Abstract
Aberrant promoter methylation of tumor relevant genes frequently occurs in early steps of carcinogenesis and during tumor progression. Epigenetic alterations could be used as potential biomarkers for early detection and for prediction of prognosis and therapy response in lung cancer. The present study quantitatively analyzed the methylation status of known and potential gatekeeper and tumor suppressor genes [O-6-methylguanine-DNA methyltransferase (MGMT), Ras association domain family member 1A (RASSF1A), Ras protein activator like 1 (RASAL1), programmed cell death 4 (PDCD4), metastasis suppressor 1 (MTSS1) and tumor suppressor candidate 3 (TUSC3)] in 42 lung cancers and in corresponding non-malignant bronchus and lung tissue using bisulfite-conversion independent methylation-quantification of endonuclease-resistant DNA (MethyQESD). Methylation status was associated with clinical and pathological parameters. No methylation was found in the promoter regions of PDCD4 and MTSS1 of either compartment. MGMT, RASSF1A and RASAL1 showed sporadic (up to 26.2%) promoter methylation. The promoter of TUSC3, however, was frequently methylated in the tumor (59.5%), benign bronchus (67.9%) and alveolar lung (31.0%) tissues from each tumor patient. The methylation status of TUSC3 was significantly associated with smaller tumor size (P=0.008) and a longer overall survival (P=0.013). Pooled blood DNA of healthy individuals did not show any methylation of either gene. Therefore, methylation of TUSC3 shows prognostic and pathobiological relevance in lung cancer. Furthermore, quantitative detection of TUSC3 promoter methylation appears to be a promising tool for early detection and prediction of prognosis in lung cancer. However, additional studies are required to confirm this finding.
Keywords: tumor suppressor candidate 3, lung cancer, prognosis, survival, quantitative methylation analysis
Introduction
Of all types of cancer, lung cancer is the malignancy with the greatest incidence and associated mortality, worldwide (GLOBOCAN 2012, v1.0). The poor chances of survival for lung cancer patients are often due to a late diagnosis (1). A variety of genetic and epigenetic factors contributing to the development of lung cancer have been discovered, yet a valid marker set for the early detection of disease has not been established. Methylation of promoter regions is a known cause for transcriptional repression (2) and can, therefore, contribute to carcinogenesis and tumor progression (2). This epigenetic mechanism of gene silencing has already been described in lung cancer for several genes, including cyclin-dependent kinase inhibitor 2A, O-6-methylguanine-DNA methyltransferase (MGMT), Ras association domain family member 1A (RASSF1A), retinoic acid receptor β (3–5), mutL homolog 1 (6), fragile histidine triad, death associated protein kinase, runt related transcription factor 3, TIMP metallopeptidase inhibitor 3 (4) or adenomatous polyposis coli (5). Certain studies even report promoter methylation in the bronchial lavage (7) or blood samples (8) of lung cancer patients, whereas others elucidate the increasing risk of developing this tumor with the accumulation of this epigenetic change in sputum (9). In future, the knowledge of the methylation status of tumor related genes could be helpful for the identification of persons at risk, the early detection of lung cancer and for the prediction of prognosis and therapeutic success. Certain studies suspect an association between promoter methylation and smoking habits (10) or exposure to environmental and industrial factors, such as smoky coal emission or chromate exposure (6,11).
To identify potential marker genes in lung cancer and in possible precursor lesions, the methylation status of MGMT, RASSF1A, Ras protein activator like 1 (RASAL1), programmed cell death 4 (PDCD4), metastasis suppressor 1 (MTSS1) and tumor suppressor candidate 3 (TUSC3) in lung tumors and corresponding non-malignant bronchus and lung tissue were quantitatively assessed using methylation-quantification of endonuclease-resistant DNA (MethyQESD). MethyQESD is a reliable bisulfite-conversion independent quantitative methylation sensitive polymerase chain reaction (qPCR) method. These six genes were chosen as they all appear to be relevant in tumor development and progression.
