Abstract
EDP-239, a potent and selective hepatitis C virus (HCV) nonstructural protein 5A (NS5A) inhibitor developed for the treatment of HCV infection, has been investigated in vitro and in vivo. This study sought to characterize genotypic changes in the HCV NS5A sequence of genotype 1 (GT1) replicons and to compare those changes to GT1 viral RNA mutations isolated from clinical trial patients. Resistance selection experiments in vitro using a subgenomic replicon identified resistance-associated mutations (RAMs) at GT1a NS5A amino acid positions 24, 28, 30, 31, and 93 that confer various degrees of resistance to EDP-239. Key RAMs were similarly identified in GT1b NS5A at amino acid positions 31 and 93. Mutations F36L in GT1a and A92V in GT1b do not confer resistance to EDP-239 individually but were found to enhance the resistance of GT1a K24R and GT1b Y93H. RAMs were identified in GT1 patients at baseline or after dosing with EDP-239 that were similar to those detected in vitro. Baseline RAMs identified at NS5A position 93 in GT1, or positions 28 or 30 in GT1a only, correlated with a reduced treatment response. RAMs at additional positions were also detected and may have contributed to reduced EDP-239 efficacy. The most common GT1a and GT1b RAMs found to persist up to weeks 12, 24, or 48 were those at NS5A positions 28, 30, 31, 58 (GT1a only), and 93. Those RAMs persisting at the highest frequencies up to weeks 24 or 48 were L31M and Q30H/R for GT1a and L31M and Y93H for GT1b. (This study has been registered at ClinicalTrials.gov under identifier NCT01856426.)
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) is a significant global health challenge, with an estimated 170 million individuals infected worldwide (1). Chronic HCV infection can lead to cirrhosis of the liver and hepatocellular carcinoma and contributes to the deaths of 700,000 individuals each year (2).
In recent years, several direct-acting antiviral (DAA) agents that inhibit the NS3 protease, nonstructural 5A (NS5A), or NS5B polymerase proteins of HCV have been approved to treat patients. Combinations of DAAs have been used as a strategy to mitigate the risk for viral resistance selection (3) since clinical trials of compounds targeting any one of these proteins have revealed the rapid emergence of resistant variants (4). Because of this, there continues to be an unmet medical need for additional approved DAA agents to be used in combinations in order to suppress the development of antiviral resistance. In order to address this unmet need, the small-molecule inhibitor EDP-239 was designed to target the NS5A protein of HCV.
NS5A is an HCV protein with no known enzymatic activities and is necessary for the replication and assembly of virions (5). EDP-239 is a novel NS5A inhibitor with in vitro 50% effective concentrations (EC50s) of 34 pM and 4 pM against genotype 1a (GT1a) and GT1b replicons, respectively (6). Inhibition of the NS5A protein of HCV has been clinically validated and leads to rapid decreases in HCV RNA in patients (7, 8). However, NS5A inhibitors are also known to possess a relatively low barrier to resistance, allowing resistant variants to be rapidly selected in monotherapy clinical studies (7, 8). Furthermore, NS5A resistance-associated mutations (RAMs) have been demonstrated to persist for up to 6 months in patients following treatment cessation (9).
Here, we sought to characterize the preclinical in vitro resistance profile of EDP-239 in GT1a and GT1b replicons. Insights gained from the in vitro resistance analyses presented here were used to guide further clinical development of EDP-239. A double-blind, randomized, placebo-controlled, multicenter, dose ranging, proof-of-concept study of a single dose of EDP-239 was initiated. Baseline or treatment-emergent changes in the NS5A genetic sequence were monitored over time using deep-sequencing methods. When possible, preexisting baseline RAMs were correlated with changes in viral RNA concentrations in individual patients in each dosing cohort. In addition, NS5A sequence changes were analyzed in conjunction with phenotypic testing of clinical isolate susceptibility to EDP-239 in vitro. The changes observed in the NS5A amino acid sequences of GT1a and GT1b viruses up to day 14 and their persistence up to weeks 12 to 48 are also reported.
MATERIALS AND METHODS
HCV inhibitors.
EDP-239 was synthesized by the chemistry department at Enanta Pharmaceuticals, and dimethyl sulfoxide (DMSO) was used as the solvent for all in vitro analyses. The clinical supply of EDP-239 was synthesized by the chemistry department at Novartis.
Cell culture and replicons.
Human hepatoma cells (Huh7 Lunet and derivatives) were obtained from ReBLikon GmbH and maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% Glutamax (Life Technologies), 1% penicillin-streptomycin (Life Technologies), and 1% nonessential amino acids solution (MEM NEAA; Life Technologies). Huh7 Lunet cells, described previously (10), were used as host cells for replicon transient transfections. Stably transfected subgenomic replicon cell lines En5-3 (11) (GT1a H77; Stan Lemon, UNC) and Huh SGR11-7 (10, 12) (GT1b Con1; ReBLikon GmbH) were used for in vitro resistance selection in cell culture and were maintained in complete DMEM additionally supplemented with 0.25 and 0.75 mg/ml Geneticin, respectively.
Single point mutations were engineered into GT1a and GT1b replicon clones for use in transient-transfection phenotypic assays. First, the XbaI site was removed from the luc-ubi-neo cassette of the GT1b replicon construct I389luc-ubi-neo/NS3-3′/ET (ReBLikon GmbH) prior to exchanging it with the Htat2ANeo cassette of the Htat2ANeo/QR/VI/KR/KR5A/SI construct (11). The resulting GT1a LucUbiNeo/QR/VI/KR/KR5A/SI plasmid construct and the GT1b I389luc-ubi-neo/NS3-3′/ET constructs were used for site-directed mutagenesis (described below). All sequences were confirmed by Sanger DNA sequencing analysis.
In vitro resistance selection.
Resistance selection experiments were performed by seeding 1.0 × 106 En5-3 (GT1a) or 0.5 × 106 Huh SGR11-7 (GT1b) cells in 175-cm2 cell culture plates and growing them in the presence of G418 (0.25 and 0.75 mg/ml) and EDP-239. Concentrations of EDP-239 at 10-fold, 20-fold, and 50-fold above the EC50 were used for GT1a, and concentrations of 10-fold and 20-fold above the EC50 were used for GT1b. Cells were passaged three times and, on the third passage, were transferred to 150-mm tissue culture dishes. Following the third passage, a majority of replicon cells were cleared of replicon RNA and were unable to survive in the medium containing G418. The cells expressing resistant replicon variants survived and formed colonies. These colonies were picked and expanded in 6-well plates at one colony per well. In order to characterize the sequence changes of each clone, cells on 6-well plates were trypsinized from each well, and half the culture was cryo-stored while the other half was pelleted by centrifugation. Cell pellets were stored at −80°C until all colonies had been successfully expanded and stored. Total RNA was extracted from each cell pellet using an RNAqueous-96 kit (Ambion). The NS5A coding region was amplified from each RNA sample using a two-step reverse transcription (RT)-PCR. A cDNA was generated from each RNA sample using a high-capacity cDNA reverse transcription kit (Thermo Fisher), followed by PCR from the cDNA templates using Pfu polymerase and the following primer pairs: for GT1a, 5′-ACCACCCAATGCAGTGGTTCCTGGCTAAGGGACATCTG-3′ and 5′-GCGTGATCAGTGCGCCTGTCCAGGAATAAGACATT-3′; for GT1b, 5′-ATCGCTGGAGCGGCTGTTG-3′ and 5′-GCTGTCTCCCACGCAGCC-3′. The amplified PCR products were purified using a Qiagen PCR purification kit, sequenced by Sequetech Corporation, and analyzed for mutations using Vector NTI software.
Preclinical transient resistance testing.
