Abstract
Hypervirulent Klebsiella pneumoniae (hvKP) is traditionally defined by hypermucoviscosity, but data based on genetic background are limited. Antimicrobial-resistant hvKP has been increasingly reported but has not yet been systematically studied. K. pneumoniae isolates from bloodstream infections, hospital-acquired pneumonia, and intra-abdominal infections were collected from 10 cities in China during February to July 2013. Clinical data were collected from medical records. All K. pneumoniae isolates were investigated by antimicrobial susceptibility testing, string test, extended-spectrum β-lactamase (ESBL) gene detection, capsular serotypes, virulence gene profiles, and multilocus sequence typing. hvKP was defined by aerobactin detection. Of 230 K. pneumoniae isolates, 37.8% were hvKP. The prevalence of hvKP varied among different cities, with the highest rate in Wuhan (73.9%) and the lowest in Zhejiang (8.3%). Hypermucoviscosity and the presence of K1, K2, K20, and rmpA genes were strongly associated with hvKP (P < 0.001). A significantly higher incidence of liver abscess (P = 0.026), sepsis (P = 0.038), and invasive infections (P = 0.043) was caused by hvKP. Cancer (odds ratio [OR], 2.285) and diabetes mellitus (OR, 2.256) appeared to be independent variables associated with hvKP infections by multivariate analysis. Importantly, 12.6% of hvKP isolates produced ESBLs, and most of them carried blaCTX-M genes. Patients with neutropenia (37.5% versus 5.6%; P = 0.020), history of systemic steroid therapy (37.5% versus 5.6%; P = 0.020), and combination therapy (62.5% versus 16.7%; P = 0.009) were more likely to be infected with ESBL-producing hvKP. The prevalence of hvKP is high in China and has a varied geographic distribution. ESBL-producing hvKP is emerging, suggesting an urgent need to enhance clinical awareness, especially for immunocompromised patients receiving combination therapy.
INTRODUCTION
Over the past few decades, increasing rates of hypervirulent Klebsiella pneumoniae (hvKP) infection have been reported worldwide (1–4). Such strains are notorious for their capacity to cause serious and metastatic infections in young and healthy individuals, such as pyogenic liver abscesses and endophthalmitis (5). Hypermucoviscosity is an important in vitro parameter for identification of hvKP (6, 7). However, several controversies have arisen regarding the association of hypermucoviscosity phenotype and virulence (8, 9). Hypermucoviscosity-negative strains are more prone to cause severe infections and have a higher mortality rate in diabetic mice than hypermucoviscous K. pneumoniae (8). Our previous study also demonstrated that one of the five hypermucoviscous K. pneumoniae isolates showed high virulence in both in vitro and in vivo assays (9). Therefore, it is apparent that hvKP cannot be defined by string test alone (10). Aerobactin accounts for increased siderophore production and is a major virulence determinant and new defining trait for hvKP based on genetic background (11). Some advances have been made recently in the epidemiology of hvKP isolates in China (6, 7, 12), although most of the studies defined hvKP by hypermucoviscosity phenotype rather than genotype, which may have led to biased results. In the present study, hvKP strains were defined on the basis of aerobactin detection.
Although hypervirulent and antimicrobial-resistant populations of K. pneumoniae were largely nonoverlapping (13), some isolates with combined virulence and resistance were detected (9, 12, 14–16). More importantly, hypervirulent carbapenem-resistant K. pneumoniae isolates have emerged in clinical settings (9, 12, 16). The confluence of multidrug resistance and enhanced virulence has the potential to be the next clinical crisis (5). So far, research on antimicrobial-resistant hvKP strains has been limited and mainly based on case reports (9, 12). A systematic study on the molecular and clinical characteristics of antimicrobial-resistant hvKP infections is still lacking.
To reduce the bias caused by subjective identification methods and to investigate the recent pattern of hvKP in China, we conducted a nationwide study and systematically analyzed the clinical and molecular characteristics and antimicrobial resistance of hvKP isolates.
MATERIALS AND METHODS
Clinical K. pneumoniae isolates.