The inactivation and, therefore, loss of expression of MGMT and RASSF1A in lung cancer has been described previously (3–6,8,12–17). The reduced expression of RASAL1 was observed in several malignant tumors, including brain, head and neck, bladder, breast, colorectal, hepatocellular and thyroid carcinoma (18–21), and appears to be associated with neoplastic progression (19). In addition, certain studies provide evidence of an association between promoter RASAL1 methylation and loss of expression (18–21), while treatment with the DNA methyltransferase inhibitor 5′-azacitidine restored the expression of RASAL1 (18). PDCD4 is also suspected to act as tumor suppressor gene (22). Reduction or loss of expression has also been observed in lung cancer (23,24), and may be associated with tumor progression and a poorer prognosis (23). The impact of MTSS1 on malignant tumors is not yet well defined. Certain studies provide evidence that MTSS1 promotes tumor initiation and progression in early stages (25), whereas MTSS1 appears to have a tumor suppressive effect in advanced stages and in metastasis (25–27). Furthermore, the reduction and loss of expression of MTSS1 has been observed in malignancies (26,27). Other studies contradict this finding, and found increased levels of MTSS1 in aggressive and metastatic tumors and tumor cell lines (28). However, in lung cancer cell lines, MTSS1 was downregulated compared with benign human bronchial epithelial cell lines (29). Finally, in a number of studies, genetic and epigenetic changes and a loss of expression of TUSC3 have been detected in several malignancies, including prostate, ovarian, colorectal, larynx and pharynx carcinoma and acute lymphoblastic leukemia (30–34). In addition, TUSC3 involvement has already been observed in non-small cell lung cancer (NSCLC) downregulation (24) and promoter methylation (35). Therefore, it is likely that TUSC3 functions as a tumor suppressor gene.
Materials and methods
Sample collection
In total, 42 patients who had been diagnosed with primary lung cancer at the University Hospital of Regensburg (Regensburg, Germany) were selected for the present study. In particular, 42 primary lung tumors (17 adenocarcinomas, 20 squamous carcinomas, 3 large cell carcinomas and 2 neuroendocrine carcinomas) as well as corresponding normal lung tissue (n=42) and non-malignant bronchus tissues (n=28 for MGMT and TUSC3, n=29 for RASSF1A, n=27 for RASAL1 and PDCD4 and n=24 for MTSS1) were retrospectively analyzed. Archival tissue samples were obtained from the Institute of Pathology at the University Hospital of Regensburg, of which the Institutional Review Board approved the study in January 1997. All lung cancer patients underwent surgical resection between January 2000 and November 2002 at the Department of Thoracic Surgery, University Hospital of Regensburg and all histological data were provided by the Department of Pathology. Tumor staging was performed according to the 7th edition of the TNM Classification of Malignant Tumours. The age of the patients at diagnosis ranged between 38 and 77 years (mean, 59 years). In total, 33 patients were males (78.6%) and 9 were females (21.4%). Two patients received neoadjuvant therapy. Clinical and histopathological data are given in Table I.
Table I.
Clinicopathological characteristics of 42 non-small cell lung cancer patients.
| Characteristics | No. of patients (%) |
|---|---|
| Total | 42 (100.0) |
| Gender | |
| Male | 33 (78.6) |
| Female | 9 (21.4) |
| Age at diagnosis, years | |
| ≤60 | 23 (54.8) |
| >60 | 19 (45.2) |
| Survival | |
| Yes | 13 (31.0) |
| No | 28 (66.7) |
| Unknown | 1 (2.4) |
| Smoking status | |
| Smoker | 37 (88.1) |
| Non-smoker | 5 (11.9) |
| Histology | |
| Adenocarcinoma | 17 (40.5) |
| Squamous carcinoma | 20 (47.6) |
| Large cell carcinoma | 3 (7.1) |
| Neuroendocrine carcinoma | 2 (4.8) |
| T category | |
| T1a | 8 (19.0) |
| T1b | 2 (4.8) |
| T2a | 16 (38.1) |
| T2b | 8 (19.0) |
| T3 | 3 (7.1) |
| T4 | 5 (11.9) |
| N category | |
| N0 | 28 (66.7) |
| N1 | 11 (26.2) |
| N2 | 3 (7.1) |
| M category | |
| M0 | 38 (90.5) |
| M1 | 3 (7.1) |
| Unknown | 1 (2.4) |
| Grading | |
| G2 | 24 (57.1) |
| G3 | 18 (42.9) |
| Resection boundaries | |
| R0 | 38 (90.5) |
| R1 | 3 (7.1) |
| R2 | 1 (2.4) |
| Stage | |
| I | 19 (45.2) |
| II | 10 (23.8) |
| III | 9 (21.4) |
| IV | 3 (7.1) |
| Unknown | 1 (2.4) |
T, tumor; N, node; M, metastasis.