Individual amino acid mutations suspected of conferring resistance to EDP-239 (>5% frequency) were created by introducing substitutions into the GT1a (H77, LucUbiNeo/QR/VI/KR/KR5A/SI) and GT1b (Con1, I389luc-ubi-neo/NS3-3′/ET) HCV subgenomic bicistronic replicon vectors using site-directed mutagenesis (Stratagene QuikChange XL II) and standard cloning methods. The presence of each mutation was confirmed by sequence analysis. Prior to in vitro transcription of replicon RNA with a Megascript T7 kit (Ambion), each GT1a and GT1b replicon-encoding plasmid was linearized by digestion with XbaI or both AseI and ScaI, respectively. The HCV subgenomic RNA was transiently transfected via chemical transfection into Huh7 Lunet cells using the DMRIE-C (Invitrogen) reagent as described previously (13), with modifications. Briefly, Huh7 Lunet cells were seeded at 106 cells per 10-cm tissue culture plate in transfection seed medium (DMEM supplemented with 10% charcoal dextran-stripped FBS and 1% MEM NEAA) and incubated overnight at 37°C, in 5% CO2. Replicon RNA (15 μg) was diluted in 6 ml DMEM (no supplements) plus 60 μl DMRIE-C reagent. Each overnight 10-cm cell culture was washed twice in DMEM, and each diluted RNA sample was added to one plate and incubated for 3.5 h at 37°C, in 5% CO2. The plates were subsequently washed twice with DMEM, followed by the addition of 10 ml complete DMEM (described above). Transfected cells were incubated overnight at 37°C, in 5% CO2, and seeded the following day at 5,000 cells/well on white 96-well plates (assay plate) in 100 μl/well assay medium (assay medium is DMEM without phenol red, supplemented with 10% FBS, 1% penicillin-streptomycin, 1% MEM NEAA, and 1% Gluta MAX-1). Assay plates were incubated overnight at 37°C, in 5% CO2. The following day, compounds previously titrated in DMSO were diluted 1,000-fold when added to each assay plate (0.1% DMSO final concentration). Each assay plate was subsequently incubated for 72 h at 37°C, in 5% CO2, and then 100 μl/well BriteLite Plus (PerkinElmer) was added and the luminescence was read with an EnVision Excite multilabel plate reader (PerkinElmer). Inhibition of luciferase expression, a measure of viral replication, was determined, and half-maximal effective concentrations (EC50s) were calculated by fitting the inhibition data to a four-parameter logistical model using XLfit.
Clinical study design.
EDP-239 was investigated in a randomized, double-blinded, placebo-controlled, parallel-cohort, multicentered study (ClinicalTrials registration no. NCT01856426). The study was approved by the institutional review boards in all study centers and conducted in accordance with good clinical practice and ethical principles. Twenty-eight male and female patients with chronic GT1 HCV infection were enrolled. The study was stratified by equal numbers of subjects infected with HCV GT1a and HCV GT1b and randomized into 1 of 5 treatment arms to receive single doses of EDP-239 or placebo. In all patients, a single dose of EDP-239 or placebo was administered (time zero, day 1). Doses of EDP-239 in individual treatment arms were as follows: 10, 30, 100, and 200 mg. Each EDP-239 treatment cohort had 6 patients, with two patients from each cohort receiving placebo. The HCV genotype of one patient (patient 5407) was incorrectly identified as GT1b prior to receipt of the first dose, but subsequent sequencing analysis and confirmatory genotyping near the end of the study indicated that the patient was infected with GT1a HCV. Plasma samples for determining HCV RNA levels were drawn at screening, prior to the first dose on day 1 (baseline), at 0.5, 1, 1.5, 2, 3, 4, 5, 6, 8, 12, 24, 48, 72, 96, 120, 216, and 312 h after the initial dose, and on weeks 12, 24, and 48. HCV RNA levels were measured by Covance, Inc., using the Roche COBAS TaqMan HighPure HCV v2.0 assay, with a lower limit of quantification of 25 IU/ml.
Deep sequencing of clinical specimens.
Deep sequencing of the HCV NS5A coding region was performed on the Illumina MiSeq platform by WuXi AppTec, Inc., for patient plasma samples if the level of HCV RNA in plasma was ≥1,000 IU/ml. Sequencing was conducted on viral RNA extracted from plasma samples drawn prior to treatment on day 1 (time zero, baseline), at 1, 2, 4, 8, 12, 24, 48, 72, 96, 120, 216, and 312 h after treatment, and on weeks 12, 24, and 48. NS5A amino acid changes from the reference were determined for baseline samples, and changes from baseline were determined for samples collected postdosing. RNA was purified from patient plasma samples using Qiagen QIAmp viral RNA minikits, according to the manufacturer's directions. The full-length region encoding NS5A was amplified by RT-PCR using nested primer pairs for GT1a (5′-CGAGGACTGCTCTACGCCATGCTCCGGYTCCTGGCTAAGGGAC-3′ and 5′-CCTGTCCATGTGTACGACATCGAGCAGCACACGACATCYTCC-3′) or GT1b (5′-CACTCAGCTGCTTAAGMGGCTHCAYCAGTGGAT-3′ and 5′-AGCCTCCTCGCTTAAGGTAGACCAAGAC-3′) and either Phusion polymerase (New England BioLabs) or Platinum Taq high-fidelity DNA polymerase (Invitrogen). Patient cDNA was subsequently deep sequenced using MiSeq reagent kits (Illumina) and a MiSeq platform (PE150). Statistical summaries and analyses of viral nucleotide deep-sequencing raw data for all GT1a patients (except patient 5407) and two GT1b patients (patients 5401 and 5402) were performed by Novartis using proprietary software. All GT1b analyses (except those for patients 5401 and 5402) and GT1a analyses for patient 5407 were conducted by WuXi AppTec, Inc., using Python scripting language and proprietary algorithms.
Phenotypic analysis of clinical specimens.
Chimeric GT1b (Con1) replicons expressing GT1a and GT1b NS5A proteins from patient viral isolates were cloned from clinical samples at baseline and nadir viral load postdosing time points. All GT1a samples (except that from misidentified patient 5407) were cloned and tested by Novartis, and all GT1b samples (including that from GT1a patient 5407) were cloned and tested by WuXi AppTec. These chimeric replicons consisted of pools of amplified NS5A sequences from the GT1a and GT1b HCV quasispecies present in each patient's plasma sample in a GT1b (Con1) backbone and were assessed for their susceptibility to EDP-239. Nadir viral load sequences for patients 5413 and 5414 could not be amplified due to low viral RNA levels, but amplification was successful at day 14 and h 72 time points, respectively.
The entire NS5A coding region was amplified from the patient-derived NS5A amplicons described above. Following PCR, NS5A amplicons were gel purified using the Qiagen MinElute gel extraction kit according to the manufacturer's instructions. Purified GT1a amplicons were cloned by Novartis into a prepared GT1b (Con1) NS5A deletion replicon shuttle vector with bicistronic expression of Renilla luciferase using In-Fusion cloning (Clontech), according to the manufacturer's instruction. GT1b NS5A amplicons (and one GT1a) were cloned by WuXi AppTec into a similar GT1b (Con1) bicistronic replicon shuttle vector using a ClonExpress one-step cloning kit (Vazyme).
Huh7.5 cells were transfected with in vitro-transcribed HCV replicon RNA and subsequently seeded in 96-well plates. To determine the EC50 of EDP-239 in the transfected chimeric replicons, compounds were serially diluted 1:3 in DMSO and further diluted 1:100 in complete cell culture medium for all GT1a samples (except that for patient 5407). WuXi AppTec serially diluted compounds 1:5 in DMSO and transferred compounds to assay plates at a 1:200 dilution in complete cell culture medium. These compound dilutions were added to cells 24 h posttransfection, resulting in a final DMSO concentration of 0.5% or 0.1% in all wells. Two top concentrations of EDP-239 were used for GT1a testing (2 μM and 2 nM), while a top concentration of 5 nM was used for GT1b testing. Cells were incubated for 72 h prior to assaying for Renilla luciferase signal on an EnVision or PHERAstar FS multimode plate reader. Percent inhibition of HCV replication was calculated in comparison to untreated cells.