A total of 230 consecutive cases of K. pneumoniae infection were collected from a national prospective surveillance program (February to July 2013 from the Chinese Antimicrobial Resistance Surveillance of Nosocomial Infections, including Beijing [n = 45], Tianjin [n = 22], Shenyang [n = 22], Jinan [n = 14], Xi'an [n = 15], Wuhan [n = 23], Changsha [n = 17], Zhejiang [n = 12], Shanghai [n = 21], and Guangzhou [n = 39]). Each center was asked to enroll the cases and collect strains isolated from hospital-acquired pneumonia, intra-abdominal infections, and bloodstream infections. Duplicate isolates from the same individual were excluded. All isolates were stored at −80°C until use. Confirmation of the K. pneumoniae isolates was performed by 16S rRNA sequencing.
Clinical data collection.
Clinical data and patient information were obtained from medical records and included demographic characteristics, underlying medical conditions, clinical presentations, antimicrobial therapy administration, and outcomes. The study was approved by the Research Ethics Board at Peking University People's Hospital (Beijing, China).
Antimicrobial susceptibility testing and phenotypic confirmation of ESBL.
Susceptibility testing was determined by the agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (17). The following antimicrobial agents were tested: cefoxitin, cefotaxime, ceftriaxone, ceftazidime, cefepime, piperacillin-tazobactam, imipenem, meropenem, amikacin, ciprofloxacin, minocycline, and tigecycline. The results were interpreted according to CLSI breakpoints (18). The interpretive criteria for tigecycline were based on the Food and Drug Administration (FDA) breakpoints (≤2 mg/liter was susceptible, 4 mg/liter was intermediate, and ≥8 mg/liter was resistant). Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as controls for antimicrobial susceptibility testing. Extended-spectrum β-lactamase (ESBL) production was confirmed by agar dilution test using cefotaxime and ceftazidime along with clavulanate, in accordance with CLSI guidelines (17).
Detection of virulence-associated features and cps genotyping.
Hypermucoviscosity was identified by the string test as previously described (5). The formation of a viscous string of >5 mm was considered positive. We conducted cps genotyping of K serotype-specific alleles for K1, K2, K5, K20, K54, and K57 by PCR as previously described (19). Virulence-associated factors, such as the exopolysaccharide synthesis regulator gene (rmpA) and an iron uptake system (aerobactin), were determined by PCR (20, 21). Previously reported primers used for PCR were the following: aerobactin forward, 5′-GCATAGGCGGATACGAACAT-3′; aerobactin reverse, 5′-CACAGGGCAATTGCTTACCT-3′ (21). The reaction mixture was kept at 95°C for 5 min, followed by 30 cycles of 95°C for 1 min, 50°C for 1 min, 72°C for 1 min, and 72°C for 10 min. The PCR products were visualized and analyzed by agarose gel electrophoresis and sequencing.
β-Lactamase gene identification.
PCR was used to determine the prevalence of β-lactamase genes, including CTX-M, SHV, and TEM β-lactamase, as previously described (22). The blaCTX-M gene was first identified with the PAN-CTX-M primer pair as previously described (forward, 5′-TTTGCGATGTGCAGTACCAGTAA-3′; reverse, 5′-CGATATCGTTGGTGGTGCCATA-3′), followed by the group-specific primers (22). The products were submitted for sequencing.
MLST.
Multilocus sequence typing (MLST) was performed as described on the Pasteur Institute MLST website (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html), including DNA sequencing analysis of the seven housekeeping genes. New alleles and STs were submitted to the MLST website and were approved.
Statistical analysis.
Statistical analysis was performed using IBM SPSS Statistics version 20.0 and Graphpad Prism version 5. We used the χ2 or Fisher's exact test for categorical variables. P < 0.05 was considered statistically significant. Logistic regression was used to identify variables associated with hvKP and ESBL-producing hvKP infections. All variables with P values of <0.1 were included in the multivariate model.
RESULTS
Virulence-associated features of K. pneumoniae isolates.