DNA extraction
Formalin-fixed paraffin-embedded 4-µm thick slides were deparaffinized after incubation at 70°C for 30 min in xylene, and then rehydrated in ethanol (graded series, 100, 96 and 70%) and deionized water. Subsequently, the slides were stained with 0.01% methylene blue. Microdissections of the bronchus, lung and tumor tissues were performed with a stereo microscope (magnification, ×40). DNA was isolated using the MagNA Pure LC DNA-Isolation Kit II (Roche Diagnostics, Mannheim, Germany), according to the manufacturer's recommendations, and quantified photometrically. Samples with low DNA content were concentrated with Amicon® Ultra 0.5 ml Centrifugal Filters (Merck Millipore, Darmstadt, Germany).
Quantitative methylation analysis
Methylation analysis was performed using MethyQESD (36), a combination of methylation-sensitive digestion and qPCR. A methylation specific quantification digestion (MQD) containing 40 units (U) of the methylation-sensitive endonuclease, Hin6I (Fermentas; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and a calibrator digestion (CalD) containing the methylation-independent endonucleases, XBaI (20 U) and DraI (20 U) (Fermentas; Thermo Fisher Scientific, Inc.), were set up for each sample. Each digestion, with a total reaction volume of 20 µl, was performed using 5 µl DNA (minimum concentration, 20 ng/µl) and 2 µl of 10x Buffer Tango™ (Fermentas; Thermo Fisher Scientific, Inc.). Cell-line DNA served as a positive control (SW48 for MGMT, RASSF1A, MTSS1 and TUSC3; HT29 for RASAL1; none for PDCD4) whereas pooled blood DNA from healthy individuals and a sample containing no DNA were used as negative controls. Following incubation at 37°C overnight, the reaction was stopped at 70°C for 20 min. qPCR was performed using the LightCycler 1.0 (Roche Diagnostics) for MGMT, RASSF1A, RASAL1, PDCD4 and MTSS1 and the LightCycler® 480 (Roche Diagnostics) for TUSC3. The total reaction volume contained 10 µl QuantiTect® SYBR® Green PCR Kit (Qiagen GmbH, Hilden, Germany), 0.5 µM Primer and 3 µl digested DNA. The primers sequences were as follows: MGMT forward: 5′-CCCGGATATGCTGGGACAG-3′; MGMT reverse: 5′-CCCAGACACTCACCAAGTCG-3′; RASSF1A forward: 5′-GCTGGGCGCGCTGGGAAG-3′; RASSF1A reverse: 5′-CAGGGACCAGCTGCCGTGT-3′; RASAL1 forward: 5′-CTCCAGACGCCTCGGCAAGAG-3′; RASAL1 reverse: 5′-AGCGCCCGTCCGGACTCTAC-3′; PDCD4 forward: 5′-CCAGTCCCAGGAGCCACAT-3′; PDCD4 reverse: 5′-GAGGAAAAGGGAGAGGAGTGA-3′; MTSS1 forward: 5′-GAGCCCAGCCAGAGCGAGC-3′; MTSS1 reverse: 5′-CGGCGTCCGGATCTGTTGCT-3′; TUSC3 forward: 5′-TACCGCGCGTGGAGGAGACA-3′; TUSC3 reverse: 5′-GTGGGCAGGTACCGCAGCC-3′. After an initial denaturation of 15 min at 95°C 45 cycles of amplification followed: Denaturation at 95°C for 10 sec (MGMT, RASSF1A, PDCD4) or 15 sec (RASAL1, MTSS1, TUSC3) and annealing at 60 °C (MGMT, PDCD4, TUSC3), 65 °C (RASSF1A) and 66 °C (RASAL1, MTSS1) for 17 sec (MGMT, RASSF1A, PDCD4), 20 sec (MTSS1) and 34 sec (TUSC3). For RASAL1 a two-step PCR was performed at 66°C for 20 sec. For the melting point analysis, PCR products were heated from 55 to 