Abstract
Staphylococcus aureus biofilms are extremely difficult to treat. They provide a protected niche for the bacteria, rendering them highly recalcitrant toward host defenses as well as antibiotic treatment. Bacteria within a biofilm are shielded from the immune system by the formation of an extracellular polymeric matrix, composed of polysaccharides, extracellular DNA (eDNA), and proteins. Many antibiotics do not readily penetrate biofilms, resulting in the presence of subinhibitory concentrations of antibiotics. Here, we show that subinhibitory concentrations of clindamycin triggered a transcriptional stress response in S. aureus via the alternative sigma factor B (σB) and upregulated the expression of the major biofilm-associated genes atlA, lrgA, agrA, the psm genes, fnbA, and fnbB. Our data suggest that subinhibitory concentrations of clindamycin alter the ability of S. aureus to form biofilms and shift the composition of the biofilm matrix toward higher eDNA content. An understanding of the molecular mechanisms underlying biofilm assembly and dispersal in response to subinhibitory concentrations of clinically relevant antibiotics such as clindamycin is critical to further optimize antibiotic treatment strategies of biofilm-associated S. aureus infections.
INTRODUCTION
Staphylococcus aureus is a major cause of both health care-related and community-associated (CA) infections. The Gram-positive human-pathogenic bacterium produces and secretes a range of toxins and enzymes leading to acute infections such as bacteremia and skin abscesses (1, 2). In addition, most S. aureus strains are capable of biofilm formation and can persist in host tissues such as the bone, leading to chronic osteomyelitis, or on implanted medical devices such as vascular catheters, vascular grafts, heart valves, and prosthetic joints (3–5). Biofilm-associated infections are extremely difficult to treat, and these chronic or relapsing infections typically require prolonged antibiotic treatment or removal of the device (6–8). Antibiotic resistance of bacteria within a biofilm may result from slow growth, phenotypic heterogeneity, persister cell formation, and inactivation or reduced penetration of the antibiotic (9, 10). Diffusion of the antibiotic through biofilm cell clusters is dependent on the thickness and the composition of the extracellular polymeric matrix (9, 11). The slow transport within biofilms suggests that the bacteria may encounter subinhibitory concentrations of antibiotics. Previous studies have shown that low doses of different antibiotics trigger biofilm formation (12, 13) and lead to dramatic alterations in bacterial gene expression in S. aureus (14).
Biofilm formation proceeds in at least three phases: initial attachment, biofilm maturation, and dispersal (15, 16). Initial surface attachment is dependent on bacterial surface molecules such as the S. aureus murein hydrolase AtlA, teichoic acids, and fibronectin-binding proteins (FnBPs) (17–20). After attachment to the surface, the bacteria multiply and produce the extracellular polymeric matrix. Various studies showed that the biofilm matrix of many laboratory and clinical staphylococcal isolates is composed of substantial quantities of extracellular DNA (eDNA) (21–23) which is counterbalanced by the effect of the bacterial nuclease Nuc1 (24, 25). The negatively charged DNA plays an important role during attachment and biofilm maturation, potentially through its electrostatic properties (26). The release of eDNA into the biofilm matrix is dependent on murein hydrolases (19, 27). AtlA, the major autolytic enzyme in S. aureus, is required for cell division, cell wall turnover, and bacterial lysis (28, 29). Autolysis in S. aureus biofilms is also regulated by the activity of the Cid/Lrg holin-antiholin system. CidA oligomerizes in the bacterial cell membrane and increases the activity of murein hydrolases, possibly by exporting them into the extracellular space (22, 30, 31). In the dispersal phase, bacteria detach from the biofilm and spread to new infection sites. A quorum-sensing system (Agr) is responsible for the switch between biofilm formation and detachment (32, 33). Recently, it was shown that the quorum-sensing-controlled phenol-soluble modulins (PSMs) (34, 35) are present in biofilms as large amyloid fibers which, under certain conditions, stabilize S. aureus biofilms (36) but also play a role in biofilm dispersal due to their surfactant-like properties (34, 37).
There is growing evidence that S. aureus specifically responds to subinhibitory antibiotic concentrations by inducing biofilm formation and changing its biofilm matrix composition (38). Clindamycin (CLI), a protein synthesis inhibitor used in the treatment of Gram-positive bacterial infections, modulates the production of several toxins and virulence factors in S. aureus (14, 39–43). Recently, it was also shown that the production and expression of PSMs are significantly increased in planktonic S. aureus cultures by subinhibitory concentrations of clindamycin (44, 45). Clindamycin achieves high intracellular levels in bones and phagocytic cells and potentiates opsonization and phagocytosis at subinhibitory concentrations (46, 47). However, the impact of subinhibitory concentrations of clindamycin on the biofilm formation and matrix composition of clindamycin-sensitive S. aureus has not been investigated so far.
It is evident that during antibiotic treatment, host tissues, wounds, and biofilm-infected medical devices are exposed to a range of antibiotic concentrations. Here, we examined whether the exposure of S. aureus to subinhibitory concentrations of clindamycin modulates biofilm formation and changes the matrix composition in the community-acquired methicillin-resistant S. aureus (CA-MRSA) USA300 LAC strain (LAC wild type [wt]) and its isogenic mutants as well as clinical isolates (CIs). This study provides a deeper understanding of the modulation and regulation of S. aureus matrix components in response to subinhibitory concentrations of clindamycin, a commonly used antibiotic in clinical settings.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Tables S1 to S3 in the supplemental material. Bacterial strains were grown on blood agar plates (Columbia agar plus 5% sheep blood; bioMérieux SA). Brain heart infusion (BHI) broth (BD Difco) supplemented with 0.4% glucose (wt/vol; BHIg) was used for all overnight cultures (220 rpm, 37°C) and static biofilm assays with clindamycin hydrochloride (AppliChem GmbH) or amoxicillin (Sigma-Aldrich). The MIC of clindamycin in BHIg broth was determined as previously described (41). The MIC for the LAC wt, the isogenic mutants, and the clinical isolates CI-1 to CI-8, CI-11, and CI-12 was 0.125 mg/liter in BHIg broth. The MIC for CI-9 was 0.06 mg/liter, and for CI-10 it was 0.03 mg/liter. All clindamycin-resistant clinical isolates (CI-13 to CI-18) showed visible growth up to 256 mg/liter of clindamycin in BHIg broth (MIC > 256 mg/liter). Antibiotics were used at the following concentrations: erythromycin at 10 μg/ml (Erm), chloramphenicol at 10 μg/ml (Cm), and ampicillin at 100 μg/ml (Amp).
To assess any effect that might be due to the genetic background of the strains, sequence typing of the polymorphic repeat region of the protein A gene (spa) (48) was performed using Ridom StaphType software (Ridom GmbH, Würzburg, Germany).
Construction of S. aureus mutants.
