Abstract
Encephalomyocarditis virus (EMCV), like hepatitis C virus (HCV), requires phosphatidylinositol 4-kinase IIIα (PI4KA) for genome replication. Here, we demonstrate that tyrphostin AG1478, a known epidermal growth factor receptor (EGFR) inhibitor, also inhibits PI4KA activity, both in vitro and in cells. AG1478 impaired replication of EMCV and HCV but not that of an EMCV mutant previously shown to escape PI4KA inhibition. This work uncovers novel cellular and antiviral properties of AG1478, a compound previously regarded only as a cancer chemotherapy agent.
TEXT
Tyrphostin AG1478 [N-(3-chlorophenyl)-6,7-dimethoxy-4-quinazolinamine] is best known as a potent and specific inhibitor of epidermal growth factor receptor (EGFR) signaling. AG1478 inhibits EGFR by blocking its protein tyrosine kinase activity and by promoting the formation of inactive EGFR dimers (1, 2). A previous study demonstrated that AG1478 induces Golgi apparatus dispersal and proposed GBF1 (Golgi-specific brefeldin A resistance factor 1) as the target (3). GBF1 is an essential host factor for genome replication of picornaviruses from the genus Enterovirus, such as poliovirus (PV) and coxsackievirus B3 (CVB3) (4–6). GBF1 inhibitors such as brefeldin A (BFA) or golgicide A (GCA) completely block enterovirus replication (7). However, we demonstrated that AG1478 does not have any effect on enterovirus replication, thus questioning the validity of AG1478 as a bona fide GBF1 inhibitor (7). Notably, AG1478 contains a 4-anilinoquinazoline core, similar to AL-9 (Fig. 1A), an established inhibitor of phosphatidylinositol 4-kinase type III isoform α (PI4KA) (8). PI4KA is one of the four mammalian PI4K isoforms that generate phosphatidylinositol 4-phosphate (PI4P) from PI (9–11). PI4KA activity was shown to be indispensable for the replication of the hepatitis C virus (HCV) of the Flaviviridae family (12–18), and more recently, we demonstrated that PI4KA activity was also essential for encephalomyocarditis virus (EMCV), a picornavirus of the genus Cardiovirus (19). These considerations prompted us to investigate whether AG1478 might be a PI4KA inhibitor.
FIG 1.
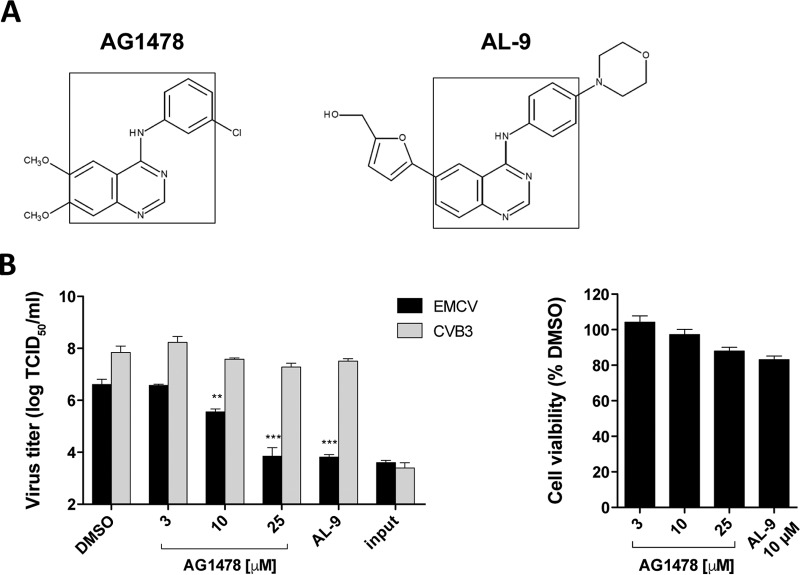
Tyrphostin AG1478 inhibits EMCV replication. (A) Chemical structures of AG1478 and the PI4KA inhibitor AL-9. Outlined is the common 4-anilino-quinazoline core. (B) Sensitivities of EMCV and CVB3 to AG1478 treatment. HeLa cells were infected with for 30 min with EMCV or CVB3 at a multiplicity of infection (MOI) of 1, after which virus-containing medium was replaced with fresh (compound-containing) medium. Cells were freeze-thawed to release intracellular virus particles, and the total virus titers at 0.5 (input) and 8 h postinfection (hpi) were determined by endpoint dilution. The cytotoxicity of the treatment was measured separately in a cell viability assay. Shown are mean values ± standard errors of the means (SEM). Means were statistically compared using the unpaired t test. TCID50, 50% tissue culture infective dose; **, P < 0.01; ***, P < 0.001.