98°C with an increase of 0.2°C/sec (MGMT, RASSF1A, RASAL1, PDCD4, MTSS1) or 0.11°C/sec (TUSC3). Fluorescence was measured continuously. Methylation was quantified according to the formula: Methylation (%) = E(CtCalD - CtMQD)x100. (Ct = Ct value; E = PCR efficiency). For calculation of the Ct-Value LightCycler Software Version 3.5 (Roche Diagnostics) (MGMT, RASSF1A, RASAL1, PDCD4 and MTSS1) and LightCycler® 480 Software 1.5.0 (Roche Diagnostics) (TUSC3) were used. PCR-efficiency was obtained by standard curves for MQD and CalD with the dilution levels 1:4, 1:16, 1:64, 1:256, 1:1024, 1:4096 and 1:16384, respectively. LightCycler Software ver. 3.5 and LightCycler® 480 Software ver. 1.5.0 was used to calculate PCR efficiencies, which were 1.97 for MGMT, RASSF1A, RASAL1, PDCD4 and MTSS1 and 1.94 for TUSC3. The cut-off value for positive methylation was >4%.
Statistical analysis
The association between two variables was analyzed using Fisher's exact test (two-sided). Survival was estimated according to Kaplan-Meier, and comparisons between differences in survival were performed with the log-rank test. P<0.05 was assumed to indicate a statistically significant difference.
Results
Methylation frequencies
The promoter methylation frequencies of MGMT, RASSF1A, RASAL1, PDCD4, MTSS1 and TUSC3 in lung tumor and corresponding non-malignant bronchus and lung tissues were quantitatively assessed, with a cut-off value of >4 per cent for positive methylation applied (Table II). No methylation was identified for PDCD4 and MTSS1 in any tissue types, and only sporadically for RASAL1 [bronchus, 0.0% (0/27); lung, 2.4% (1/42); tumor, 4.8% (2/42)]. MGMT showed methylation in 7.1% of benign bronchus (2/28) and tumor (3/42) samples, as well as in 2.4% (1/42) of non-malignant lung tissue. Simultaneous methylation in two tissue types occurred only in one case (tumor and lung). RASSF1A was scarcely methylated in bronchial tissue (3.4%; 1/29), not at all in normal lung (0.0%; 0/42) and in 26.2% of tumor tissues (11/42). However, in the one case of bronchus methylation, no other tissue type was affected. The highest methylation frequencies were detected for TUSC3 in all three tissue types: Bronchus, 67.9% (19/28); lung tissue, 31.0% (13/42); and tumor tissue, 59.5% (25/42). Notably, 5 cases demonstrated TUSC3 methylation in all three tissue types, 8 cases demonstrated TUSC3 methylation in the bronchus and tumor tissues, 3 cases in the lung and tumor tissues, and 9 cases demonstrated TUSC3 methylation in the tumor tissues only. Of the 3 cases with methylation in the lung and tumor and from 6/9 cases with tumor methylation, bronchial material was not available for examination. In addition, 12 patients showed methylation of >1 gene in the three tissue types. MGMT, RASSF1A and TUSC3 were methylated more often in the bronchus compared with in the lung tissue: MGMT, 7.1% (2/28) vs. 2.4% (1/42); RASSF1A, 3.4% (1/29) vs. 0.0% (0/42); and TUSC3, 67.9% (10/28) vs. 31.0% (13/42). No methylation of any gene was detected in pooled blood DNA of healthy individuals.