RESULTS
In vitro resistance selection of NS5A mutations and analyses.
As with any hepatitis C viral protein target, the development of drug resistance is a major concern. A natural consequence of the high replication rate and low-fidelity polymerase of HCV is that resistant variant viruses with decreased susceptibility to antiviral compounds preexist and can be selected during therapy. Therefore, studies were conducted to identify resistance mutations selected when replicon cells were grown in the presence of EDP-239 and G418 (0.25 mg/ml) for approximately 3 weeks. Only those cells expressing replicating resistant replicons survived to grow into a colony. In this study, 84 GT1a colonies and 64 GT1b colonies were picked and expanded for genotypic characterization.
Resistant mutations selected by NS5A inhibitors have been reported to localize to the first 100 amino acids of NS5A (13). A similar observation was made in this study, and the predominant changes noted in both GT1a and GT1b NS5A after selection with EDP-239 are reported in Table 1. The data also suggest that the pattern of mutations differed between the GT1a and GT1b genetic backgrounds. Subsequent phenotypic analyses of these mutations determined that GT1a NS5A changes conferring resistance to EDP-239 were identified at positions 24, 28, 30, 31, and 93 and a combination of positions 24 and 36. The greatest resistance to EDP-239 (>100-fold) was conferred by M28T (334-fold), Q30R (538-fold), Y93C (478-fold), Y93H (16,140-fold), and Y93N (32,078-fold) in comparison to the wild type. Of these changes, all except Q30R conferred a decrease in replication fitness. Additionally, K24G/R, M28V, Q30H, and L31M conferred less resistance (3- to 79-fold) to EDP-239, with a modest impact on replication fitness for M28V (93%) and Q30H (69%). While the greatest resistance to EDP-239 was observed for RAMs at position 93, they were only identified among GT1a clones at frequencies of <7% and were associated with very low replication efficiencies (≤6%).
TABLE 1.
RAM frequencies in vitro and potency of EDP-239 against NS5A mutations in transient GT1 replicons
| HCV genotype | Wild type or mutant replicona | No. (%) of clones with substitutionb | EDP-239 EC50 (pM) (mean ± SD)c | Fold resistanced | Replication efficiency (%)e |
|---|---|---|---|---|---|
| 1a (H77c) | Wild type | 3.6 ± 0.4 | 100 | ||
| K24G | 21 (25) | 177.4 ± 41.7 | 50 | 179 | |
| K24R | 34 (41) | 10.8 ± 2.0 | 3 | 135 | |
| K24T | 5 (6.0) | 5.1 ± 1.8 | 1 | 121 | |
| M28T | 10 (12) | 1,191 ± 222 | 331 | 26 | |
| M28V | 6 (7.1) | 24.0 ± 3.3 | 7 | 93 | |
| Q30R | 19 (23) | 1,938 ± 522 | 538 | 106 | |
| Q30H | 13 (15) | 137.3 ± 30.6 | 38 | 69 | |
| Q30P | 2 (2.4) | 4.9 ± 2.8 | 1 | 183 | |
| L31M | 3 (3.6) | 284 ± 65 | 79 | 114 | |
| F36L | 58 (69) | 4.4 ± 1.2 | 1 | 173 | |
| Y93C | 2 (2.4) | 1,908 ± 600 | 530 | 6 | |
| Y93H | 6 (7.1) | 58,103 ± 25,595 | 16,140 | 1 | |
| Y93N | 2 (2.4) | 115,479 ± 40,071 | 32,078 | 2 | |
| K24R F36L | 2 (2.4) | 116.6 ± 48.7 | 32 | 21 | |
| 1b (Con1) | Wild type | 2.7 ± 1.0 | 1 | 100 | |
| L31M | 3 (4.7) | 5.6 ± 0.5 | 2 | 240 | |
| L31V | 22 (34) | 54.6 ± 9.0 | 20 | 338 | |
| A92V | 14 (22) | 3.2 ± 0.7 | 1 | 200 | |
| Y93H | 42 (66) | 58.5 ± 7.1 | 22 | 33 | |
| A92V Y93H | 10 (16) | 1,917 ± 232 | 710 | 40 | |
| L31V Y93H | 1 (1.6) | 172,012 ± 16,253 | 63,708 | 48 | |
| L31V A92V Y93H | 1 (1.6) | 476,379 ± 61,883 | 176,437 | 14 |
NS5A mutations selected by EDP-239 and cloned into transiently transfected GT1 replicons.
Number of clones and frequency at which mutations were identified out of the total number of colonies sequenced for genotypes 1a (84 total) and 1b (64 total).
Averages for at least 6 biological replicates.
Calculated from the ratio of the EC50s for the mutant to that of the wild-type replicon.
Calculated as a percentage of mutant replicon replication relative to replication of the wild-type replicon.
In GT1b in vitro selection experiments, the predominant mutations noted were L31V, A92V, Y93H, and A92V Y93H. Of these, the L31V mutation was 3-fold more fit than wild-type GT1b while conferring 20-fold resistance to EDP-239. The Y93H mutation conferred a similar fold loss in potency to EDP-239 but was 33% as fit as the wild type. Only GT1b replicons with changes at NS5A amino acid position 93 or multiple positions experienced a decrease in replication fitness (14% to 48%).
GT1a F36L and GT1b A92V do not individually confer resistance to EDP-239 in replicons carrying these mutations. However, each enhances the resistance conferred by GT1a K24R or GT1b Y93H, respectively, when coexpressed. In each example, resistance to EDP-239 was greater for the double mutant than for the K24R (32- versus 3-fold) or Y93H (710- versus 22-fold) single mutants (Table 1).
Additional combinations of mutations were tested in the GT1b replicon in order to assess fold resistance. Only 1 clone each was identified with either a double mutation of L31V Y93H or a triple mutation of L31V A92V Y93H. While these RAMs, when coexpressed, are highly resistant to EDP-239, it is likely that their poor fitness and the lower probability of selecting multiple RAMs within the same protein contributed to the selection of so few clones. Additional changes were noted throughout the NS5A proteins of both GT1a and GT1b, but subsequent testing demonstrated that these changes do not confer resistance to EDP-239 (data not shown). It is possible that the additional changes offered some improvement to replicon fitness.
Characterization of baseline clinical isolate NS5A sequences.
We sought to determine whether the in vitro resistance profile of EDP-239 correlates with mutations identified in viral populations isolated from clinical trial patients. We conducted a placebo-controlled, proof-of-concept study in four cohorts comprised of 6 GT1 HCV-infected patients each. Administration of a single dose of EDP-239 resulted in rapid reductions in GT1a and GT1b HCV RNA levels in patients receiving 10, 30, 100, and 200 mg (14). Deep sequencing of the NS5A gene from GT1a and GT1b clinical viral isolates at baseline discovered many known NS5A RAMs similar to those reported here or elsewhere (15). NS5A amino acid mutations at a frequency of ≥1% were analyzed from all patient plasma samples with HCV RNA levels of >1,000 IU/ml. Prior to patients' receiving a dose of EDP-239 at baseline, preexisting resistance-associated mutations were detected at NS5A amino acid positions 24, 28, 30, 36, 58, 62, or 93 in 9 of 13 (69%) GT1a patients and at positions 30, 58, 62, or 93 in 5 of 11 (45%) GT1b patients (Table 2). The most prevalent preexisting resistance-associated mutation identified at baseline in GT1a patients was at position 28 in proportions ranging from 2% to 43% of each patient's viral population; however, in GT1b patients, baseline RAMs were identified in fewer numbers of patients but, when present, were typically found at higher frequencies among the viral population.