Phage ϕ11 was used to transduce all knockout mutations into the LAC wt background. The agr::tet(M) mutation was transduced from S. aureus strain RN6911 (49) into LAC. The generated strain LAC agr::tet(M) had a complete deletion of the agr locus, which was confirmed by sequencing using the primers agrlocus-F and agrlocus-R (see Table S2 in the supplemental material). The strains NE1438, NE1692, and NE460 from the Nebraska Transposon Mutant Library were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) repository (50). The mutants carry the bursa aurealis mariner-based erythromycin resistance-expressing transposon (Tn) within the lrgA (SAUSA300_0256), cidA (SAUSA300_2479), or atlA (SAUSA300_0955) gene and were used to introduce mutations into LAC wt by phage transduction. To delete the erm cassette of the S. aureus LAC transposon mutants, we used allelic exchange via homologous recombination of the bursa aurealis Tn insertions (51). Briefly, the transposon mutants were transformed with the plasmid pTnT carrying a 500-bp homologous region 5′ and 3′ of the bursa aurealis Tn and a chloramphenicol resistance marker. Strains carrying the plasmid were incubated overnight on chloramphenicol (Cm) agar plates. The next day the colonies were restreaked, and after an additional overnight incubation on Cm agar plates at 44°C, the colonies representing single recombinants were used for further subculturing steps at 30°C without antibiotic. Anhydrotetracycline (ATc; 100 ng/ml) was used as counterselection because it inhibits growth of cells still carrying the plasmid. Colonies which had lost the plasmid (Cm sensitive) were screened on Erm agar plates for double recombination. The erm cassette deletion was confirmed by PCR using the primer pairs cidA/Buster, atlA/Upstream, and lrgA/lrgA-R and by sequencing with the primers listed in Table S2 in the supplemental material.
Biofilm experiments.
Bacterial strains were grown overnight on fresh blood agar plates, resuspended in phosphate-buffered saline (PBS; Kantonsapotheke Zurich, Switzerland), and diluted in BHIg broth. A total of 180 μl of BHIg broth with or without clindamycin or amoxicillin was transferred to a polystyrene 96-well plate (Cellstar; Greiner), and 20 μl of diluted bacteria (final concentration, about 5 × 105 CFU/ml) or medium (blank) was added before incubation for 18 h at 37°C in a wet chamber unless otherwise indicated. After incubation, the wells were rinsed three times with 200 μl of PBS, air dried, and stained with 0.1% crystal violet for 30 min. Unbound dye was removed by three washes with distilled water. The adhering dye was dissolved with 30% acetic acid, and the absorption was measured at 570 nm with a VersaMax microplate reader and Soft Max Pro, version 4.3.1 LS, software (Molecular Devices). Biofilm assays in 12-well polystyrene plates (Cellstar; Greiner Bio-One) used for RNA isolation, viable bacteria count (CFU/ml), and eDNA analysis were done as described above with 7.5-fold more volume per well.
qRT-PCR analysis.
Transcripts from LAC wt and atlA mutant biofilms cultured with or without subinhibitory concentrations of clindamycin were assayed by quantitative reverse transcription-PCR (qRT-PCR). After 18 h of incubation in a 12-well polystyrene plate, S. aureus biofilm of LAC wt was washed three times with PBS and scraped off with a cell scraper. An aliquot was used for determination of CFU counts per milliliter. The biomass from two wells was pooled and spun down at 12,000 × g at 4°C for 5 min. RNA isolation and qRT-PCR were performed as previously described (41). All primers used for qRT-PCR are listed in Table S2 in the supplemental material. Differences in mRNA levels were calculated as follows: the ΔCT (where CT is cycle threshold) values of gyrB and the genes of interests were calculated by normalization of the mean CT from each biological replicate to the median of the biological replicates without CLI. For each sample, the fold change was calculated according to the Pfaffl analysis method (52). PCR experiments were performed using at least three biological replicates, each tested in technical triplicates.
Isolation and quantification of extracellular DNA.
Bacteria were cultured in 12-well plates in BHIg broth with or without subinhibitory concentrations of clindamycin (0.06 μg/ml). After 18 h of incubation, the supernatant was removed, and the biofilm was washed three times with PBS to remove planktonic cells. Five hundred microliters of PBS was added, and the cells were scraped from the surface using a cell scraper. The cells were transferred to a 1.5-ml microcentrifuge tube and sonicated in an ultrasonic bath (XUBA3; Grant). To degrade components of the biofilm matrix to release eDNA, the samples were incubated for 30 min at 37°C with 1 μl of peptide-N-glycosidase F ([PNGase F] 500 U/μl; NEB) and then with proteinase K (final concentration, 5 μg/ml; Sigma-Aldrich). An aliquot was used for determination of the CFU count per milliliter. After filtration (0.22-μm-pore-size polyvinylidene difluoride [PVDF] filter; Millipore), 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1; Sigma-Aldrich) was added, and the mixture was vortexed and centrifuged at 10,000 × g for 10 min. The upper phase was collected, and eDNA was precipitated by addition of 1/10 volume of 3 M sodium acetate (Sigma-Aldrich) and 2.5 volumes of 100% ethanol (Sigma-Aldrich). After overnight precipitation at −20°C, centrifugation at 20,000 × g for 10 min at 4°C, and washes with ice-cold 70% ethanol, the eDNA pellet was air dried, resuspended in Tris-EDTA (10:1) buffer, and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The eDNA amount was calculated relative to the mean biofilm mass of a 12-well polystyrene plate. eDNA was separated on a 0.8% agarose gel (0.5 μg/ml ethidium bromide) and visualized with a Benchtop UV Transilluminator (UVP, Axon Lab) and TS Image software (UVP).
Zymogram assays.
S. aureus LAC wt and atlA overnight cultures grown in BHIg broth without or with (0.03 and 0.06 μg/ml) clindamycin were set to the same optical densities at 600 nm (OD600) and centrifuged at 4,000 × g at 4°C for 10 min. The supernatant was filtered (0.45-μm-pore-size PVDF filter; Millipore) and stored at −80°C. Pellets were resuspended in PBS (pH 8) containing protease inhibitor (cOmplete protease inhibitor cocktail tablet; Roche). After incubation at 4°C for 1 h, cells were pelleted as described above and resuspended in 200 μl of PBS (pH 8) plus 2% SDS. Cells were incubated at 37°C for 20 min, followed by centrifugation (10,000 × g, 4°C). Protein concentrations of the crude cell wall extracts (SDS extracts) were determined by a bicinchoninic acid (BCA) protein assay (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. Murein hydrolase activities in the crude cell wall extract and the supernatant were analyzed by zymographic analysis. Ten micrograms of total protein from crude cell wall extracts and 13 μg of total protein from supernatants were separated using a 10% acrylamide gel containing about 100 mg of pellet (wet weight) of heat-inactivated Micrococcus luteus or S. aureus RN4220 with an electrophoresis system (Bio-Rad). After electrophoresis, the gels were incubated in the reaction buffer (25 mM Tris-HCl [pH 8], 1% Triton X-100) for at least 18 h, statically at 37°C. Gels were imaged with an Epson Perfection V700 photo scanner (Epson). Contrast and brightness were enhanced with IrfanView software.
Extraction of staphyloxanthin.
LAC wt and clinical isolates were grown in BHIg broth at 37°C in petri dishes (94 by 16 mm, Cellstar; Greiner) for 18 h. The supernatant was removed and after washes with PBS, the biofilm was scraped off with a cell scraper, followed by centrifugation (10,000 × g for 10 min) and washing with double-distilled H2O (ddH2O). Pictures of pelleted bacteria were enhanced for contrast and brightness with IrfanView software. Methanol (Sigma-Aldrich) extraction of staphyloxanthin was performed as described by Morikawa et al. (53). Absorption was measured at 465 nm with a VersaMax microplate reader and Soft Max Pro, version 4.3.1 LS, software (Molecular Devices).
Surfactant activity.
A drop-collapse assay was used as semiquantitative measurement of surfactant activity. The reduction of surface tension causes a collapse of the drop on a hydrophobic surface (Parafilm). Bacteria (LAC wt, agr, and atlA strains) were cultured with shaking or under static conditions in a 96-well plate in BHIg broth with or without clindamycin (0.03 and 0.06 μg/ml) at 37°C for different time periods. After incubation, shaking cultures or the supernatant of the 96-well plate were spun down at 8,000 × g for 15 min. Fifty-microliter drops were spotted on Parafilm and imaged with an Epson Perfection V700 photo scanner (Epson).