We examined whether AG1478 inhibits EMCV replication in a single-cycle assay. To this end, HeLa cells were infected with either EMCV or CVB3 for 30 min, after which virus-containing medium was replaced with compound-containing medium. Eight hours later, cells were lysed by freeze-thawing to determine the total virus titers by endpoint dilution. As we have previously shown (7), AG1478 did not perturb CVB3 replication (Fig. 1B). However, EMCV was inhibited by AG1478 in a dose-dependent manner, with a complete inhibition at 25 μM. The inhibition observed with AG1478 was comparable to that obtained with 10 μM AL-9 (Fig. 1B). In parallel, a cell viability assay was performed to verify that the antiviral activity of AG1478 was not due to cytotoxic effects (Fig. 1B).
Having established that AG1478 impairs EMCV replication and shares structural similarities with the known PI4KA inhibitor AL-9, we next wondered whether AG1478 targets PI4KA. To address this question, we first investigated if AG1478 can directly inhibit the lipid kinase activity of PI4KA in an in vitro assay. Using a previously described protocol (20), we measured the in vitro activities of immunoprecipitated PI4KA or commercially available purified PI4KA, in the absence or presence of increasing concentrations of AG1478 or the established PI4KA inhibitor “compound A” (21). Both compounds reduced PI4KA activity, although AG1478 was less potent than compound A (Fig. 2A).
FIG 2.
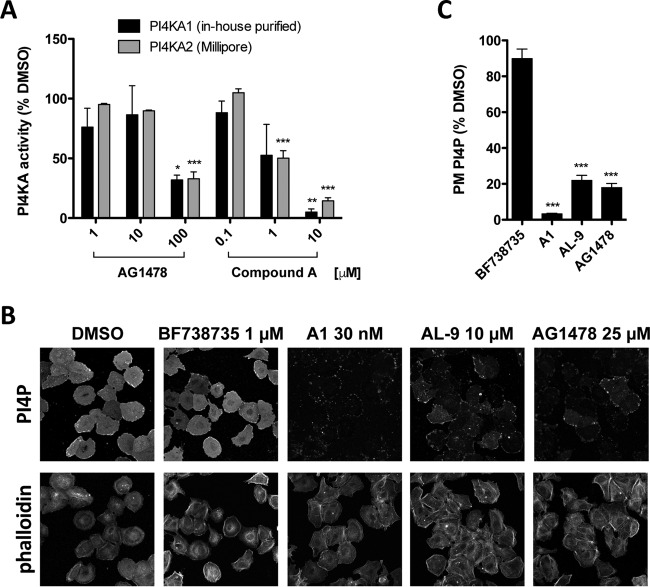
AG1478 targets PI4KA activity. (A) PI4KA lipid kinase in vitro assay. Hemagglutinin (HA)-tagged PI4KA purified by immunoprecipitation as described previously (20) or purified commercially available PI4KA (Merck Millipore, Darmstadt, Germany) was preincubated for 1 h with the indicated concentrations of AG1478 or compound A before the lipid kinase reaction was started as previously described (20). PI4KA activity was measured as incorporation of 32P and is expressed as a percentage of the DMSO control level (100%). Shown are mean values ± SEM. (B) AG1478 inhibits PI4KA-dependent PI4P production in intact cells. Huh7-Lunet/T7 cells were treated for 3 h with the indicated concentrations of AG1478 or PI4KA inhibitors AL-9 and A1 or the PI4KB inhibitor BF738735, included here as a negative control. Following compound treatment, cells were fixed with paraformaldehyde and further subjected to immunofluorescence analysis to visualize plasma membrane PI4P (22). Alexa Fluor 488-phalloidin was used as a counterstain. (C) The intensities of PI4P signals given in panel B were quantified using ImageJ, as previously described (19). Shown are mean values ± SEM for at least 40 cells per condition, expressed as percentages of the DMSO control level (100%). Means were statistically compared using the unpaired t test. ***, P < 0.001.