TABLE 2.
NS5A RAMs detected at baseline in GT1 patients by deep sequencing
| HCV genotype | Dose (mg) | Patient | Baseline RAM | RAM frequency (%) | Maximum RNA reduction (log10 IU/ml) |
|
|---|---|---|---|---|---|---|
| Patient | Cohort mean | |||||
| 1a | 10 | 5306 | M28V | 2 | 2.91 | 2.08 |
| 10 | 5310 | K24R | 2 | 2.54 | ||
| M28V | 2 | |||||
| 10 | 5314 | K24R | 4 | 1.21 | ||
| Q30H | 44 | |||||
| 10 | 5407a | M28V | 23 | 1.68 | ||
| H58R | 2 | |||||
| Y93F | 28 | |||||
| 30 | 5313 | Q30R | 4 | 1.42 | 2.32 | |
| 100 | 5308 | M28V | 2 | 2.74 | 3.58 | |
| 200 | 5304 | M28V | 29 | 3.24 | 3.39b | |
| 200 | 5305 | K24N | 3 | 0.56 | ||
| K24R | 3 | |||||
| K24S | 1 | |||||
| M28T | 90 | |||||
| Q30H | 56 | |||||
| Q30L | 2 | |||||
| Q30R | 13 | |||||
| F36L | 99 | |||||
| H58D | 1 | |||||
| H58R | 1 | |||||
| 200 | 5309 | M28V | 43 | 3.58 | ||
| E62V | 25 | |||||
| 1b | 10 | 5404 | Y93H | 4 | 4.14 | 3.90 |
| 30 | 5414 | Y93H | 99 | 2.79 | 3.50 | |
| 100 | 5402 | R30Q | 76 | 4.37 | 4.36 | |
| 200 | 5403 | P58S | 99 | 4.02 | 3.80 | |
| 200 | 5411 | R30Q | 100 | 2.99 | ||
| Q62R | 100 | |||||
Incorrectly identified as infected with GT1b HCV at screening.
Results for patient 5305 excluded from mean calculation due to viral RNA level of <105 IU/ml at baseline.
Preexisting mutations detected at baseline may help to explain the virologic response in some patients following treatment. Table 2 lists individual patients with RAMs detected at baseline and the viral RNA reduction for each patient compared to the dose cohort maximum mean reduction. It is particularly noteworthy that one patient (patient 5305) passed the inclusion criteria for this study at screening with a plasma viral RNA concentration of 4.4 × 105 IU/ml. However, prior to receiving a single dose of EDP-239 on day 1, this patient's viral load at baseline had decreased by more than 15-fold to 2.5 × 104 IU/ml, below the limit imposed for inclusion (data not shown). In addition to the low plasma viral RNA levels, patient 5305 also possessed several NS5A RAMs at baseline (Table 2). Most notably, the RAMs identified with the highest frequencies in patient 5305 were M28T (90%) and Q30H/L/R (71%). Given that the baseline viral load of this patient was effectively below the inclusion limit, and that NS5A RAMs were present at unusually high percentages even prior to the commencement of EDP-239 treatment, the results for this patient were excluded from all mean calculations pertinent to the GT1a 200-mg dose cohort. However, the presence of baseline RAMs at high proportions in this patient also presented an opportunity to assess their impact on EDP-239 efficacy.
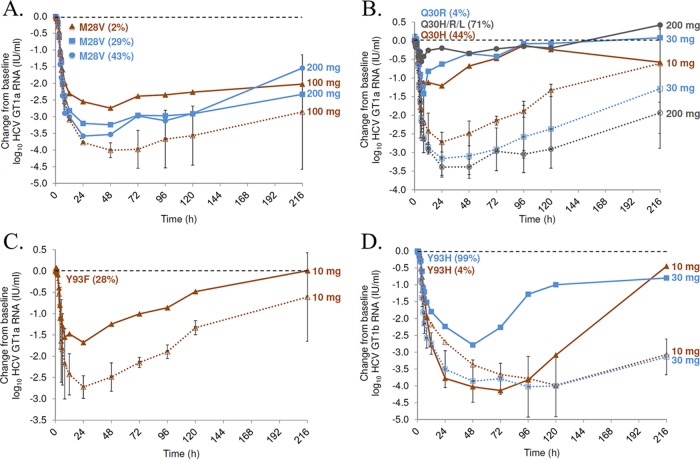
M28, Q30, and Y93 baseline RAMs were found to correlate with reduced potency of EDP-239 in GT1a patients (Fig. 1; Table 2), a finding that is in direct agreement with the in vitro data reported here. In three GT1a patients receiving either 100 or 200 mg EDP-239, M28V RAMs were detected at baseline (Fig. 1A). The maximum viral load reduction for the patient receiving 100 mg was 2.7 log10 compared to the mean reduction of 4.0 log10 IU/ml for the two patients receiving 100 mg without baseline Q30 or Y93 RAMs. In patients receiving 200 mg EDP-239, M28V mutations were also detected in two remaining patients, excluding one patient with Q30 or Y93 RAMs. The presence of the M28V mutation at baseline likely contributed to the reduced potency of 200 mg EDP-239 compared to that of the 100-mg dose. The existence of an E62V mutation in addition to M28V in patient 5309 did not appear to confer a greater loss of sensitivity to EDP-239 (3.58 log10 IU/ml reduction) than that for patient 5304, who possessed only a single M28V mutation (3.24 log10 IU/ml reduction).
FIG 1.
HCV RNA IU/ml log10 change from baseline plotted over time (hours) for patients with baseline Q30, Y93, or M28 RAMs. Results for individual patients with baseline RAMs are represented by solid lines, and those for patients without Y93 or Q30 (GT1a only) RAMs are averaged by dose and represented by dotted lines. All dose cohort averages represent an n of 2, except the 10-mg dose in panel D, which represents a single patient. Error bars represent the standard deviation for each time point and dose. Results for GT1a patients are illustrated in panels A, B, and C, and results for GT1b patients are illustrated in panel D. The mutation and its percent frequency at baseline is indicated for each patient. (A) Impact of baseline M28V RAMs on EDP-239 potency. The viral population of the patient with the M28V mutation (43%) also expressed an E62V mutation (25%). (B) The patient with Q30H/R/L RAM (71%) also expressed K24N/R/S (7%), M28T (90%), and F36L (99%) mutations; the patient receiving a 10-mg dose also expressed a K24R mutation (4%); both patients in the 200-mg dose cohort without Q30 or Y93 RAMs expressed the M28V mutation (>29%). (C) The patient receiving a 10-mg dose and with a Y93F RAM also expressed an M28V mutation (23%), as did both patients receiving a 10-mg dose without baseline Q30 or Y93 RAMs. (D) Effect of baseline Y93 mutations on GT1b viral RNA reduction.
Three GT1a HCV-infected patients possessed a substitution at position 30, with a viral population frequency as low as 4% and up to 71%, alone or in combination with other RAMs (Table 2; Fig. 1B). The maximum viral RNA reduction in these patients was 0.6 to 1.4 log10 IU/ml, whereas GT1a patients without preexisting Q30 or Y93 RAMs at baseline experienced maximum mean RNA reductions of 2.7 to 3.4 log10 IU/ml, depending on the dose.
One GT1a patient was identified with a preexisting Y93F mutation. A 1.7 log10 IU/ml maximum viral RNA reduction was observed in this patient, compared to the maximum mean reduction of 2.7 log10 IU/ml in patients without Q30 or Y93 RAMs in the 10-mg cohort (Fig. 1C).