Electron microscopy.
LAC wt and the atlA mutant were grown in BHIg broth with and without clindamycin (0.06 μg/ml) for 18 h at 37°C. Bacteria were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer and postfixed in 1% osmium tetroxide and then 1% aqueous uranyl acetate. After stepwise dehydration in 70, 80, and 100% ethanol followed by propylene oxide, samples were embedded in Epon 30 and thin sectioned. Samples were poststained according to the Reynolds lead staining procedure. Images were taken with a 100 kV transmission electron microscope (TEM) with a digital charge-coupled-device (CCD) camera (TEM Philips CM100).
spa typing of clinical isolates.
Sequence typing of the polymorphic repeat region of the protein A gene (spa) (48) was performed using Ridom StaphType software (Ridom GmbH, Würzburg, Germany). Primers for spa typing are listed in Table S2 in the supplemental material.
Statistics.
Data were analyzed by using a Mann-Whitney U test for single comparisons or a Kruskal-Wallis test followed by post hoc Dunn's multiple comparisons. P values were calculated by using GraphPad Prism (version 5), and differences were considered significant for P values of <0.05.
RESULTS
Increased amount of biofilm formation and eDNA by S. aureus USA300 LAC in the presence of subinhibitory concentrations of clindamycin.
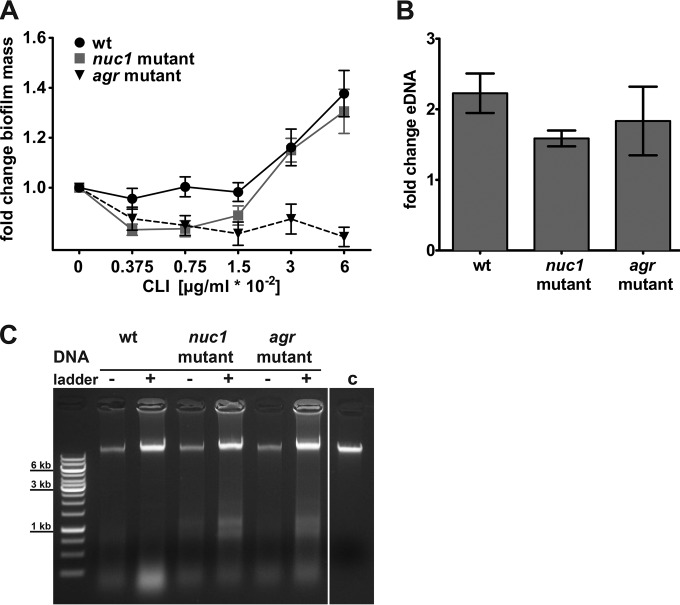
We assessed the impact of subinhibitory concentrations of clindamycin on biofilm formation. Growth of S. aureus LAC wt in the presence of subinhibitory concentrations of clindamycin led to an increase in the static biofilm mass after 18 h of growth in a microtiter plate. The most pronounced increase in biofilm mass occurred in the presence of 0.06 μg/ml clindamycin (Fig. 1A). The viable LAC wt bacteria counts (numbers of CFU per milliliter) from the biofilm mass after 18 h showed no significant difference when bacteria were grown with various subinhibitory concentrations of clindamycin (see Fig. S1A in the supplemental material). As eDNA is a major matrix component and might contribute to the increased biofilm mass, we analyzed the amount of eDNA in clindamycin-exposed biofilm cultures and compared the amount to that of controls grown without clindamycin. When normalized to the total biofilm mass, larger amounts of eDNA were detected in biofilms cultured with clindamycin than in the respective control without clindamycin (Fig. 1B). The isolated eDNA contained high-molecular-weight DNA (Fig. 1C), as described previously (12).
FIG 1.
Modulation of biofilm formation and eDNA release in S. aureus LAC in response to subinhibitory concentrations of clindamycin. (A) Quantification of the biofilm mass of the LAC wt, nuc1, and agr strains with crystal violet in the absence and presence of subinhibitory concentrations of clindamycin (CLI) after 18 h of static incubation. Biofilm mass without clindamycin was set to 1, and fold changes are presented. Results presented are average data from at least three replicates ± standard error of the mean. (B) eDNA isolated from biofilms grown for 18 h from wt and mutants (nuc1 and agr strains) grown in the presence of clindamycin were normalized to the biofilm mass, and results are presented as fold change relative to the level of the control without clindamycin. Results are average data from three replicates ± standard error of the means. (C) Representative agarose gel of isolated eDNA from biofilms incubated with (+; 0.06 μg/ml CLI) and without (−) clindamycin. Lane c, S. aureus wt chromosomal DNA control (∼ 60 ng). A Gene Ruler 1-kb DNA ladder is shown in the leftmost lane.
Since the Nuc1 nuclease and the Agr quorum-sensing system are important for biofilm development and dispersal, we also tested isogenic mutants deficient in nuc1 and the agr locus (25, 33). The nuc1 and agr mutants showed a higher total biofilm mass than the wt after 18 h of incubation (see Fig. S1B in the supplemental material). In a next step, we evaluated the influence of subinhibitory clindamycin concentrations on biofilm formation in both mutants. In the nuc1 strain, the clindamycin-induced increase in biofilm mass was similar to the level of the wt strain, whereas no increase was observed in the agr mutant (Fig. 1A). Viable bacteria counts of both mutants grown with or without clindamycin (0.06 μg/ml) remained stable (see Fig. S1C). Similar to the wt strain, both mutants had larger amounts of eDNA in their biofilms when cultured with clindamycin than the biofilms of the respective controls without clindamycin (Fig. 1B and C). As the agr mutant did not show a clindamycin-induced increase in biofilm mass in the static biofilm assay (Fig. 1A), we concluded that the observed increased biofilm mass of the wt and nuc1 mutant in response to subinhibitory concentrations of clindamycin did not result only from increased amounts of eDNA in the extracellular matrix.
Elevated biofilm formation by the S. aureus USA300 LAC autolysis mutant atlA in the presence of clindamycin.
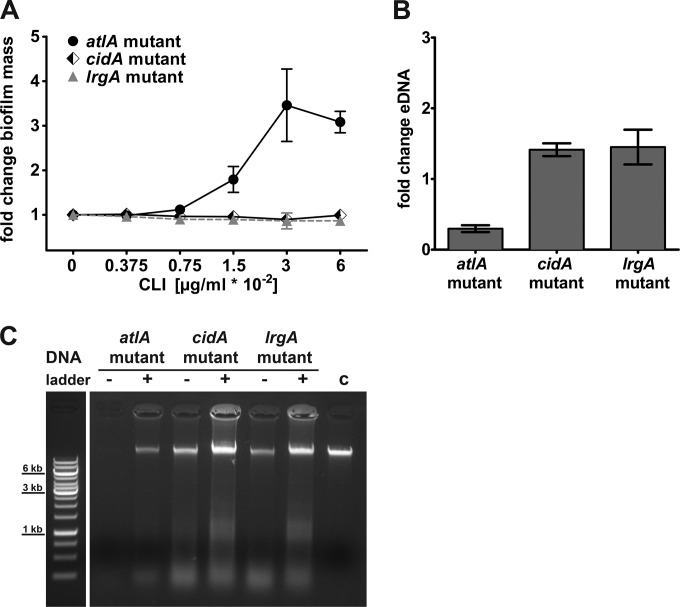
The observed increase in eDNA led us to investigate whether S. aureus autolysis systems (AtlA and CidA/LrgA) are involved in clindamycin-induced changes of the biofilm matrix. We confirmed that deletion of atlA significantly impaired biofilm formation (see Fig. S1B in the supplemental material) (19). When we cultured the isogenic atlA mutant in the presence of subinhibitory concentrations of clindamycin, we observed an up to 3.5-fold increase in the biofilm mass with 0.03 and 0.06 μg/ml of clindamycin (Fig. 2A). In addition, the atlA mutant strain cultured with clindamycin (0.06 μg/ml) showed an approximately 3-fold increase in the CFU count per milliliter in the biofilm mass (see Fig. S1C). The lrgA mutant grown without clindamycin resulted in increased biofilm accumulation compared to the level in the wt strain (see Fig. S1B). In contrast to results in a previous study by Rice et al. (22), deletion of cidA did not impair biofilm formation in our biofilm setting after 18 h but did produce an increase in biofilm mass (see Fig. S1B). No clindamycin-induced alteration of the biofilm phenotype was observed in the cidA and lrgA mutants (Fig. 2A).