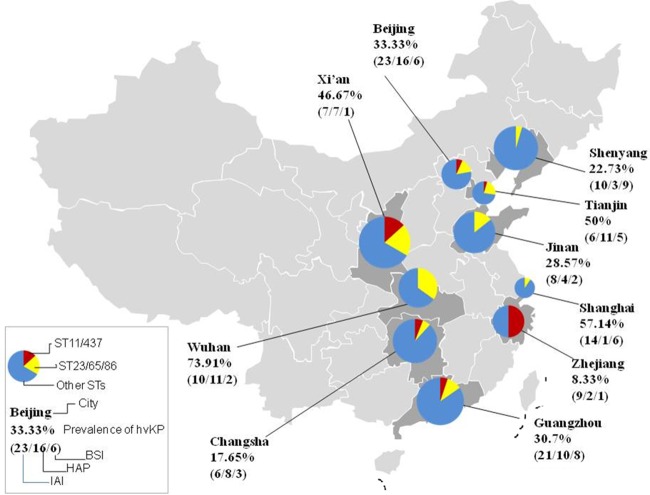
Aerobactin-positive strains were designated hvKP, which was determined by PCR. Eighty-seven of 230 (37.8%) isolates were hvKP. The prevalence of hvKP infection ranged from 8.33% to 73.9% in China, with the highest rate in Wuhan and the lowest rate in Zhejiang. The geographic distribution in China is shown in Fig. 1. Among all these strains, 74.7% (65/87) were hypermucoviscous, while six hypermucoviscous isolates were detected in the aerobactin-negative group. Hypermucoviscosity was strongly associated with hvKP (74.7% versus 4.2%; P < 0.001). A total of 71 (71/230; 30.9%) isolates were positive for K1, K2, K5, K20, and K57 serotypes, and the K54 serotype was not found in this study. K1, K2, and K20 were more likely to be identified in hvKP isolates (Table 1). The rmpA gene, a regulator of exopolysaccharide synthesis, had a strong relation to aerobactin (97.7% versus 3.5%; P < 0.001) (Table 1).
FIG 1.
Geographic distribution of hvKP infection cases in China. The prevalence of hvKP infections ranged from 8.33% to 73.91%, with a varied geographic distribution. Virulence-associated ST types (including ST23, ST65, and ST86) and resistance-associated ST types (including ST11 and ST437) demonstrated different distributions among K. pneumoniae isolates from different cities.
TABLE 1.
Microbiological and clinical characteristics of hvKP isolates
| Characteristic | Value (no. [%]; no. positive/total no.) fora: |
P value | |
|---|---|---|---|
| hvKP (n = 87) | cKP (n = 143) | ||
| Microbiological features | |||
| Hypermucoviscosity | 65 (74.7) | 6 (4.2) | <0.001 |
| K serotype | |||
| K1 | 29 (33.3) | 8 (5.6) | <0.001 |
| K2 | 15 (17.2) | 4 (2.8) | <0.001 |
| K5 | 2 (2.3) | 0 | 0.142 |
| K20 | 6 (6.9) | 0 | 0.003 |
| K57 | 4 (4.6) | 3 (2.1) | 0.431 |
| rmpA | 85 (97.7) | 5 (3.5) | <0.001 |
| Antimicrobial susceptibility | |||
| Cefoxitin | 78 (89.7) | 103 (72.0) | 0.002 |
| Cefotaxime | 75 (86.2) | 70 (49.0) | <0.001 |
| Ceftazidime | 78 (89.7) | 88 (61.5) | <0.001 |
| Cefepime | 82 (94.3) | 91 (63.6) | <0.001 |
| Piperacillin-tazobactam | 86 (98.9) | 111 (77.6) | <0.001 |
| Imipenem | 85 (97.7) | 129 (90.2) | 0.033 |
| Meropenem | 86 (98.9) | 129 (90.2) | 0.011 |
| Amikacin | 81 (93.1) | 130 (90.9) | 0.629 |
| Ciprofloxacin | 83 (95.4) | 89 (62.2) | <0.001 |
| ESBL production | 11 (12.6) | 61 (42.7) | <0.001 |
| Clinical characteristics | |||
| Male sex | 62 (77.5; 62/80) | 79 (67.5; 79/117) | 0.149 |
| Age (yr) (means ± SD) | 55.9 ± 1.5 | 51.8 ± 1.7 | 0.230 |
| Coexisting conditions | |||
| Pulmonary disease | 12 (15; 12/80) | 11 (9.4; 11/117) | 0.263 |
| Cancer | 27 (33.8; 27/80) | 22 (18.8; 22/117) | 0.020 |
| Liver disease | 8 (10; 8/80) | 14 (12.0; 14/117) | 0.819 |
| Neurological disease | 10 (12.5; 10/80) | 21 (17.9; 21/117) | 0.327 |
| Diabetes mellitus | 22 (27.5; 22/80) | 17 (14.7; 17/116) | 0.030 |
| Neutropeniab | 7 (8.8; 8/80) | 13 (11.1; 13/117) | 0.640 |
| Splenectomy | 0 | 2 (1.1; 2/117) | 0.515 |
| Surgery within 1 mo | 22 (27.5; 22/80) | 39 (33.3; 39/117) | 0.434 |