Only two GT1b patients possessed a preexisting Y93H resistance-associated mutation at baseline (Fig. 1D). The presence of this mutation in only 4% of the viral population in one patient did not appear to reduce EDP-239 efficacy, considering the maximum RNA reduction of 4.1 log10 IU/ml compared to the mean reduction of 3.9 log10 IU/ml for the 10-mg dose cohort. The Y93H mutation was present at a much higher proportion of 99% of the viral population in patient 5414 and appears to have reduced EDP-239 efficacy, given the maximum RNA reduction of only 2.8 log10 IU/ml compared to the reduction of 3.5 log10 IU/ml for the 30-mg dose cohort. Viral RNA concentrations rebounded faster in these two patients than in any other patients without preexisting Y93 RAMs in either the 10-mg or 30-mg cohorts.
The viral RNA reductions in the remaining GT1b patients with single baseline R30Q or P58S RAMs appeared unaffected by the presence of these mutations; however, patient 5411 possessed an R30Q Q62R double mutant that may have resulted in the less robust viral decay (by 0.8 log10) than the 200-mg cohort mean (Table 2). It is also apparent that preexisting RAMs were more common in the GT1a patient population than in the GT1b population.
Numbers of GT1 patients with RAMs at nadir viral RNA concentrations.
In those patients receiving EDP-239, complex mixtures of RAMs were detected and the composition of those mixtures changed over time. These changes likely reflect the outgrowth of individual RAMs and their relative fitness once the selective pressure of EDP-239 was removed. GT1a NS5A resistance-associated mutations at positions 24, 28, Q30H, 31, 58, 92, and Y93C/F/H were detected in a greater proportion of patients receiving 10 mg or 30 mg EDP-239 than in patients receiving 100-mg or 200-mg doses (Table 3). The Q30E RAM was identified in all 3 patients receiving a 200-mg dose of EDP-239 but in only one patient receiving 30 mg. These results suggest that the Q30E RAM may confer a higher degree of resistance to EDP-239 than other mutations, but its replication efficiency may not be fit enough to enable its outgrowth at lower doses. RAMs identified in the highest frequencies of patients at nadir viral loads were Q30R, Y93C/H, and M28T/V. Q30R mutations were detected in equal numbers of GT1a patients across all dosing cohorts, suggesting that this specific RAM confers appreciable resistance to EDP-239 without adversely impacting viral replication efficiency, as observed in vitro. Y93 mutations were the next most prevalent RAMs identified across all dosing cohorts, followed by M28T/V mutations. These data support the findings from the in vitro studies identifying these RAMs as the primary GT1a mutations that confer the greatest resistance to EDP-239.
TABLE 3.
Numbers of patients with NS5A RAMs detected by deep sequencing across dosing cohorts at nadir viral RNA level
| Genotype | Mutation | No. of GT1a or GT1b patients receiving indicated dose of EDP-239 with NS5A RAMs at nadir |
|||
|---|---|---|---|---|---|
| 10 mg | 30 mg | 100 mg | 200 mg | ||
| 1a | K24R | 2 | 1 | ||
| K24N | 1 | ||||
| M28A | 1 | ||||
| M28T | 2 | 3 | 2 | 1 | |
| M28V | 3 | 1 | 1 | 2 | |
| Q30H | 4 | 3 | 1 | ||
| Q30E | 1 | 3 | |||
| Q30R | 3 | 3 | 3 | 3 | |
| L31M | 3 | ||||
| L31V | 1 | ||||
| F36L | 1 | 1 | |||
| H58D | 3 | 2 | 1 | ||
| H58L | 1 | ||||
| H58C | 1 | ||||
| A92T | 2 | ||||
| Y93C | 2 | 2 | 1 | 1 | |
| Y93F | 1 | ||||
| Y93H | 2 | 2 | 1 | 2 | |
| Y93N | 1 | 1 | |||
| Y93S | 1 | ||||
| 1b | L28M | 1 | |||
| R30Q | 1 | 1 | 1 | ||
| R30H | 1 | ||||
| L31F | 1 | ||||
| L31V | 2 | ||||
| L31M | 1 | ||||
| P58S | 1 | 1 | |||
| Q62P | 1 | ||||
| Q62R | 1 | ||||
| A92K | 1 | ||||
| Y93C | 1 | 1 | 1 | ||
| Y93H | 1 | 2 | 1 | ||
| Y93N | 1 | ||||
The most common resistance-associated mutations found in GT1b patients at nadir viral load included changes at NS5A positions 30, 31, and 93 (Table 3). L31 mutations were detected in 3 of 5 patients receiving 10 mg or 30 mg of EDP-239 but not in patients receiving 100 or 200 mg. The R30 and Y93 RAMs were identified in patients receiving EDP-239 doses of 30 mg or higher, suggesting a higher level of resistance imparted by these mutations. Y93 RAMs were the most prevalent, detected in 6 of 11 patients at nadir viral load across all dosing cohorts but more often in patients receiving 30 mg or higher. These data are consistent with in vitro GT1b replicon selection experiments identifying mutations at position Y93 as EDP-239 RAMs.
Genotypic analysis of clinical viral isolates at nadir viral load.
Known resistance-associated mutations were found at nadir viral load in 13 of 13 (100%) GT1a patients and 7 of 11 (64%) GT1b patients (Tables 4 and 5). RAMs at positions 28, 30, and 93 were the predominant treatment-emergent mutations detected in GT1a patients receiving EDP-239 (Table 4). Of the four GT1a patients with L31 mutations, ≤8% at nadir, none preexisted at baseline, but expansion was limited over time, suggesting poor replication fitness for viral variants with these mutations. F36L, an enhancer of K24 mutation resistance in vitro, was identified in the viral populations of only two patients without coincident frequency expansion of K24 RAMs, suggesting that K24R/N and F36L contribute little to EDP-239 resistance in vivo. While not previously selected in vitro in GT1a replicons, H58 RAMs were found in all patients except one, but at frequencies of less than 10% of the viral population, suggesting that this position may not contribute as much to EDP-239 resistance as mutations at other NS5A positions. Three GT1a patients were identified with preexisting Q30 RAMs that were predominant at nadir viral RNA levels (Table 4). In one of these three patients, the baseline frequency of Q30R was only at 4% of the viral population; however, substitutions at Q30 rapidly expanded to a frequency of 98% within 48 h after patients received a dose of EDP-239, consistent with the in vitro data demonstrating robust replication efficiency and resistance for the Q30R mutation. In general, Y93 RAMs at nadir viral load made up less than 25% of each patient's viral population, with one exception (45% in patient 5304), suggesting that the fitness of these mutations may not be as robust as that of Q30 RAMs. A mutation at position 92, demonstrated to enhance GT1b Y93 resistance in vitro, was identified at very low percentages (≤3%) in two GT1a patients receiving 10 mg EDP-239 and did not increase in frequency over time.
TABLE 4.