FIG 2.
Subinhibitory concentrations of clindamycin affect an S. aureus autolysin mutant. (A) Quantification of the biofilm mass of the LAC atlA, cidA, and lrgA mutants with crystal violet in the absence and presence of subinhibitory concentrations of clindamycin (CLI) after 18 h of static incubation. Biofilm without clindamycin was set to 1, and fold changes are presented. Results presented are average data from at least three replicates ± standard error of the means. (B) eDNA isolated from biofilms grown for 18 h from LAC mutants (atlA, cidA, and lrgA strains) grown in the presence of clindamycin were normalized to the biofilm mass and presented as fold change relative to the control without clindamycin. Results are average data from three replicates ± standard error of the mean. (C) Representative agarose gel of isolated eDNA from biofilms incubated with (+; 0.06 μg/ml CLI) and without (−) clindamycin. Lane c, S. aureus wt chromosomal DNA control (∼ 60 ng). A Gene Ruler 1-kb DNA ladder is shown in the leftmost lane.
When we calculated the amount of eDNA relative to the biofilm mass, the atlA mutant showed a negative fold change of eDNA after exposure to subinhibitory concentrations of clindamycin compared to the level in the control (Fig. 2B). In contrast, a modest increase in eDNA in the extracellular biofilm matrix was observed in the lrgA and cidA mutants (Fig. 2B), and the isolated eDNA of all three mutant strains contained high-molecular-weight DNA (Fig. 2C). We concluded that CidA and LrgA are not essential for clindamycin-induced eDNA release in our biofilm model.
Mlynek and colleagues showed that subinhibitory concentrations of the β-lactam antibiotic amoxicillin induced biofilm formation in the USA300 wt and a nuclease-deficient mutant, while no alteration of biofilm formation was observed in an atlA mutant (54). To compare our biofilm model with these previously published data on antibiotic-induced biofilm alterations, we also tested the influence of amoxicillin on biofilm formation. In agreement with Mlynek and colleagues, we observed amoxicillin-mediated induction of biofilm formation for the USA300 LAC wt as well as the nuc1 background, and no induction was observed for the atlA mutant background (see Fig. S2 in the supplemental material).
Upregulation of genes involved in stress response, attachment, and eDNA release by clindamycin.
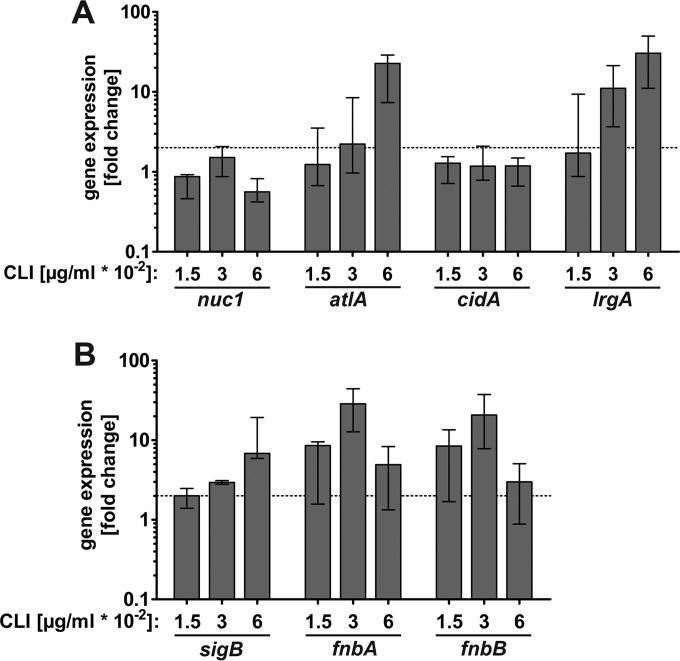
To corroborate our functional biofilm assay, we investigated the effect of subinhibitory clindamycin concentrations on the expression of genes involved in autolysis, stress response, and attachment. cDNA levels derived from RNA of 18-h biofilms grown with and without clindamycin were quantified by real-time PCR (qRT-PCR). As shown in Fig. 3A, expression of atlA and lrgA was upregulated in response to subinhibitory concentrations of clindamycin while cidA and nuc1 expression was not affected.
FIG 3.
Impact of subinhibitory concentrations of clindamycin on the expression of biofilm-associated genes. Relative transcript levels of genes important for eDNA and autolysis (A) or stress response and adhesion (B) derived from LAC wt biofilm mass with and without clindamycin after 18 h as determined by qRT-PCR. Fold change ratios were calculated by normalizing cDNA levels of the gene of interest against the housekeeping gene gyrB. Data are presented as fold change compared to levels in biofilms grown without clindamycin (set to 1), and error bars indicate interquartile range of at least three independent experiments. The dotted line indicates a 2-fold upregulation.
As subinhibitory concentrations of antibiotics can induce a general stress response (38, 55), gene expression of the alternative sigma factor B (σB) was investigated in our biofilm model and showed an upregulation of 3- and 7-fold in response to 0.03 and 0.06 μg/ml clindamycin, respectively (Fig. 3B). Since the S. aureus carotenoid pigment staphyloxanthin is positively regulated by σB (56, 57), we quantified the amount of staphyloxanthin of the LAC wt in response to subinhibitory concentrations of clindamycin and amoxicillin in our biofilm model. We found that the amount of staphyloxanthin increased with subinhibitory concentrations of clindamycin but not with addition of amoxicillin (see Fig. S3A and B in the supplemental material). As σB upregulates adhesion factors (58–60), we investigated the expression of the genes encoding the fibronectin binding proteins A (FnbA) and B (FnbB). Transcript levels of fnbA and fnbB were clearly higher in biofilms grown in the presence of 0.03 μg/ml of clindamycin (Fig. 3B).
Subinhibitory concentrations of clindamycin induce psm gene expression and surfactant activity.