Table II.
Frequencies of methylation in lung cancer and corresponding non-malignant lung tissues from the same patients.
| Bronchus tissue | Lung tissue | Tumor tissue | ||||
|---|---|---|---|---|---|---|
| Analyzed gene | % patients | n/ntotal | % patients | n/ntotal | % patients | n/ntotal |
| MGMT | 7.1 | 2/28 | 2.4 | 1/42 | 7.1 | 3/42 |
| RASSF1A | 3.4 | 1/29 | 0.0 | 0/42 | 26.2 | 11/42 |
| RASAL1 | 0.0 | 0/27 | 2.4 | 1/42 | 4.8 | 2/42 |
| PDCD4 | 0.0 | 0/27 | 0.0 | 0/42 | 0.0 | 0/42 |
| MTSS1 | 0.0 | 0/24 | 0.0 | 0/42 | 0.0 | 0/42 |
| TUSC3 | 67.9 | 19/28 | 31.0 | 13/42 | 59.5 | 25/42 |
MGMT, O-6-methylguanine-DNA methyltransferase; RASSF1A, Ras association domain family member 1A; RASAL1, Ras protein activator like 1; PDCD4, programmed cell death 4; MTSS1, metastasis suppressor 1; TUSC3, tumor suppressor candidate 3.
Survival and clinicopathological parameters
There was no significant association between survival time and gender (P=0.864), smoking habits (P=0.322), tumor histology (P=0.788) and grading (P=0.301). However, patients diagnosed with lung cancer that were older than 60 years of age lived significantly longer (P=0.034) compared with patients that were younger than 60 years of age at the time of diagnosis. Furthermore, the association between survival time and detailed T-(tumor; detailed: Stadium subdivided in a and b), N (node)-, and M (metastasis)-stadium (P=0.019; P=0.000; P=0.000), tumor stage (P=0.000) and R-classification (P=0.017) reached statistical significance.
Promoter methylation and clinicopathological parameters
No statistically significant associations between promoter methylation and gender, age at diagnosis, smoking habits, lymph node and distant metastasis, grading, R-classification and tumor histology were observed for MGMT, RASSF1A, RASAL1, PDCD4, MTSS1 and TUSC3 and bronchus, lung and tumor tissue. However, tumor promoter methylation of TUSC3 significantly associated with R0-resection (R0 vs. R1 and R2, P=0.021) and smaller tumor size (T1a, T1b and T2a vs. T2b, T3 and T4; P=0.008; Table III). The latter effect, however, disappeared when dismissing the subdivision of T-Stadiums in a and b (T1 and T2 vs. T3 and T4, P=0.235). TUSC3 was also significantly more often methylated in bronchus tissues in lower tumor stages (stage I and II vs stage III and IV, P=0.035). In addition, a slight tendency for higher methylation frequencies of TUSC3 in bronchus (10 of 17 vs.9 of 11 patients) and lesser in tumor (13 of 23 vs. 12 of 19 patients) can be observed in patients >60 years at time of diagnosis.
Table III.
Number of patients with positive methylation status and clinicopathological data.