| Systemic steroid therapy | 7 (8.8; 7/80) | 16 (13.7; 16/117) | 0.369 |
| Use of immunosuppressant | 7 (8.8; 7/80) | 10 (8.6; 10/116) | 1.000 |
| Hospitalization within last 90 days | 20 (25; 20/80) | 53 (45.3; 53/117) | 0.004 |
| Transferred from another hospital | 12 (15; 12/80) | 17 (14.5; 17/117) | 1.000 |
| Receipt of antibiotics 30 days before infection | 25 (31.3; 25/80) | 52 (45.2; 52/115) | 0.054 |
| Use of invasive devices | |||
| Artery cannula | 4 (8.5; 4/47) | 3 (3.6; 3/83) | 0.253 |
| Presence of central venous catheters | 9 (19.1; 9/47) | 12 (14.6; 12/82) | 0.621 |
| Tracheal cannula | 9 (19.1; 9/47) | 11 (13.3; 11/83) | 0.450 |
| Tracheotomy | 3 (6.4; 3/47) | 4 (4.8; 4/83) | 0.703 |
| Presence of Foley catheter | 15 (31.9; 15/47) | 23 (27.7; 23/83) | 0.689 |
| Clinical presentation | |||
| Bacteremia | 24 (30; 24/80) | 35 (30.2; 35/116) | 1.000 |
| Abdominal infection | 16 (20; 16/80) | 27 (23.3; 27/116) | 0.604 |
| Hospital-acquired pneumonia | 36 (45; 36/80) | 43 (37.1; 43/116) | 0.301 |
| Liver abscess | 10 (11.5; 10/87) | 5 (3.5; 5/143) | 0.026 |
| Septic shock | 6 (6.9; 6/87) | 13 (9.1; 13/143) | 0.629 |
| Sepsis | 43 (49.4; 43/87) | 50 (35.0; 50/143) | 0.038 |
| Invasive infectionc | 11 (12.6; 11/87) | 7 (4.9; 7/143) | 0.043 |
| Infection occurred in ICU | 11 (13.8; 11/80) | 31 (26.9; 31/115) | 0.033 |
| Surgical treatment | 13 (18.3; 13/71) | 24 (21.6; 24/111) | 0.706 |
| Outcomes | |||
| Clinical improvement without modification of initial treatment | 50 (70.4; 50/71) | 86 (76.1; 86/113) | 0.395 |
| Change of initial antibiotics due to clinical worsening | 17 (23.9; 17/71) | 17 (15.0; 17/113) | 0.172 |
| Relapse | 2 (2.8; 2/71) | 1 (0.9; 1/113) | 0.560 |
| In-hospital mortality | 2 (2.8; 2/71) | 9 (8.0; 9/113) | 0.208 |
| Bacterial clearance after 72-h treatment | 64 (83.1; 64/77) | 95 (81.9; 95/116) | 1.000 |
| Persistent infection after 72-h treatment | 7 (9.1; 7/77) | 18 (15.5; 18/116) | 0.274 |
K. pneumoniae strains carrying aerobactin were designated hvKP, while cKP was defined by aerobactin negativity. Values in boldface are statistically significant.
Absolute neutrophil count of <500 cells/μl.
Invasive infections include liver abscess and meningitis.
Clinical characteristics of hvKP infection.
Table 1 shows the baseline characteristics of patients with hvKP infections. Compared with the classic K. pneumoniae (cKP) group, more patients with hvKP infections had cancer (33.8% versus 18.8%; P = 0.02) and diabetes mellitus (27.5% versus 14.7%; P = 0.03) as their underlying diseases. A significantly higher number of patients presenting with liver abscess (11.5% versus 3.5%; P = 0.026), sepsis (49.4% versus 35%; P = 0.038), and invasive infections (12.6% versus 4.9%; P = 0.043) were infected by hvKP. It was also noted that patients with a history of hospitalization within the previous 90 days (25% versus 45.3%; P = 0.004) and intensive care unit (ICU) stay (13.8% versus 26.9%; P = 0.033) were more likely to have a cKP infection.