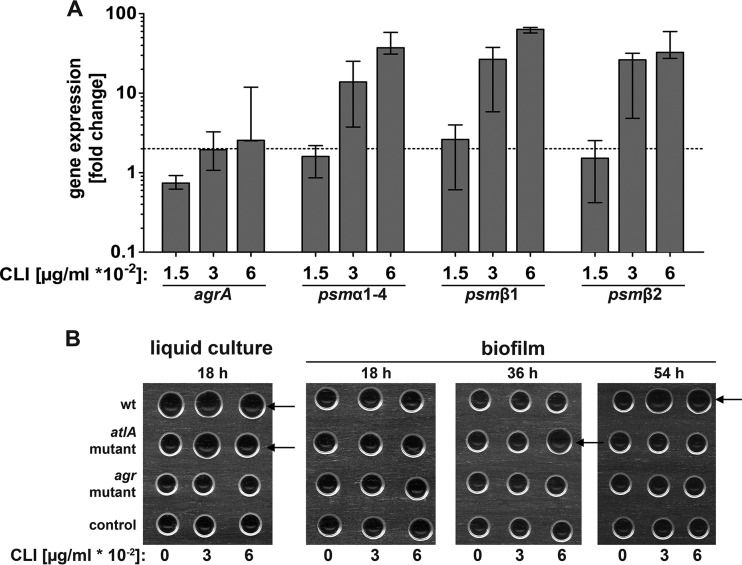
In addition to eDNA and the FnBPs, the recently discovered PSMs are important biofilm matrix components which are able to stabilize S. aureus biofilms but also play a role in biofilm dispersal (34, 36, 37). Transcription of the psm genes is under the control of the AgrA DNA binding protein (35). Thus, we studied the effect of subinhibitory concentrations of clindamycin on gene expression of agrA, the psmα operon (psmα1-psmα4), and the two genes of the psmβ operon (psmβ1 and psmβ2) in our biofilm system. The expression of agrA was upregulated 2.5-fold in the presence of clindamycin (0.06 μg/ml), and the upregulation of psmα1-psmα4 as well as of psmβ1 and psmβ2 was even more pronounced (up to 60-fold) (Fig. 4A). We also tested the induction of psm transcription in an atlA mutant background, which showed the same clindamycin-induced upregulation of psm genes as observed in the wt strain (see Fig. S4 in the supplemental material). PSMs were shown to exhibit surfactant-like activities (61), which lower the surface tension of liquids. In order to assess the functional effect of the highly upregulated psm transcription, we used a drop-collapse assay to illustrate the differences in surface tension of supernatants from biofilms or bacterial cultures grown with and without clindamycin. Surface tension was low in supernatants from bacterial overnight cultures of LAC wt and the atlA mutant, irrespective of clindamycin treatment (Fig. 4B). In contrast, surface tension was high in biofilm supernatants after 18 h. After 36 h the biofilm supernatant of the atlA mutant showed a decrease in surface tension in response to 0.06 μg/ml of clindamycin. In the supernatant of a 54-h old biofilm, the surface tension of the wt strain was clearly decreased in response to clindamycin compared to that of the control. As PSM transcription was highly induced in the wt strain and the atlA mutant by clindamycin, the observed decrease in surface tension in these two strains might derive from PSM overproduction. No surfactant activity was detected in the supernatants of the agr mutant strain, indicating that the observed decrease in surface tension is dependent on the quorum-sensing system.
FIG 4.
Overexpression of the psm genes and increase in surfactant activity in S. aureus biofilms in response to clindamycin. (A) Relative transcript levels of agrA and the psm genes derived from LAC wt biofilm mass after 18 h with and without clindamycin as determined by qRT-PCR. Fold change ratios were calculated by normalizing cDNA levels of the gene of interest against the housekeeping gene gyrB. Data are presented as fold change compared to biofilms grown without clindamycin (set to 1), and error bars indicate interquartile range of at least three independent experiments. The dotted line indicates a 2-fold upregulation. (B) Surface tension of supernatants of a bacterial liquid culture and of biofilms grown for indicated time with and without clindamycin is illustrated by a drop-collapse assay. If surfactants were present in the drop, the surface tension was reduced, and the drops spread or collapsed (indicated by arrows). Surfactant activity was high in the LAC wt and the atlA mutant in shaking cultures after 18 h. When grown as a biofilm, the atlA mutant showed an increase in surfactant activity in response to clindamycin after 36 h while the LAC wt showed a similar effect after 54 h. Representative images of three independent experiments are shown.
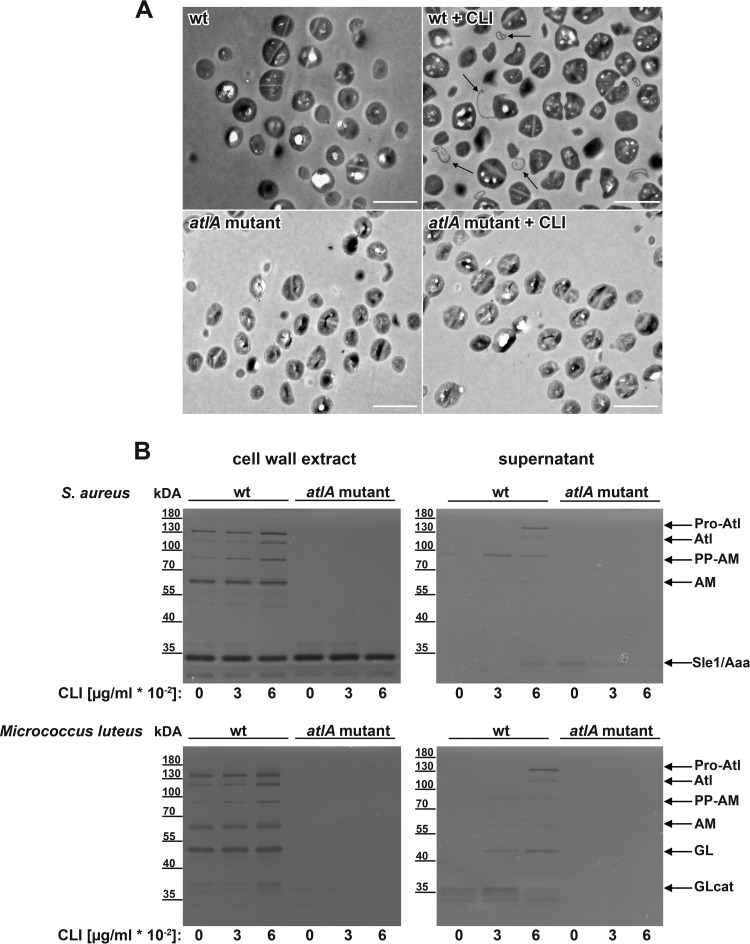
Difference in S. aureus USA300 LAC morphology due to subinhibitory concentrations of clindamycin.
Previous studies showed that the cell walls of staphylococci are thickened when exposed to high concentrations of clindamycin (62, 63). Therefore, we examined the effect of subinhibitory concentrations of clindamycin on S. aureus cell morphology by transmission electron microscopy (TEM). Cells grown in the presence of clindamycin (0.06 μg/ml) lost their spherical cell shape and appeared inflated, potentially reflecting a stress response phenotype. We did not see a thickening of the cell wall after 18 h of incubation with clindamycin. However, we observed a “peeling off” of the cell wall along with membrane-like fibers protruding from the bacterial cells (Fig. 5A). As AtlA was shown to play an important role in cell division and separation (28, 29), clindamycin-induced atlA expression (Fig. 3A) may contribute to the observed phenotype. Therefore, we also examined the cell morphology of the atlA mutant grown with and without clindamycin by TEM. In the atlA mutant no apparent peeling-off phenotype was observed in response to clindamycin (Fig. 5A). In conclusion, the AtlA upregulation by clindamycin might be responsible for the observed peeling-off phenotype of the cell wall. To investigate the influence of clindamycin on AtlA hydrolase activity in more detail, we performed zymogram analysis of the wt and the atlA mutant in response to clindamycin. Micrococcus luteus or Staphylococcus aureus cells were used to differentiate between glucosaminidase (GL) and amidase (AM) enzyme activity, respectively. Several autolytic bands were seen with the wt strain because AtlA undergoes multiple proteolytic cleavage events which are absent in the atlA mutant (Fig. 5B). The additional 32-kDa murein hydrolase Sle1 (64), also referred to as Aaa (65), was present in the wt as well as in the atlA mutant, as seen when Staphylococcus aureus was used as a substrate within the gel. The crude cell wall extracts showed minimal enhanced autolytic activity in response to clindamycin (0.03 and 0.06 μg/ml) compared to that of the control (Fig. 5B, left panel). A more profound effect of clindamycin was observed for the extracellular autolytic activity in the culture supernatants, in particular, the GL activity seen with Micrococcus as a substrate (Fig. 5B, right panel).