| MGMT | RASSF1A | RASAL1 | TUSC3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | B | L | T | B | L | T | B | L | T | B | L | T |
| Gender | ||||||||||||
| Female | 2/22 | 1/33 | 3/33 | 1/23 | 0/33 | 8/33 | 0/21 | 1/33 | 2/33 | 17/22 | 12/33 | 20/33 |
| Male | 0/6 | 0/9 | 0/9 | 0/6 | 0/9 | 0/9 | 0/6 | 0/9 | 0/9 | 2/6 | 1/9 | 5/9 |
| Age | ||||||||||||
| ≤60 | 2/18 | 1/23 | 2/23 | 0/18 | 0/23 | 6/23 | 0/17 | 1/23 | 2/23 | 10/17 | 7/23 | 13/23 |
| >60 | 0/10 | 0/19 | 1/19 | 1/11 | 0/19 | 5/19 | 0/10 | 0/19 | 0/19 | 9/11 | 6/19 | 12/19 |
| Survival | ||||||||||||
| Yes | 1/8 | 0/13 | 1/13 | 0/9 | 0/13 | 2/13 | 0/8 | 0/13 | 0/13 | 7/9 | 6/13 | 11/13a |
| No | 1/20 | 1/28 | 2/28 | 1/20 | 0/28 | 9/28 | 0/19 | 1/28 | 2/28 | 12/19 | 7/28 | 13/28a |
| Smoking | ||||||||||||
| Yes | 2/26 | 1/37 | 3/37 | 1/27 | 0/37 | 9/37 | 0/27 | 1/37 | 2/37 | 19/26 | 11/37 | 22/37 |
| No | 0/2 | 0/5 | 0/5 | 0/2 | 0/5 | 2/5 | 0/2 | 0/5 | 0/5 | 0/2 | 2/5 | 3/5 |
| T category | ||||||||||||
| T1a | 0/2 | 1/8 | 1/8 | 0/3 | 0/8 | 2/8 | 0/2 | 1/8 | 1/8 | 3/3 | 2/8 | 6/8b |
| T1b | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 1/2 | 0/2 | 0/2 | 0/2 | 0/2 | 2/2 | 0/2b |
| T2a | 1/10 | 0/16 | 1/16 | 1/10 | 0/16 | 3/16 | 0/10 | 0/16 | 0/16 | 8/10 | 5/16 | 14/16b |
| T2b | 0/7 | 0/8 | 1/8 | 0/7 | 0/8 | 4/8 | 0/6 | 0/8 | 1/8 | 5/6 | 3/8 | 2/8b |
| T3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 | 1/3b |
| T4 | 1/4 | 0/5 | 0/5 | 0/4 | 0/5 | 0/5 | 0/4 | 0/5 | 0/5 | 2/4 | 1/5 | 2/5b |
| N category | ||||||||||||
| N0 | 2/18 | 0/28 | 1/28 | 0/19 | 0/28 | 7/28 | 0/17 | 0/28 | 1/28 | 14/18 | 9/28 | 18/28 |
| N1 | 0/8 | 1/11 | 2/11 | 1/8 | 0/11 | 3/11 | 0/8 | 1/11 | 1/11 | 4/8 | 2/11 | 5/11 |
| N2 | 0/2 | 0/3 | 0/3 | 0/2 | 0/3 | 1/3 | 0/2 | 0/3 | 0/3 | 1/2 | 2/3 | 2/3 |
| M category | ||||||||||||
| M0 | 2/26 | 1/38 | 3/38 | 1/27 | 0/38 | 10/38 | 0/25 | 1/38 | 2/38 | 19/26 | 11/38 | 23/38 |
| M1 | 0/2 | 0/3 | 0/3 | 0/2 | 0/3 | 1/3 | 0/2 | 0/3 | 0/3 | 0/2 | 2/3 | 1/3 |
| Grading | ||||||||||||
| G1 | 1/15 | 1/24 | 1/24 | 0/16 | 0/24 | 6/24 | 0/15 | 1/24 | 2/24 | 11/16 | 7/24 | 15/24 |
| G2 | 1/13 | 0/18 | 2/18 | 1/13 | 0/18 | 5/18 | 0/12 | 0/18 | 0/18 | 8/12 | 6/18 | 10/18 |
| Resection boundaries | ||||||||||||
| R0 | 2/26 | 1/38 | 2/38 | 1/26 | 0/38 | 11/38 | 0/25 | 1/38 | 2/38 | 18/26 | 11/38 | 25/38c |