Variables associated with hvKP infection.
Univariate regression analysis revealed that cancer (odds ratio [OR], 1.963) and diabetes mellitus (OR, 2.086) were statistically significant variables associated with hvKP infections (Table 2). In addition, cancer (OR, 2.285) and diabetes mellitus (OR, 2.256) were independent variables associated with hvKP infections, while hospitalization within the previous 90 days (OR, 0.345) and infection in the ICU (OR, 0.412) were independent protective factors for hvKP by multivariate analysis.
TABLE 2.
Regression analysis of variables associated with hvKP infections
| Variable | Analysis result |
|||
|---|---|---|---|---|
| Univariate |
Multivariate |
|||
| OR (95% CIa) | P value | OR (95% CI) | P value | |
| Cancer | 1.963 (1.012–3.806) | 0.046 | 2.285 (1.102–4.738) | 0.026 |
| Diabetes mellitus | 2.086 (1.034–4.210) | 0.040 | 2.256 (1.050–4.846) | 0.037 |
| Hospitalization within last 90 days | 0.403 (0.216–0.751) | 0.004 | 0.345 (0.171–0.695) | 0.003 |
| Receipt of antibiotics 30 days before infection | 0.551 (0.303–1.002) | 0.051 | 0.549 (0.279–1.078) | 0.081 |
| Infection occurred in ICU | 0.432 (0.202–0.922) | 0.030 | 0.412 (0.183–0.928) | 0.032 |
CI, confidence interval.
Antimicrobial susceptibility and prevalence of ESBL genes among K. pneumoniae isolates.
The antimicrobial resistance rate of cKP was significantly higher than that of hvKP strains (Table 1). ESBL was identified in 72 isolates (72/230; 31.3%) and was less common in the hvKP group (12.6% versus 42.7%; P < 0.001). Among 87 hvKP isolates, 12.6% (11/87) produced ESBLs, and one of them exhibited resistance to carbapenem.
The designation of the 11 ESBL-producing hvKP strains was by patient number. The demographics, virulence-associated features, and clinical characteristics of the 11 patients are summarized in Table 3. A total of six patients developed septicemia or bacteremia, and most of the strains were isolated from blood samples. All of these isolates harbored CTX-M, SHV, and TEM β-lactamase, and most of them had one or two genes that encoded ESBLs.
TABLE 3.
Clinical and microbiological characteristics of ESBL-producing hvKP isolates
| Clinical characteristic | Value for patient isolate: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Age (yr) | 76 | 76 | 49 | 43 | 60 | 53 | 20 | 53 | 62 | 48 | 1 mo |
| Gender | Male | Male | Male | Female | Female | Male | Female | Male | Female | Female | Male |
| City | Shanghai | Guangzhou | Guangzhou | Shenyang | Shenyang | Shenyang | Tianjin | Xi'an | Beijing | Jinan | Wuhan |
| Specimen type | Blood | Sputum | Sputum | Bile | Bile | Blood | Blood | Blood | Blood | Blood | Blood |
| Date of specimen collection (yr/mo/day) | 2013/4/13 | 2013/4/2 | 2013/3/6 | 2013/1/31 | 2013/2/18 | 2013/1/29 | 2013/3/17 | 2013/2/25 | 2013/7/23 | 2013/7/26 | 2013/3/26 |
| Infection type | Septicemia | Pneumonia | Pneumonia | Liver abscess | Abdominal infection | Liver abscess | Septicemia | Bacteremia | Septicemia | Bacteremia | Septicemia |
| Clinical outcome | Survived | Survived | Survived | Unknown | Unknown | Unknown | Survived | Survived | Survived | Survived | Survived |
| β-Lactamase(s) | SHV-75, CTX-M-55 | SHV-11, TEM-1, CTX-M-55 | SHV-11, TEM-1, CTX-M-55 | SHV-11, TEM-1, CTX-M-like | SHV-11, TEM-1, CTX-M-like | SHV-11, TEM-1, CTX-M-like | SHV-148 | CTX-M-14 | SHV-11, CTX-M-14 | CTX-M-14 | SHV-11, TEM-53 |
| Virulence-associated features | |||||||||||
| String test | + | − | + | + | + | + | + | + | + | − | + |
| cps genotyping | K20 | K1 | K1 | K1 | K20 | K20 | K2 | K1 | K nontypeable | K nontypeable | K2 |
| rmpA gene | + | + | + | + | + | + | + | + | + | + | + |
| MLST | ST420 | ST23 | ST23 | ST268 | ST268 | ST268 | ST1658 | ST367 | ST17 | ST35 | ST65 |
Patient characteristics and variables associated with hvKP isolates with and without ESBL production.