FIG 5.
Subinhibitory concentrations of clindamycin change S. aureus cell morphology. (A) Representative transmission electron images of the LAC wt and atlA mutant from cultures grown without and with clindamycin (0.06 μg/ml CLI). Arrows highlight membrane-like fibers. Scale bar, 2 μm. (B) Zymogram analysis of the LAC wt and atlA mutant autolytic activity from cultures grown for 18 h without and with clindamycin (0.03 to 0.06 μg/ml CLI). Crude cell wall extract (left panel) or filtered supernatants (right panel) were loaded on gels containing S. aureus RN4220 or Micrococcus luteus as the substrate. Bacteriolytic activity is indicated by the dark bands on the opaque gel. Bands are assigned as follows (27, 77): Pro-Atl (∼138 kDa), Atl with full-length propeptide; Atl (∼115 kDa), amidase plus glucosaminidase; PP-AM (∼ 84 kDa), amidase with propeptide; AM (62 to 65 kDa), amidase; GL (<55 kDa), glucosaminidase; Atl-GLcat (∼36 kDa and 38 kDa), glucosaminidase catalytic domain lacking repeat domain; Sle1/Aaa (∼ 32 kDa), S. aureus autolysin. Band size was estimated based on the PageRuler prestained protein ladder. Representative gels of at least three independent experiments are shown.
Alteration of biofilm formation by clindamycin in clinical isolates.
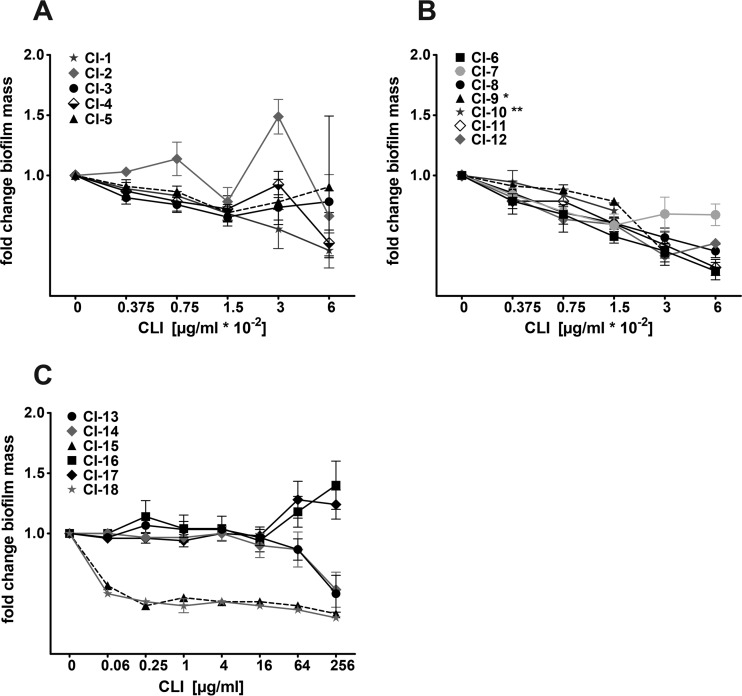
Since clindamycin is routinely used in clinics to treat S. aureus infections, we investigated whether subinhibitory concentrations of clindamycin also altered biofilm formation in S. aureus clinical isolates (strains are listed in Table S3 in the supplemental material) in our static biofilm model. All 18 tested clinical isolates formed a static biofilm (see Fig. S5). One out of five clindamycin-sensitive, methicillin-sensitive S. aureus (MSSA) strains (CI-1 to CI-5) showed a slight induction of biofilm formation when subjected to clindamycin at subinhibitory concentrations (CI-2) (Fig. 6A). This strain was isolated from a foreign-body-associated infection (vascular prosthesis infection), while the other four strains were isolated from native tissue-associated infections (endocarditis, panaritium, abscess, and wound infection). All seven clindamycin-sensitive MRSA strains (CI-6 to CI-12) showed a reduction of biofilm-forming capacity upon clindamycin exposure (Fig. 6B). We also tested six clindamycin-resistant MRSA strains for their biofilm phenotype when subjected to clindamycin at concentrations just below their MICs. Two out of six strains showed clindamycin-dependent biofilm induction when challenged with 64 and 256 μg/ml clindamycin (Fig. 6C). These two clinical isolates (CI-16 and CI-17) were USA300 clones (spa type t008) (66), like our model strain LAC wt, and had been isolated from skin and soft tissue infections. The other four clindamycin-resistant strains were of spa types t586 (n = 2) and t032 (n = 2) and showed no clindamycin-dependent biofilm induction, but strains from each of the two spa types showed similar behavior in response to clindamycin. This indicates that the clindamycin-dependent biofilm phenotype might be more pronounced in certain S. aureus subgroups, like the USA300 clonal lineage.
FIG 6.
Impact of subinhibitory concentrations of clindamycin on S. aureus clinical isolates. Biofilm mass quantification of MSSA clinical isolates CI-1 to CI-5 (A), clindamycin-sensitive MRSA clinical isolates CI-6 to CI-12 (B), and clindamycin-resistant MRSA clinical isolates CI-13 to CI-18 (C) with crystal violet in the absence and presence of subinhibitory concentrations of clindamycin (CLI) after 18 h of static incubation. Biofilm without clindamycin was set to 1, and fold changes are presented. Results are average data from at least three replicates ± standard error of the mean. *, highest tested clindamycin concentration, 0.03 μg/ml (MIC, 0.06 μg/ml); **, highest tested clindamycin concentration, 0.015 μg/ml (MIC, 0.03 μg/ml).
We next investigated whether the biofilm phenotype of the clinical isolates in response to clindamycin correlated with staphyloxanthin production and was thereby associated with a σB response to clindamycin. We chose five representative strains to quantify staphyloxanthin production, including two MSSA strains, one showing a clindamycin-induced biofilm phenotype (CI-2) and one which did not show a response to clindamycin (CI-4). Furthermore, we tested two clindamycin-sensitive MRSA strains, neither of which showed biofilm induction by clindamycin (CI-6 and CI-8), and one clindamycin-resistant MRSA strain which showed induction of biofilm formation upon clindamycin exposure (CI-16). As shown in Fig. S3C and D in the supplemental material, four out of five tested clinical isolates showed an increase in staphyloxanthin production. Strains CI-4 and CI-6 showed clear induction of staphyloxanthin production upon clindamycin exposure but did not show an increase in biofilm formation. The clindamycin-resistant MRSA CI-16 (spa type t008; USA300) showed the same biofilm and staphyloxanthin phenotype as our model strain LAC. These data indicate that the observed clindamycin-induced biofilm phenotype of our tested clinical isolates correlates with a σB response to clindamycin in the USA300 background but not in the other tested clinical isolates that had distinct spa types.
DISCUSSION
In this study, we show that subinhibitory concentrations of the frequently clinically used protein synthesis inhibitor clindamycin induced biofilm formation and altered the biofilm matrix composition of the CA-MRSA strain USA300 LAC. Although previous studies have investigated the effect of several antibiotics at subinhibitory concentrations on staphylococcal biofilm formation and matrix composition (12, 13, 38, 54, 67), the effect of subinhibitory concentrations of clindamycin, an important antibiotic to treat S. aureus infections, had not been investigated so far. Clindamycin-mediated biofilm induction was observed in the wt strain as well as in the nuc1 and atlA mutants. In addition, clindamycin had no effect on nuc1 expression. Taking these findings together, this shows that the observed biofilm induction by clindamycin is not due to transcriptional nuc1 repression. The atlA mutant was deficient in biofilm formation, which is in agreement with the role of AtlA in primary attachment (19, 28), and showed a 3.5-fold induction of biofilm formation by clindamycin. The presence of clindamycin also resulted in an increase of the viable bacteria count in the atlA mutant. Therefore, the clindamycin-induced increase in biofilm mass might derive from a combination of the higher viable bacteria count in the biofilm and an increase in primary attachment to the plastic surface.