Our in vitro data suggested that AG1478 is a direct PI4KA inhibitor with lower potency than other PI4KA inhibitors. We next examined whether AG1478 inhibits PI4KA activity in living cells also and how this compares to inhibition by known PI4KA inhibitors. PI4KA is responsible for the production of the plasma membrane (PM) pool of PI4P; thus, monitoring the PM PI4P levels provides a direct and reliable measure of the PI4KA activity in cells. Using a previously established quantitative immunofluorescence staining protocol (19, 22), we measured the intensity of PM PI4P signals in Huh7 cells. Unlike HeLa cells, Huh7 cells are relatively flat and exhibit a continuous sheet-like PI4P pattern covering the entire cell surface, which enables imaging of the entire PM-associated signal for accurate quantification. We compared the effects of AG1478 on PM PI4P with those of AL-9 and “A1” (Fig. 2B, upper panels), the last a recently described very potent PI4KA inhibitor (11) (of note, A1 should not be confused with compound A, as they represent distinct PI4KA inhibitors). Quantification of the PI4P intensity signals (Fig. 2C) revealed that AG1478 treatment led to a significant reduction of PI4P levels (up to 83%) compared to the dimethyl sulfoxide (DMSO) control level, very similar to AL-9 treatment (79% inhibition), while the impact of A1 treatment was even greater (97%). In contrast, treatment with BF738735, an inhibitor of the PI4K type IIIβ isoform (23), responsible for generating PI4P at Golgi membranes (24, 25), did not have a significant effect on PM PI4P levels. Alexa Fluor 488-coupled phalloidin (Invitrogen) was used as a counterstain to facilitate delineation of the cell margins where the PI4P signal was almost completely lost upon treatment (Fig. 2B, lower panels). These results demonstrated that AG1478 indeed impairs PI4KA activity in intact cells.
Recently, we discovered that single point mutations in the viral protein 3A render EMCV replication resistant to PI4KA inhibitors (26). To validate that AG1478 inhibits EMCV by targeting PI4KA, we tested whether the previously identified mutation in EMCV 3A with an A-to-V change at position 32 (3A-A32V) conferring resistance to PI4KA inhibitors AL-9 and A1 would also provide cross-resistance to AG1478. Furthermore, we evaluated if AG1478 blocks EMCV at the step of viral genome replication by wild-type (WT) or 3A-A32V mutant EMCV expressing Renilla luciferase upstream of the capsid coding region (RLuc-EMCV) (26). Determining the Renilla luciferase activity in this assay provides a direct measurement of viral RNA replication. Cells were infected with WT or mutant RLuc-EMCV for 30 min, followed by treatment with AG1478 or the established EMCV replication inhibitor dipyridamole (Dip) (27). Seven hours later, cells were lysed to allow quantification of the intracellular amount of luciferase. Dip treatment completely inhibited the genome replication of both WT and mutant virus. In contrast, AG1478 blocked only the WT virus, not the mutant, which replicated to almost the full extent in the presence of AG1478 (Fig. 3). These data strongly indicated that AG1478 inhibits EMCV genome replication by targeting PI4KA.
FIG 3.