Genotypic and phenotypic analysis of GT1a patient viral isolatesa
| Dose (mg) | GT or patient | Nadir (h) | NS5A RAM(s) at baseline, first 100 aa (%) | NS5A RAM(s) at nadir, first 100 aa (%) | EDP-239 EC50 (nM)c |
||
|---|---|---|---|---|---|---|---|
| Baseline | Nadir | Fold change | |||||
| GT1a (H77) | Reference | 0.006 | |||||
| 10 | 5306 | 24 | M28V(2) | K24R(2), M/V28T(40)/V(3), Q30H(2)/R(30), L31M(7), F36L(49), H58D(3)/L(2), A92T(1), Y93C(2)/H(8) | 0.005 | 17.0 | 3,400 |
| 5310 | 24 | K24R(2), M28V(2) | K24R(2), M/V28T(7)/V(5), Q30H(5)/R(45), L31M(8), H58D(2), A92T(3), Y93C(7)/H(12)/N(1) | 0.006 | 0.29 | 48 | |
| 5314 | 24 | K24R(4), Q30H(44) | Q30H(99) | 0.004 | 7.4 | 1,850 | |
| 5407 | 24 | M28V(23), H58R(2), Y93F(28) | M28V(72), Q30H(4)/R(80), L31M(2), H/R58C(19)/D(8), Y93F(13) | 0.002 | >5 | >2,500 | |
| 30 | 5303b | 48 | M28T(17)/V(5), Q30E(2)/H(3)/R(57), H58D(6), Y93C(4)/H(3) | 0.010 | 0.043 | 4 | |
| 5307 | 48 | M28T(22), Q30H(7)/R(31), L31V(7), H58D(7), Y93C(7)/H(14) | 0.010 | 1.7 | 170 | ||
| 5313 | 24 | Q30R(4) | M28T(6), Q/R30H(9)/R(87) | 0.004 | 0.40 | 100 | |
| 100 | 5301 | 48 | Q30R(94), Y93H(6) | 0.007 | 5.6 | 800 | |
| 5308 | 48 | M28V(2) | M/V28T(12)/V(10), Q30R(64), Y93C(25) | 0.006 | 4.7 | 783 | |
| 5311 | 24 | M28T(9), Q30R(46) | 0.011 | 0.009 | 1 | ||
| 200 | 5304 | 48 | M28V(29) | M/V28A(2)/V(64), Q30E(4)/R(46), Y93C(8)/H(34)/S(3) | 0.008 | 76.2 | 9,525 |
| 5305 | 24 | K24N(3)/R(3)/S (1), M28T(90), Q30H(56)/L(2), Q30R(13), F36L(99), H58D(1)/R(1) | K24N(2), F36L(99), M28T(94), Q/H30H(78)/E(4)/R(16) | 776.3 | 222.4 | 0.3 | |
| 5309 | 24 | M28V(43), E62V(25) | M28V(63), Q30E(3)/R(58), H58D(7), Y93H(7)/N(10) | 0.008 | 5.5 | 688 | |
aa, amino acids.
Sequencing of baseline sample failed; 2-h time point used for comparison.
n = 3 independent experiments.
TABLE 5.
Genotypic and phenotypic analysis of GT1b patient viral isolatesa
| Dose (mg) | GT or patient | Nadir (h) | NS5A RAM(s) at baseline, first 100 aa (%) | NS5A RAM(s) at nadir, first 100 aa (%) | EDP-239 EC50 (nM)c |
||
|---|---|---|---|---|---|---|---|
| Baseline | Nadir | Fold change | |||||
| GT1b (Con1) | Reference | Reference | 0.003 | ||||
| 10 | 5404 | 72 | Y93H(4) | L28M(32), L31F(22), P58S(46), Y93H(98) | 0.002 | 0.454 | 272 |
| 5409 | 24 | 0.002 | 0.002 | 1 | |||
| 30 | 5405 | 12 | 0.004 | 0.004 | 1 | ||
| 5413b | 312 | L31M(34)/V(27), P58S(1), A92K(2), Y93C(6)/H(33)/N(2) | 0.003 | 0.071 | 25 | ||
| 5414b | 72 | Y93H(99) | R30Q(7), L31V(7), Y93H(99) | 1.2 | 3.2 | 3 | |
| 100 | 5402 | 24 | R30Q(76) | R30Q(17), Q62P(51), Y93C(1) | 0.004 | 0.003 | 1 |
| 5406 | 48 | 0.002 | 0.002 | 1 | |||
| 5410 | 12 | 0.002 | 0.002 | 1 | |||
| 200 | 5403 | 96 | P58S(99) | R30H(53), Y93H(99) | 0.002 | 1.8 | 809 |
| 5408 | 24 | Y93C(8) | 0.002 | 0.003 | 1 | ||
| 5411 | 12 | R30Q(100), Q62R(100) | R30Q(99), Q62R (99) | 0.001 | 0.001 | 1 | |
aa, amino acids.
The viral RNA levels at nadir viral loads for patients 5413 and 5414 were below 1,000 IU/ml, and the sequence could not be amplified; 72 h was selected for patient 5414 and day 14 for patient 5413.
n = 2 biological replicates.
In GT1b patients, nadir viral loads included fewer NS5A sequence changes. The predominant treatment-emergent changes noted were at amino acid positions 28, 30, 31, 58, 62, and 93 (Table 5). The Y93H mutation was identified in patient viral populations at some of the highest frequencies across all dosing cohorts, indicating that it is the primary GT1b resistance mutation selected by EDP-239 in vivo. R30 substitutions were found in patients receiving EDP-239 doses of 30 mg or higher, but these mutations were found at high percentages only in those patients receiving 200 mg of study drug (Table 5).
Phenotypic analysis of clinical viral isolates at nadir.
In order to assess the impact of the NS5A RAMs identified at nadir viral load on EDP-239 susceptibility, phenotypic assays were conducted for all baseline and nadir viral load samples. Pools of full-length NS5A amplicons were amplified from baseline and nadir plasma samples (HCV RNA ≥ 1,000 IU/ml) from day 2 or 3 (24 or 48 h) postdosing for GT1a and day 1 to 5 (12, 24, 48, 72, or 96 h) postdosing for GT1b, with the exception of patient 5413 (day 14). All amplicon pools were cloned into a GT1b NS5A replicon shuttle vector, in vitro transcribed, and transfected into Huh 7.5 cells. Transiently transfected cells were treated with serial dilutions of EDP-239 to determine the EC50. The predose sample on the day of dosing was designated the baseline sample, and EC50s were used for calculating fold shift of postdosing samples (Tables 4 and 5).
All recombinant replicons replicated in vitro at levels sufficient for phenotypic analysis, and cloned subject baseline EC50s were within 2- or 3-fold of the EC50s determined for the GT1a or GT1b reference replicons, with the exception of those for one GT1a patient (patient 5305) and one GT1b patient (patient 5414). GT1a patient 5305 harbored multiple known NS5A RAMs at baseline, and this correlated with reduced susceptibility of the baseline sample to EDP-239 by >129,000-fold (Table 4). Similarly, the baseline EC50 of EDP-239 in GT1b patient 5414 was significantly higher than the reference EC50 due to the presence of a preexisting Y93H RAM at a frequency of 99% (Table 5). All postbaseline GT1a samples analyzed expressed known NS5A RAMs, and most displayed reduced susceptibility to EDP-239. The one exception was from patient 5311, whose day 2 sample harbored mixtures of M28T (9%) and Q30R (46%), yet no reduced EDP-239 susceptibility was observed among three independent replicon experiments. The highest degree of reduced susceptibility was an EC50 fold change of >9,000 from baseline in the day 3 sample from patient 5304, which was likely due to the high frequency of resistance-associated mutations detected at positions 28, 30, and 93.
In contrast with the GT1a phenotypic analysis, only 4 of 11 GT1b recombinant replicons exhibited significantly reduced susceptibility to EDP-239 (Table 5). All four possessed a Y93H RAM at proportions of greater than 33% of the viral population, further confirming the importance of Y93 RAMs to GT1b resistance to EDP-239. The presence of R30Q and Q62R RAMs at frequencies >99% in patient 5411 at baseline and nadir viral load did not result in any loss of EDP-239 potency in vitro (Table 5). However, these RAMs appeared to minimize the maximum viral load reduction in vivo, in comparison with the mean maximum viral load reduction for the 200-mg dose cohort (Table 2). These results highlight the apparent differences in sensitivity between the genotypic and phenotypic assays.