Patients with neutropenia (37.5% versus 5.6%; P = 0.020), history of systemic steroid therapy (37.5% versus 5.6%; P = 0.020), and combination therapy (use of multiple antimicrobials) (62.5% versus 16.7%; P = 0.009) were more likely to be infected with ESBL-producing hvKP.
MLST genotyping.
MLST identified 100 STs among 230 K. pneumoniae isolates. The most prevalent ST in K. pneumoniae isolates was ST23 (10.9%), followed by ST11 (5.2%). ST23, ST268, ST375, ST412, and ST660 were strongly associated with hvKP, while ST11, ST15, and ST37 were more common in the cKP group. ST17, ST23, ST35, ST65, ST268, ST367, ST420, and ST1658 were identified in hvKP isolates with ESBL production. In addition, STs showed different distributions among different cities (Fig. 1). In Wuhan, ST23 was the most dominant, with a prevalence of 21.7%, while ST11 was the most common in Zhejiang, with a prevalence of 25%.
DISCUSSION
To our knowledge, this is the first systematic multicenter study on hvKP in China, and it provides the most comprehensive understanding of hvKP defined by genetic background. According to previous designation criteria for hvKP, 30.9% of K. pneumoniae isolates were identified as string test positive in our study. The proportion was lower than that reported in previous studies conducted in a single center in China, with a prevalence of 33% in Beijing (7). However, it remains unclear whether all hvKP isolates had this phenotype. Serotypes, STs, and other genomic background data should be taken into consideration when defining hvKP. Therefore, the prevalence of hvKP might be underestimated due to the lack of objective diagnostic methods.
Aerobactin-positive isolates are associated with more serious infections (11, 23). Recently, aerobactin was considered a defining genetic trait and potential antivirulence target for hvKP (24). In our study, 37.8% of K. pneumoniae isolates were defined as hvKP by aerobactin. The prevalence showed a varied geographic distribution, with the highest rate in Wuhan and the lowest in Zhejiang. In Wuhan, 34.8% of the isolates belonged to ST23, ST65, and ST86, which are closely related to high virulence (9, 25). However, resistance-associated STs, including ST11 and ST437, were prevalent in Zhejiang, with a prevalence of 50%, and no isolate belonged to ST23 (26).
The prevalence of resistance in hvKP was still lower than that in cKP in the present study, particularly with regard to ESBLs. Contrary to the previous view that the prevalence of antimicrobial-resistant hvKP is low, a high percentage (12.6%) of ESBL production was found among hvKP in our study. Most of them carried blaCTX-M genes, indicating the compatibility of plasmids containing blaCTX-M genes with hvKP strains. Even more disturbing is that CTX-M-type ESBL is the most common genotype in China (27). The widespread dissemination of the CTX-M plasmid into hypervirulent variants from different STs is a frightening prospect in the future. Such isolates could produce various broad β-lactamases and ESBLs with a high level of resistance to multiple antimicrobials. Recently, carbapenemase-producing hvKP isolates have been identified in clinical settings, causing fatal infections (12, 16). K. pneumoniae carbapenemase-producing plasmid could be transferred into hvKP strains successfully (28). The situation calls for an immediate response to multidrug-resistant hvKP infections.
hvKP is clinically important because it can cause severe infections, and it should be given priority in clinical settings. Our results also demonstrate that hvKP is more likely to cause liver abscess, sepsis, and invasive infections than cKP, especially for patients in non-ICU departments and with no hospitalization prior to infection. The risk of ESBL-producing hvKP infection increased with neutropenia, systemic steroid therapy, and combination therapy. Therefore, compromised immunity is an important factor for antimicrobial-resistant hvKP infections.
There were some limitations to our study. Genotypic identification did not necessarily imply the expression of aerobactin; thus, further studies are needed to better define the hvKP strains. In addition, K. pneumoniae isolates were collected only in China, and whole-genome framework was not investigated. Therefore, little is known about the lineage diversification in our isolates, and it is unclear that our results can be generalized to hvKP outside China (29, 30). A study that includes more isolates from other regions, especially for antimicrobial-resistant strains, is needed.