Glucose-induced biofilms in the USA300 LAC strain are mainly polysaccharide intercellular adhesin (PIA) independent (16, 68), and a major component of these biofilms is eDNA (19, 22, 23). In S. aureus biofilms, the eDNA is likely produced through the autolysis of a subpopulation of cells which is mediated by the murein hydrolase AtlA (27). Isolation of eDNA from biofilms grown with and without clindamycin confirmed the presence of a small amount of eDNA in the atlA mutant relative to the biofilm mass. In contrast to results in the atlA mutant, clindamycin increased the amount of eDNA within the biofilm matrix in the wt as well as in the other mutant strains (nuc1, agr, cidA, and lrgA strains) relative to the biofilm mass. This suggests that the increase in eDNA of the wt and nuc1 mutant only partially contributes to the increase in biofilm formation. Clindamycin had no effect on biofilm formation of the cidA and the lrgA mutants. In the absence of clindamycin, we observed increased biofilm formation in the lrgA mutant compared to that in the wt strain, as previously reported for an S. aureus UAMS-1 lrgAB mutant (24). Expression of lrgAB is induced by the LytSR two-component system, which senses a reduction in the membrane potential (ΔΨ) (69). Although in an earlier study a cidA mutant in S. aureus UAMS-1 exhibited decreased lysis resulting in smaller amounts of eDNA and impaired biofilm formation (22, 24), the LAC cidA mutant showed the same biofilm phenotype as the wt strain in our biofilm model after 18 h. Transcription of cidA in UAMS-1 biofilms steadily increased over time (up to 90-fold after 72 h) (70). Therefore, its effect on biofilm formation might play only a minor role in our 18-h-old biofilm system while LrgAB might be activated by changes in the membrane potential at an earlier time point.
The atlA gene expression was approximately 25-fold upregulated in response to subinhibitory concentrations of clindamycin, which is the most likely cause of the observed increase of eDNA in the biofilm. We did not detect a difference in cidA expression but an upregulation of agrA and lrgA in response to clindamycin. The lrgAB operon is positively regulated by the Agr system and by the transcriptional regulator SarA (71), which was shown to be upregulated in response to clindamycin (14). Therefore, the observed upregulation of lrgA is likely due to the clindamycin-derived upregulation of these regulators. Since subinhibitory concentrations of clindamycin highly induced biofilm formation in the atlA mutant strain, we speculate that additional factors, likely fibronectin-binding proteins (FnBPs), are upregulated that might compensate for the impaired attachment of the atlA mutant strain. FnBPs mediate intercellular cell-cell adhesion via homophilic bonds (72) and are required for primary attachment and biofilm accumulation in the USA300 LAC strain (20). Corroborating this, we observed an upregulation of fnbA and fnbB expression by clindamycin in the biofilm. It has previously been shown that the S. aureus strain Newman showed increased amounts of fnbB transcripts due to selective mRNA stabilization in response to clindamycin (73).
The observed alteration in the expression profile in response to subinhibitory concentrations of clindamycin might be due to the activation of a general stress response via the alternative sigma factor B (σB). We confirmed that sigB expression was upregulated and that significantly larger amounts of σB-dependent staphyloxanthin were isolated in response to clindamycin. σB controls a large regulon which includes the expression of fnbA and fnbB (58–60) as well as atlA in certain S. aureus strains (19).
In contrast to the results obtained with our model strain USA300 LAC, the σB-dependent staphyloxanthin production correlated with the observed biofilm phenotypes in only two out of five selected clinical isolates. This indicates that the clindamycin-induced σB stress response does not necessarily translate into elevated biofilm formation. In addition, the observed amoxicillin-dependent biofilm induction was not associated with staphyloxanthin production. These experiments indicate that antibiotic-induced biofilm formation might derive from a σB-dependent stress response in only certain clonal lineages (USA300). At the same time, a σB-dependent stress response is not a necessary prerequisite for antibiotic-induced biofilm formation.
Other important matrix components which were recently discovered to play a role in S. aureus biofilm formation and dispersal are PSMs (74). The formation of amyloid fibers is thought to contribute to S. aureus biofilm integrity under certain conditions and might serve as a storage mechanism until the surfactant activity of the monomeric PSM peptides is required (36, 37). The psmα and psmβ genes are highly induced by subinhibitory concentrations of clindamycin in our biofilm model. In addition, surfactant activity increased in the supernatant of a 36-h-old atlA mutant biofilm and after 54 h also in the wt strain in response to clindamycin. The presence of eDNA in the biofilm matrix facilitates PSMs to form amyloid fibers (75). As we observed induction of psm gene expression and eDNA release in our biofilm model, we speculate that the formation of amyloid fibers might be increased at early time points (18 h) when we observed a thick biofilm with large amounts of eDNA. At later time points, degradation of eDNA and decomposition of the fibers might lead to biofilm dispersal via surfactant activity of the PSM monomers. Deficient release of eDNA in the atlA mutant might therefore hinder eDNA-mediated amyloid fiber formation of PSMs and thereby contribute to the observed surfactant activity at early time points in the mutant.
In summary, we showed that subinhibitory concentrations of clindamycin have diverse effects on S. aureus biofilm formation which highly depend on the strain background. In-depth analysis of the model strain LAC wt from the USA300 clonal lineage showed that subinhibitory concentrations of clindamycin alter the biofilm matrix composition by changing autolysis and eDNA release, increasing adhesion factors and secreted proteins, which likely leads to an interaction between the matrix components. Previous studies showed that biofilm matrix components bind to each other and act as an electrostatic network by connecting cells within the biofilm (26, 76). We hypothesize that the clindamycin-induced matrix components such as eDNA, PSMs, and FnbBs are highly interconnected within the biofilm matrix, which might result in a more compact and stable biofilm. Since the increase in biofilm formation and alteration of the matrix composition might impact S. aureus biofilm-associated infections, clindamycin should be dosed as high as possible in order to prevent subinhibitory clindamycin concentrations in affected tissues and biofilms.
Supplementary Material
ACKNOWLEDGMENTS
Imaging was performed with the support of the Center for Microscopy and Image Analysis, University of Zurich.
The authors have no potential conflicts to report.
Funding Statement
This work was supported by grants from the Swiss National Science Foundation (310030_146295/1 to A.S.Z.), the Hartmann Müller-Stiftung (to A.S.Z.), and the Promedica Stiftung (1080/A to B.H.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00463-16.