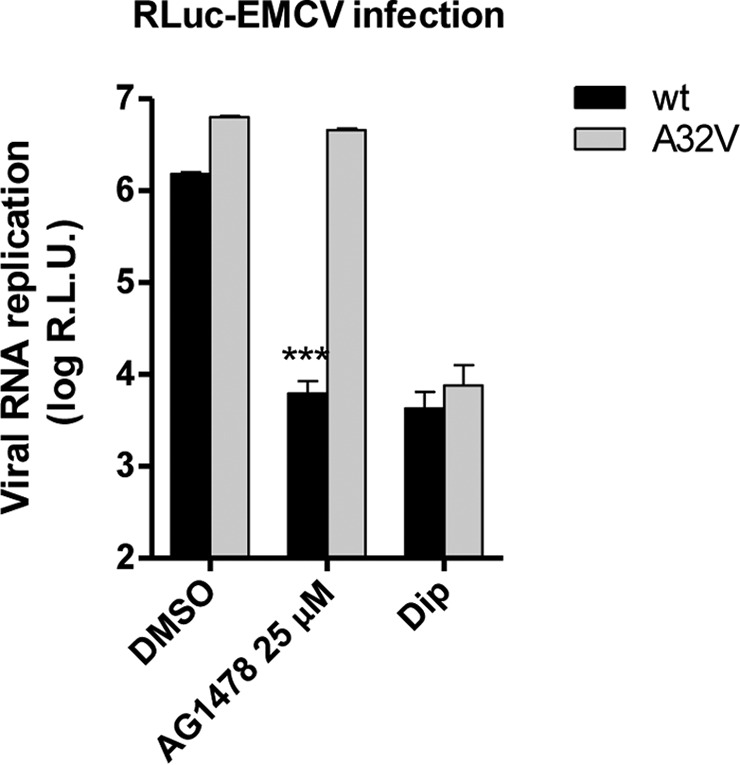
The 3A-A32V EMCV PI4KA escape mutant is cross-resistant to AG1478. HeLa cells were infected with virus at an MOI of 1 for 30 min. After removal of the inoculum, fresh (compound-containing) medium was added to the cells. After 8 h, cells were lysed to determine the intracellular Renilla luciferase activity as a measure of viral RNA replication. Shown are mean values ± SEM. Means were statistically compared using the unpaired t test. RLU, relative light units; ***, P < 0.001.
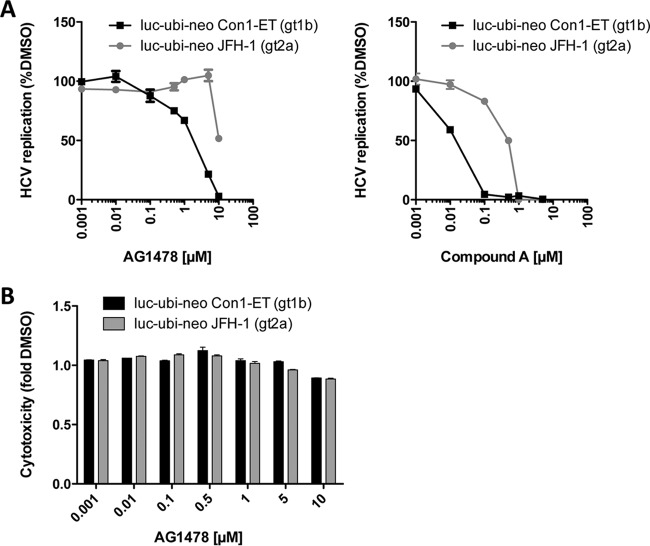
Finally, we tested whether AG1478 also exerts antiviral activity against HCV. To this end, we compared the effects of AG1478 with those of compound A in Huh7 cells stably expressing genotype 1b (gt1b; Con1-ET) or 2a (gt2a; JFH-1) subgenomic HCV replicons (28–30) by measuring viral RNA replication after 3 days of treatment. Similar to compound A treatment, AG1478 treatment had greater effects on gt1b than on gt2a (Fig. 4A). The observed antiviral effects of AG1478 on HCV were not due to cytotoxicity, as demonstrated by a cell viability assay (Fig. 4B). Unfortunately, we were unable to test concentrations of AG1478 higher than 10 μM in this assay, because these were cytotoxic (data not shown), impairing further assessment of the antiviral effects on gt2a. Notably, AG1478 was less potent than compound A in inhibiting HCV, which correlates with the potencies of the two compounds in inhibiting PI4KA activity observed in the in vitro kinase assay (Fig. 2A). These results suggested that AG1478 likely inhibits HCV by targeting PI4KA.