In conclusion, the prevalence of hvKP was high in China, with a varied geographic distribution. Such strains were more likely to cause serious infections, such as liver abscess and sepsis in clinical settings. Our study highlights the urgent need to enhance clinical awareness and management of hvKP infections. Given the high prevalence of CTX-M ESBLs in China, the propensity for the acquisition of resistance genes among hvKP isolates from different STs, especially for immunocompromised patients with multiple antimicrobials treatment, should increase our concern.
ACKNOWLEDGMENTS
We thank the institutions that participated in the Chinese Antimicrobial Resistance Surveillance of Nosocomial Infections network in China. We acknowledge the following: Chunmei Zhou, Zhongshan Hospital Fudan University; Kang Liao, The First Affiliated Hospital Sun Yat-Sen University; Danhong Su, Guangzhou Institute of Respiratory Disease; Yunzhuo Chu, The First Hospital of China Medical University; Zhidong Hu, General Hospital of Tianjin Medical University; Xiuli Xu, Xijing Hospital; Rong Zhang, The Second Affiliated Hospital of Zhejiang University School of Medicine; Wenen Liu, Xiangya Hospital of Central South University; Yanping Luo, Chinese PLA General Hospital; Yingmei Liu, Beijing Chao-Yang Hospital of Capital Medical University; Ji Zeng, Union Hospital of Huazhong University of Science and Technology; and Yan Jin, Shandong Province Hospital.
REFERENCES
- 1.Chang L, Bastian I, Warner M. 2013. Survey of Klebsiella pneumoniae bacteraemia in two South Australian hospitals and detection of hypermucoviscous phenotype and magA/rmpA genotypes in K. pneumoniae isolates. Infection 41:559–563. doi: 10.1007/s15010-012-0374-y. [DOI] [PubMed] [Google Scholar]
- 2.Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. 2012. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 3.Decré D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, Offenstadt G, Maury E, Brisse S, Arlet G. 2011. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 49:3012–3014. doi: 10.1128/JCM.00676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, Ip M. 2011. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, Cao B. 2014. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother 58:5379–5385. doi: 10.1128/AAC.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Sun GZ, Yu YH, Li N, Chen M, Jin R, Jiao Y, Wu H. 2014. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 58:225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 8.Lin YC, Lu MC, Tang HL, Liu HC, Chen CH, Liu KS, Lin C, Chiou CS, Chiang MK, Chen CM, Lai YC. 2011. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol 11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YW, Zeng J, Liu WE, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Cao B, Wang H. 2015. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Alcántar-Curiel MD, Girón JA. 2015. Klebsiella pneumoniae and the pyogenic liver abscess: implications and association of the presence of rmpA genes and expression of hypermucoviscosity. Virulence 6:407–409. doi: 10.1080/21505594.2015.1030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae (hvKP) strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su SC, Siu LK, Ma L, Yeh KM, Fung CP, Lin JC, Chang FY. 2008. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum beta-lactamase. Antimicrob Agents Chemother 52:804–805. doi: 10.1128/AAC.01269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 55:930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Li XY, Wan LG, Jiang WY, Yang JH, Li FQ. 2014. Virulence and transferability of resistance determinants in a novel Klebsiella pneumoniae sequence type 1137 in China. Microb Drug Resist 20:150–155. doi: 10.1089/mdr.2013.0107. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2015. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 10th ed CLSI, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed Approved standard M100S. CLSI, Wayne, PA. [Google Scholar]
- 19.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 20.Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decré D. 2014. Multiplex PCR for the detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 52:4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JC, Koh TH, Lee N, Fung CP, Chang FY, Tsai YK, Ip M, Siu LK. 2014. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog 6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis JS II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother 51:4015–4021. doi: 10.1128/AAC.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassif X, Sansonetti PJ. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. 2015. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrade LN, Curiao T, Ferreira JC, Longo JM, Clímaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Cantón R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, Ma Y. 2007. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect 54:53–57. doi: 10.1016/j.jinf.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Siu LK, Huang DB, Chiang T. 2014. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis 14:176. doi: 10.1186/1471-2334-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hennequin C, Robin F. 2016. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]