Zhang WJ, Chu S, Wang XM, Dai L, Hua X, Dong Z, Schwarz S, Liu S. 2015. Novel plasmid-borne multidrug resistance gene cluster including lsa(E) from a linezolid-resistant Enterococcus faecium isolate of swine origin. Antimicrob Agents Chemother 59:7113–7116. doi: 10.1128/AAC.01394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmieri C, Magi G, Mingoia M, Bagnarelli P, Ripa S, Varaldo PE, Facinelli B. 2012. Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob Agents Chemother 56:4697–4702. doi: 10.1128/AAC.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang A, Yang M, Hu P, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. 2011. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics 12:523. doi: 10.1186/1471-2164-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies MR, Shera J, Van Domselaar GH, Sriprakash KS, McMillan DJ. 2009. A novel integrative conjugative element mediates genetic transfer from group G streptococcus to other β-hemolytic streptococci. J Bacteriol 191:2257–2265. doi: 10.1128/JB.01624-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Q, Wendlandt S, Li H, Li J, Wu C, Shen J, Schwarz S, Wang Y. 2014. Identification of the novel lincosamide resistance gene lnu(E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: de novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob Agents Chemother 58:1785–1788. doi: 10.1128/AAC.02007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.