Persistence of NS5A RAMs.
NS5A inhibitors are characterized by their relatively low barriers to development of HCV resistance. In a previous report, it has been shown that NS5A inhibitor RAMs persist for up to at least 6 months in patients treated with daclatasvir in a 14-day monotherapy clinical trial (9). In an effort to minimize the extent of RAM selection in this proof-of-concept study, patients were given only a single dose of EDP-239. Deep sequencing of patient viral isolates was used to identify RAMs, their frequency over time, and their continued persistence up to 48 weeks postdosing in some patients (Table 6).
TABLE 6.
Persistence of NS5A RAMs through postdose week 12, 24, or 48
| GT | Dose | Patient | RAM(s) persistent up to wk 48 (%)a |
|---|---|---|---|
| 1a | 10 | 5306 | UNK |
| 10 | 5310 | Q30H(1)/R(37), L31M(43), H58D(2), Y93N(7)/C(8) | |
| 10 | 5314 | Q30H(56)/R(13), F36L(1) | |
| 10 | 5407b | K24R(2), M28V(66), Q30H(8)/R(80), L31M(2), H58C(15)/D(2), Y93F(12) | |
| 30 | 5303 | Q30H(14)/R(79), H58D(3) | |
| 30 | 5307 | ND | |
| 30 | 5313 | UNK | |
| 100 | 5301 | M28T(2), L31M(77) | |
| 100 | 5308 | M28T(5)/V(4), Q30H(2)/R(78), L31M (1), Y93C(2) | |
| 100 | 5311c | M28T(63), Q30R(53) | |
| 200 | 5304 | UNK | |
| 200 | 5305 | UNK | |
| 200 | 5309 | UNK | |
| 1b | 10 | 5404 | UNK |
| 10 | 5409 | ND | |
| 30 | 5405 | Y93H(3) | |
| 30 | 5413c | ND | |
| 30 | 5414 | Y93H(99) | |
| 100 | 5402 | L28 M(6), R30Q(16), L31 M(35), Y93H(37)/N(45) | |
| 100 | 5406b | R30G(3), L31V(46)/M(2), Y93H(4) | |
| 100 | 5410 | L31M(12), Y93H(10) | |
| 200 | 5403 | UNK | |
| 200 | 5408c | L31V(2)/M(97), Y93H(92) | |
| 200 | 5411b | L28 M(58), R30Q(5)/H(95) |
UNK, no data for week 12, 24, or 48; ND, no RAMs detected.
Week 12 persistence.
Week 24 persistence.
Of the 13 GT1a patients who received EDP-239, plasma samples from 6 patients were deep sequenced at week 48 postdosing, while a sample from one patient was sequenced up to week 12 and one up to week 24. Deep sequencing was performed up to week 48 for 5 of 11 GT1b patients, to week 24 for 2 patients, and to week 12 for 2 patients. RAMs were not detected in only 1 of 8 (12.5%) GT1a patients and in only 2 of 9 (22.2%) GT1b patients whose plasma samples were sequenced at week 12, 24, or 48.
In 5 of the 6 (83%) GT1a patients whose samples were deep sequenced at week 48, previously identified NS5A resistance-associated mutations at positions 28, 30, 31, 58, and 93 persisted at proportions as low as 1% and as high as 79%. Q30R/H RAMs, in particular, persisted at the highest viral population frequencies (>38%) at week 48 in 4 of 6 patients. Y93 RAMs were found at frequencies of <8% and only in 2 patients, further supporting the contention that the fitness of a GT1a virus is compromised when this mutation is expressed. While M28 RAMs persisted at weeks 12 and 24 at proportions of >60%, none of the viral populations sequenced at week 48 expressed resistance-associated mutations at this position at >5% and were identified only in 2 of 6 (33%) GT1a patients. Conversely, the frequency of the L31M substitution was expanded over time in 2 patients to >40% once the selective pressure of EDP-239 had diminished.
Of the 9 GT1b patients whose plasma samples were sequenced at week 12, 24, or 48, the most frequently detected resistance-associated mutations were identified at positions 31 and 93 in 4 (44%) and 6 (67%) patients, respectively. Of the five GT1b patients whose samples were sequenced at week 48, four patients (80%) expressed resistance-associated mutations at positions 31 and 93 at the highest frequencies. Moreover, Y93 RAMs persisted in all four of these patients at frequencies as low as 3% and as high as 99%, suggesting that mutations at NS5A amino acid position 93 in GT1b are more fit than those in GT1a. RAMs at position 31 were detected in the week 48 samples from 2 of 5 patients at frequencies of >12%.
DISCUSSION
This is the first report of HCV resistance to EDP-239 in vitro and in vivo. These analyses were conducted in order to identify GT1a and 1b NS5A RAMs selected with EDP-239 in vitro, to determine whether similar baseline or treatment-emergent RAMs could be detected in patients and whether their presence correlates with reduced efficacy of EDP-239 monotherapy in vivo.
Resistance selection studies with EDP-239 in vitro identified clones with substitutions within the first 100 amino acids of GT1 NS5A. Predominant GT1a resistance mutations (>10%) were identified at positions 24, 28, 30, and 93. Of these mutations, the greatest fold shifts in EDP-239 potency were conferred by M28T, Q30R, and Y93C/H/N. However, the fitness of M28T and Y93 RAMs was not as robust as the wild type. A Q30R mutation conferred a >500-fold decrease in sensitivity to EDP-239, and its replication efficiency was determined to be comparable to that of the wild type by using transiently transfected replicons in vitro. These results indicated that Q30 mutations were likely to be observed in patients and may limit the efficacy of EDP-239 in the clinic. The poor replication fitness of Y93 RAMs relative to other mutations is likely the reason for the lower frequencies selected. In addition, an F36L mutation was identified as a secondary substitution that, when linked with primary resistance mutation K24R, markedly enhanced resistance to EDP-239 but decreased replicon replication fitness.
RAMs identified in this study differed substantially between the two GT1 subtypes, consistent with previous reports of NS5A inhibitors investigated in vitro and in vivo (7, 13, 15–17). The reduction in EDP-239 potency was also observed to be greater for GT1a RAMs than for GT1b RAMs. Predominant GT1b RAMs selected in vitro (>10%) were L31V, Y93H, and A92V Y93H, which resulted in EDP-239 potency losses ranging from 3- to 1,100-fold compared to a transiently transfected wild-type replicon. While A92V did not confer resistance alone to EDP-239 or negatively affect replication fitness, it enhanced the resistance of the Y93H RAM to EDP-239. The mutations identified in vitro in this study have been observed in patients that have failed to achieve a sustained virologic response (SVR) following therapy with other NS5A inhibitors (4). However, there was no evidence that these mutations are cross-resistant to other DAA compounds (data not shown) or that previously reported RAMs arising from treatment with DAA compounds are cross-resistant to EDP-239 (14).
There was a dynamic change in the NS5A sequence observed over time among patients receiving EDP-239. These changes likely reflect the outgrowth of individual RAMs and their relative fitness once the selective pressure of EDP-239 was removed. The sample size in this in vivo clinical study for each escalating dose of EDP-239 administered was small, making it difficult to interpret many findings with a high degree of confidence; however, the intent was to identify mutations of interest to anticipate in future studies with longer durations of EDP-239 monotherapy. Given that only a single dose of compound was used in this study, it is not possible to determine whether the mutations that did increase in frequency following treatment would have been selected had repeated doses of EDP-239 been given. Subsequent clinical studies with multiple doses of EDP-239 are expected to provide clarity on this question.