FIG 4.
AG1478 inhibits HCV replication in a genotype-dependent manner. Stable Huh7 cell lines harboring luciferase HCV reporter replicons (luc-ubi-neo) of either genotype Con1-ET (gt1b) or genotype JFH-1 (gt2a) were incubated with the indicated amounts of compound for 72 h, after which firefly luciferase activity was determined as a measure of virus RNA replication (A). Cytotoxic effects of the AG1478 treatment were evaluated separately in a cell viability assay (B). Shown are mean values ± SEM.
Summarizing, we here identify PI4KA as a novel cellular target of tyrphostin AG1478, a compound previously recognized only as an EGFR inhibitor and a Golgi apparatus-dispersing agent. We reveal that AG1478 exerts antiviral properties against EMCV and HCV and demonstrate that its mode of action involves inhibition of PI4KA activity. Our in vitro data suggested that AG1478 is a direct inhibitor of PI4KA; however, we cannot exclude the possibility that AG1478 targets PI4KA activity indirectly or a combination of both. The antiviral properties of AG1478 are most likely not linked to its effects on EGFR signaling, since AG1478 was shown to inhibit EGFR in the low nanomolar range (31), whereas inhibition of virus replication (and PI4KA activity) requires micromolar concentrations. Although it is unlikely that EGFR inhibition accounts for the antiviral activity of AG1478, it would be interesting to investigate in the future whether AL-9 (and other structurally related inhibitors) may also exhibit anti-EGFR properties. In conclusion, our study uncovers important cellular effects and antiviral properties of tyrphostin AG1478, a compound proposed earlier as a promising treatment in cancer chemotherapy.
ACKNOWLEDGMENTS
We are grateful to R. De Francesco for providing AL-9, T. Balla for providing A1, Rob Bleumink for providing phalloidin, and S. Breitfelder for providing compound A.
Funding Statement
This work was supported by research grants from The Netherlands Organization for Scientific Research (NWO-VENI-863.12.005 to H.M.V.D.S., NWO-VENI-722.012.006 to J.R.P.M.S., NWO-ALW-820.02.018 to F.J.M.V.K., and NWO-VICI-91812628 to F.J.M.V.K.), the European Union 7th Framework (EUVIRNA Marie Curie Initial Training Network, grant agreement number 264286, to F.J.M.V.K.), and grants from the Deutsche Forschungsgemeinschaft (LO 1556/4-1 to V.L.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Levitzki A, Gazit A. 1995. Tyrosine kinase inhibition: an approach to drug development. Science 267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 2.Arteaga CL, Ramsey TT, Shawver LK, Guyer CA. 1997. Unliganded epidermal growth factor receptor dimerization induced by direct Interaction of quinazolines with the ATP binding site. J Biol Chem 272:23247–23254. doi: 10.1074/jbc.272.37.23247. [DOI] [PubMed] [Google Scholar]
- 3.Pan H, Yu J, Zhang L, Carpenter A, Zhu H, Li L, Ma D, Yuan J. 2008. A novel small molecule regulator of guanine nucleotide exchange activity of the ADP-ribosylation factor and Golgi membrane trafficking. J Biol Chem 283:31087–31096. doi: 10.1074/jbc.M806592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanke KHW, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJM. 2009. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol 83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorobantu CM, Ford-Siltz LA, Sittig SP, Lanke KHW, Belov GA, van Kuppeveld FJM, van der Schaar HM. 2015. GBF1- and ACBD3-independent recruitment of PI4KIIIβ to replication sites by rhinovirus 3A proteins. J Virol 89:1913–1918. doi: 10.1128/JVI.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Linden L, van der Schaar HM, Lanke KHW, Neyts J, van Kuppeveld FJM. 2010. Differential effects of the putative GBF1 inhibitors golgicide A and AG1478 on enterovirus replication. J Virol 84:7535–7542. doi: 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P, De Francesco R. 2012. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog 8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balla A, Tuymetova G, Tsiomenko A, Várnai P, Balla T. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell 16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, De Camilli P. 2012. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol 199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, Harrison S, Moore C, Balla T. 2014. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem 289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet M-S, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 14.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A 106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai AW, Benita Y, Peng LF, Kim S-S, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, CornellaTaracido I, Baryza J, Tallarico J, Joberty G, Bantscheff M, Schirle M, Bouwmeester T, Mathy JE, Lin K, Compton T, Labow M, Wiedmann B, Gaither LA. 2009. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol 83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset F-L, Gripon P, Le Seyec J. 2009. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J 23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorobantu CM, Albulescu L, Harak C, Feng Q, van Kampen M, Strating JRPM, Gorbalenya AE, Lohmann V, van der Schaar HM, van Kuppeveld FJM. 2015. Modulation of the host lipid landscape to promote RNA virus replication: the picornavirus encephalomyocarditis virus converges on the pathway used by hepatitis C virus. PLoS Pathog 11:e1005185. doi: 10.1371/journal.ppat.1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harak C, Radujkovic D, Taveneau C, Reiss S, Klein R, Bressanelli S, Lohmann V. 2014. Mapping of functional domains of the lipid kinase phosphatidylinositol 4-kinase type III alpha involved in enzymatic activity and hepatitis C virus replication. J Virol 88:9909–9926. doi: 10.1128/JVI.01063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaillancourt FH, Brault M, Pilote L, Uyttersprot N, Gaillard ET, Stoltz JH, Knight BL, Pantages L, McFarland M, Breitfelder S, Chiu TT, Mahrouche L, Faucher A-M, Cartier M, Cordingley MG, Bethell RC, Jiang H, White PW, Kukolj G. 2012. Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target. J Virol 86:11595–11607. doi: 10.1128/JVI.01320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond GRV, Schiavo G, Irvine RF. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem J 422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Schaar HM, Leyssen P, Thibaut HJ, de Palma A, van der Linden L, Lanke KHW, Lacroix C, Verbeken E, Conrath K, Macleod AM, Mitchell DR, Palmer NJ, van de Poël H, Andrews M, Neyts J, van Kuppeveld FJM. 2013. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIβ. Antimicrob Agents Chemother 57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol 1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 25.Boura E, Nencka R. 2015. Phosphatidylinositol 4-kinases: Function, structure, and inhibition. Exp Cell Res 337:136–145. doi: 10.1016/j.yexcr.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Dorobantu CM, Albulescu L, Lyoo H, van Kampen M, De Francesco R, Lohmann V, Harak C, van der Schaar HM, Strating JRPM, Gorbalenya AE, van Kuppeveld FJM. 2016. Mutations in encephalomyocarditis virus 3A protein uncouple the dependency of genome replication on host factors phosphatidylinositol 4-kinase IIIα and oxysterol-binding protein. mSphere 1:e00068-16. doi: 10.1128/mSphere.00068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fata-Hartley CL, Palmenberg AC. 2005. Dipyridamole reversibly inhibits mengovirus RNA replication. J Virol 79:11062–11070. doi: 10.1128/JVI.79.17.11062-11070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jo J, Aichele U, Kersting N, Klein R, Aichele P, Bisse E, Sewell AK, Blum HE, Bartenschlager R, Lohmann V, Thimme R. 2009. Analysis of CD8+ T-cell-mediated inhibition of hepatitis C virus replication using a novel immunological model. Gastroenterology 136:1391–1401. doi: 10.1053/j.gastro.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol 77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J Virol Methods 110:201–209. doi: 10.1016/S0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- 31.Gazit A, Chen J, App H, McMahon G, Hirth P, Chen I, Levitzki A. 1996. Tyrphostins IV—highly potent inhibitors of EGF receptor kinase. Structure-activity relationship study of 4-anilidoquinazolines. Bioorg Med Chem 4:1203–1207. [DOI] [PubMed] [Google Scholar]