REFERENCES
- 1.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 5.Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM. 2005. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am J Infect Control 33:385–391. doi: 10.1016/j.ajic.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 7.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Donlan RM. 2001. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis 33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 9.Doroshenko N, Tseng BS, Howlin RP, Deacon J, Wharton JA, Thurner PJ, Gilmore BF, Parsek MR, Stoodley P. 2014. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob Agents Chemother 58:7273–7282. doi: 10.1128/AAC.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart PS. 2003. Diffusion in biofilms. J Bacteriol 185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198–12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirani ZA, Jamil N. 2011. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microbiol 51:191–195. doi: 10.1002/jobm.201000221. [DOI] [PubMed] [Google Scholar]
- 14.Herbert S, Barry P, Novick RP. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect Immun 69:2996–3003. doi: 10.1128/IAI.69.5.2996-3003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto M. 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 16.Moormeier DE, Bose JL, Horswill AR, Bayles KW. 2014. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341–14. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross M, Cramton SE, Gotz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann C, Hussain M, Peters G, Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 19.Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCourt J, O'Halloran DP, McCarthy H, O'Gara JP, Geoghegan JA. 2014. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett 353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 21.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. 2010. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res 28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 22.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dengler V, Foulston L, DeFrancesco AS, Losick R. 2015. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol 197:3779–3787. doi: 10.1128/JB.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett 259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 29.Foster SJ. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol 177:5723–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjit DK, Endres JL, Bayles KW. 2011. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol 193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadykov MR, Bayles KW. 2012. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol 15:211–215. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 35.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A 109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan JB. 2011. Antibiotic-induced biofilm formation. Int J Artif Organs 34:737–751. doi: 10.5301/ijao.5000027. [DOI] [PubMed] [Google Scholar]
- 39.Dumitrescu O, Badiou C, Bes M, Reverdy ME, Vandenesch F, Etienne J, Lina G. 2008. Effect of antibiotics, alone and in combination, on Panton-Valentine leukocidin production by a Staphylococcus aureus reference strain. Clin Microbiol Infect 14:384–388. doi: 10.1111/j.1469-0691.2007.01947.x. [DOI] [PubMed] [Google Scholar]
- 40.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis 195:202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 41.Schilcher K, Andreoni F, Uchiyama S, Ogawa T, Schuepbach RA, Zinkernagel AS. 2014. Increased neutrophil extracellular trap-mediated Staphylococcus aureus clearance through inhibition of nuclease activity by clindamycin and immunoglobulin. J Infect Dis 210:473–482. doi: 10.1093/infdis/jiu091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlievert PM, Kelly JA. 1984. Clindamycin-induced suppression of toxic-shock syndrome-associated exotoxin production. J Infect Dis 149:471. doi: 10.1093/infdis/149.3.471. [DOI] [PubMed] [Google Scholar]
- 43.Ohlsen K, Ziebuhr W, Koller KP, Hell W, Wichelhaus TA, Hacker J. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 42:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo HS, Chan JL, Cheung GY, Otto M. 2010. Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:4942–4944. doi: 10.1128/AAC.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaki J, Synold T, Wong-Beringer A. 2013. Tigecycline induction of phenol-soluble modulins by invasive methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 57:4562–4565. doi: 10.1128/AAC.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veringa EM, Verhoef J. 1986. Influence of subinhibitory concentrations of clindamycin on opsonophagocytosis of Staphylococcus aureus, a protein A-dependent process. Antimicrob Agents Chemother 30:796–797. doi: 10.1128/AAC.30.5.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mader JT, Adams K, Morrison L. 1989. Comparative evaluation of cefazolin and clindamycin in the treatment of experimental Staphylococcus aureus osteomyelitis in rabbits. Antimicrob Agents Chemother 33:1760–1764. doi: 10.1128/AAC.33.10.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 37:3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morikawa K, Maruyama A, Inose Y, Higashide M, Hayashi H, Ohta T. 2001. Overexpression of sigma factor, σB, urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem Biophys Res Commun 288:385–389. doi: 10.1006/bbrc.2001.5774. [DOI] [PubMed] [Google Scholar]
- 54.Mlynek KD, Callahan MT, Shimkevitch AV, Farmer JT, Endres JL, Marchand M, Bayles KW, Horswill AR, Kaplan JB. 22 April 2016. Effects of low-dose amoxicillin on Staphylococcus aureus USA300 biofilms. Antimicrob Agents Chemother doi: 10.1128/AAC.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen HY, Chen CC, Fang CS, Hsieh YT, Lin MH, Shu JC. 2011. Vancomycin activates σB in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS One 6:e24472. doi: 10.1371/journal.pone.0024472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kullik I, Giachino P, Fuchs T. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol 180:4814–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katzif S, Lee EH, Law AB, Tzeng YL, Shafer WM. 2005. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J Bacteriol 187:8181–8184. doi: 10.1128/JB.187.23.8181-8184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D, Renzoni A, Estoppey T, Bisognano C, Francois P, Kelley WL, Lew DP, Schrenzel J, Vaudaux P. 2005. Induction of fibronectin adhesins in quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin or by sigma B transcription factor activity is mediated by two separate pathways. Antimicrob Agents Chemother 49:916–924. doi: 10.1128/AAC.49.3.916-924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nair SP, Bischoff M, Senn MM, Berger-Bachi B. 2003. The σB regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect Immun 71:4167–4170. doi: 10.1128/IAI.71.7.4167-4170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsompanidou E, Denham EL, Becher D, de Jong A, Buist G, van Oosten M, Manson WL, Back JW, van Dijl JM, Dreisbach A. 2013. Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl Environ Microbiol 79:886–895. doi: 10.1128/AEM.03157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wecke J, Johannsen L, Giesbrecht P. 1990. Reduction of wall degradability of clindamycin-treated staphylococci within macrophages. Infect Immun 58:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakao M, Kitanaka E, Ochiai K, Nakazawa S. 1972. Cell wall synthesis by Staphylococcus aureus in the presence of protein synthesis inhibitory agents. I. J Antibiot (Tokyo) 25:60–63. doi: 10.7164/antibiotics.25.60. [DOI] [PubMed] [Google Scholar]
- 64.Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi J, Komatsuzawa H, Sugai M. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol 58:1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- 65.Heilmann C, Hartleib J, Hussain MS, Peters G. 2005. The multifunctional Staphylococcus aureus autolysin Aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect Immun 73:4793–4802. doi: 10.1128/IAI.73.8.4793-4802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seidl K, Leimer N, Palheiros Marques M, Furrer A, Senn G, Holzmann-Burgel A, Matt U, Zinkernagel AS. 2014. USA300 methicillin-resistant Staphylococcus aureus in Zurich, Switzerland between 2001 and 2013. Int J Med Microbiol 304:1118–1122. doi: 10.1016/j.ijmm.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Frank KL, Reichert EJ, Piper KE, Patel R. 2007. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob Agents Chemother 51:888–895. doi: 10.1128/AAC.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patton TG, Yang SJ, Bayles KW. 2006. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol Microbiol 59:1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x. [DOI] [PubMed] [Google Scholar]
- 70.Grande R, Nistico L, Sambanthamoorthy K, Longwell M, Iannitelli A, Cellini L, Di Stefano A, Hall Stoodley L, Stoodley P. 2014. Temporal expression of agrB, cidA, and alsS in the early development of Staphylococcus aureus UAMS-1 biofilm formation and the structural role of extracellular DNA and carbohydrates. Pathog Dis 70:414–422. doi: 10.1111/2049-632X.12158. [DOI] [PubMed] [Google Scholar]
- 71.Fujimoto DF, Brunskill EW, Bayles KW. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J Bacteriol 182:4822–4828. doi: 10.1128/JB.182.17.4822-4828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herman-Bausier P, El-Kirat-Chatel S, Foster TJ, Geoghegan JA, Dufrene YF. 2015. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. mBio 6:e00413–15. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blickwede M, Wolz C, Valentin-Weigand P, Schwarz S. 2005. Influence of clindamycin on the stability of coa and fnbB transcripts and adherence properties of Staphylococcus aureus Newman. FEMS Microbiol Lett 252:73–78. doi: 10.1016/j.femsle.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 74.Otto M. 2014. Phenol-soluble modulins. Int J Med Microbiol 304:164–169. doi: 10.1016/j.ijmm.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. 2016. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol 99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. 2010. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci U S A 107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.