The use of deep-sequencing technologies in this study enabled us to identify a complex mixture of viral quasispecies in each patient, both at baseline and at later time points. In addition, the pattern of resistance mutations reported here was significantly different between GT1a and GT1b patients, a finding that is consistent with our in vitro observations as well as clinical trial data reported for other NS5A inhibitors in development (7, 8, 17). In some GT1a patients with suboptimal antiviral responses, the data presented here indicate that baseline resistance-associated mutations at NS5A amino acid position 28, 30, or 93 limited the potency of EDP-239. These findings correlate with our in vitro resistance selection experiments.
In two GT1b patients, EDP-239 was less potent in a patient expressing the Y93H mutation at a baseline frequency of 99%; however, the Y93H RAM was found at a frequency of only 4% in another patient and did not appear to negatively impact the maximum viral load decrease. Viral loads in both patients with the Y93H RAM appeared to rebound faster than patients without a mutation at Y93 at baseline, suggesting a negative correlation between the presence of baseline Y93 RAMs and EDP-239 efficacy. The finding that RAMs present at baseline can impact the antiviral response to monotherapy is not unique to EDP-239. Several other studies of NS5A inhibitors, including daclatasvir, ledipasvir, and PPI-668 (16–18), reported patient isolates harboring NS5A RAMs prior to dosing, all of which limited the antiviral response.
At nadir viral RNA concentrations, Q30 RAMs were detected at frequencies >32% of the viral population in every GT1a patient. Moreover, the Q30R substitution was found to have the broadest representation across GT1a dosing cohorts and subjects, suggesting that this mutation is a major determinant of EDP-239 resistance in GT1a patients. In all but one GT1b patient with RAMs at nadir, Y93H was the predominant resistance mutation selected by EDP-239. These data suggest that viruses possessing the Y93H substitution are better able to survive in the presence of EDP-239. Additional mutations were identified at nadir viral load at positions 28, 31, and 58 in GT1b patients but only in those receiving 10- or 30-mg doses. At 100- and 200-mg doses, mutations detected at positions 30 and 62 were more common than they were at lower doses. The contributions of these mutations to EDP-239 resistance in vitro in our replicon phenotypic analyses suggest that these changes do not confer resistance.
Phenotypic analyses of patient isolates from this study were generally consistent with many other first-generation NS5A inhibitors with substitutions at GT1a NS5A positions 28, 30, 31, and 93 or GT1b position 93, causing reduced susceptibility to EDP-239 (16, 17). In comparing nadir viral load phenotypes to that at baseline, all but one GT1a clinical isolate showed reduced susceptibility to EDP-239 and all but two GT1b clinical isolates at nadir were less susceptible. The detection of a RAM at baseline did not always correlate with reduced EDP-239 susceptibility and may be a reflection of the higher sensitivity of the sequencing assay than of the phenotypic assay, consisting of a mixture of quasispecies from a pool of chimeric NS5A replicons. Additionally, in the context of individual patient isolates, there may be additional patient-specific polymorphisms along the NS5A coding region that affect the ability to detect a phenotypic change with the cell-based assay used here.
Previous studies have demonstrated that RAMs selected by NS5A inhibitor monotherapy in patients can persist for up to 6 months (9). In this study, many RAMs were detected at weeks 12, 24, and 48 postdosing. The most prevalent GT1a resistance-associated mutations present at week 48 were at NS5A positions 30 and 31 while mutations at NS5A positions 31 and 93 were more frequently detected in GT1b. The presence of these RAMs well after the removal of EDP-239 selective pressure suggests that these mutations confer an appreciable degree of fitness to the viral population. It will be interesting to investigate in future studies whether the selection and/or persistence of these RAMs occurs following increased treatment durations with EDP-239.
ACKNOWLEDGMENTS
We thank the patients for their participation in this study. We also acknowledge the ENANTA Pharmaceuticals and Novartis chemistry departments for their compound synthesis contributions. We are grateful to the Bioinformatics teams at Novartis and WuXi AppTec for their contributions to deep-sequencing data analysis.
This study was supported by ENANTA Pharmaceuticals, Inc., and Novartis.
All authors from Enanta and Novartis were employees of Enanta and Novartis at the time the research was completed.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
For a companion article on this topic, see doi:10.1128/AAC.00808-16.
REFERENCES
- 1.Anonymous. 1999. Hepatitis C—global prevalence (update). Wkly Epidemiol Rec 74:425–427. [PubMed] [Google Scholar]
- 2.Anonymous. 2016. Hepatitis C fact sheet no. 164. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Thompson AJ, McHutchison JG. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J Viral Hepat 16:377–387. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]
- 4.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, McPhee F, Mo H, Parkin N, Pilot-Matias T, Miller V. 2015. Hepatitis C virus drug resistance-associated substitutions: state of the art summary. Hepatology 62:1623–1632. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 5.Hughes M, Griffin S, Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol 90:1329–1334. doi: 10.1099/vir.0.009332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owens CM, Brasher BB, Polemeropoulos A, Peng X, Wang C, Ying L, Cao H, Qiu Y-L, Jiang L, Or YS. 2010. EDP-239 is a novel potent NS5A inhibitor with an excellent preclinical and resistance profile, abstr P-1863. 61st Annu Meet AASLD, The Liver Meeting, Boston, MA 29 October to 2 November 2010. [Google Scholar]
- 7.Lawitz EJ, Gruener D, Hill JM, Marbury T, Moorehead L, Mathias A, Cheng G, Link JO, Wong KA, Mo H, McHutchison JG, Brainard DM. 2012. A phase 1, randomized, placebo-controlled, 3-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J Hepatol 57:24–31. doi: 10.1016/j.jhep.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Sun JH, O'Boyle DR II, Nower P, Valera L, Roberts S, Fridell RA, Gao M. 2013. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrob Agents Chemother 57:2054–2065. doi: 10.1128/AAC.02494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 11.Yi M, Lemon SM. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J Virol 78:7904–7915. doi: 10.1128/JVI.78.15.7904-7915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother 54:3641–3650. doi: 10.1128/AAC.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owens CM, Brasher BB, Polemeropoulos A, Rhodin MHJ, McAllister N, Peng X, Wang C, Ying L, Cao H, Lawitz E, Poordad F, Rondon J, Box TD, Zeuzem S, Buggisch P, Lin K, Qiu Y-L, Jiang L, Colvin R, Or YS. 2016. Preclinical profile and clinical efficacy of a novel hepatitis C virus NS5A inhibitor, EDP-239. Antimicrob Agents Chemother 60:6207–6215. doi: 10.1128/AAC.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng G, Tian Y, Doehle B, Peng B, Corsa A, Lee YJ, Gong R, Yu M, Han B, Xu S, Dvory-Sobol H, Perron M, Xu Y, Mo H, Pagratis N, Link JO, Delaney W. 2016. In vitro antiviral activity and resistance profile characterization of the HCV NS5A inhibitor ledipasvir. Antimicrob Agents Chemother 60:1847–1853. doi: 10.1128/AAC.02524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridell RA, Wang C, Sun JH, O'Boyle DR II, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 17.Wong KA, Worth A, Martin R, Svarovskaia E, Brainard DM, Lawitz E, Miller MD, Mo H. 2013. Characterization of Hepatitis C virus resistance from a multiple-dose clinical trial of the novel NS5A inhibitor GS-5885. Antimicrob Agents Chemother 57:6333–6340. doi: 10.1128/AAC.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Q, Huang N, Huq A, Lau M, Peng E, Lalezari J, Farrell G, Shah P, Lawitz E, Vig P, Brown N, Colonno R. 2013. Vast majority of detected NS5A resistant variants are not amplified in HCV patients during 3-day monotherapy with the optimized NS5A inhibitor PPI-668, abstr 1192. Abstr 48th Annu Meet Eur Assoc Study Liver, Amsterdam, The Netherlands. [Google Scholar]