| R1 | 0/1 | 0/3 | 1/3 | 0/1 | 0/3 | 0/3 | 0/1 | 0/3 | 0/3 | 1/1 | 1/3 | 0/3c |
| R2 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 0/1c |
| Histology | ||||||||||||
| AC | 1/11 | 0/17 | 0/17 | 0/11 | 0/17 | 6/17 | 0/11 | 0/17 | 0/17 | 6/11 | 3/17 | 12/17 |
| SCC | 0/13 | 1/20 | 2/20 | 1/14 | 0/20 | 4/20 | 0/12 | 1/20 | 2/20 | 10/13 | 9/20 | 11/20 |
| LCA | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 2/3 | 1/3 | 1/3 |
| Carc | 0/1 | 0/2 | 1/2 | 0/1 | 0/2 | 1/2 | 0/1 | 0/2 | 0/2 | 1/1 | 0/2 | 1/2 |
| Stage | ||||||||||||
| I | 1/11 | 0/19 | 1/19 | 0/12 | 0/19 | 4/19 | 0/11 | 0/19 | 0/19 | 9/12d | 6/19 | 15/19 |
| II | 0/7 | 1/10 | 2/10 | 1/7 | 0/10 | 4/10 | 0/6 | 1/10 | 2/10 | 6/6d | 3/10 | 4/10 |
| III | 1/8 | 0/9 | 0/9 | 0/8 | 0/9 | 2/9 | 0/8 | 0/9 | 0/9 | 4/8d | 2/9 | 4/9 |
| IV | 0/2 | 0/3 | 0/3 | 0/2 | 0/3 | 1/3 | 0/2 | 0/3 | 0/3 | 0/2d | 2/3 | 1/3 |
Data are expressed as n/ntotal. Groups with significant P-values from multiple comparison analysis:
P=0.039
P=0.005
P=0.021
P=0.041. MGMT, O-6-methylguanine-DNA methyltransferase; RASSF1A, Ras association domain family member 1; RASAL1, RAS protein activator like 1; TUSC3, tumor suppressor candidate 3; B, bronchus; L, lung; T, tumor; T/N/M category, tumor/node/metastasis category; AC, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell carcinoma; Carc, carcinoid.
Promoter methylation and survival
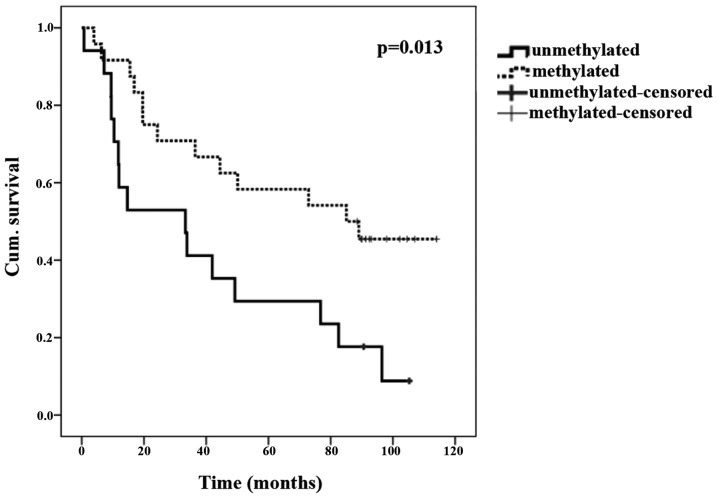
Patients with promoter methylation of TUSC3 in tumor tissues lived significantly longer compared with those without this epigenetic modification (P=0.039; Fisher's exact test). The Kaplan-Meier curve is shown in Fig. 1 (P=0.013; log-rank test).
Figure 1.
Kaplan-Meier overall survival curve. Dotted line, tumor patients with TUSC3 methylation; solid line, patients without TUSC3 methylation. TUSC3, tumor supressor candidate 3.
Discussion
In the present study, the promoter methylation status of MGMT, RASSF1A, RASAL1, PDCD4, MTSS1 and TUSC3 was quantitatively examined using a highly reliable methylation-sensitive qPCR, avoiding bisulfite-conversion.
In the patients studied, MGMT methylation was less frequently observed in lung tumor tissues compared with other studies, which describe methylation frequencies of 10.0–77.8% (3,4,6,8,12,16,17). In contrast to the present study, the majority of other studies did not analyze normal tissue; therefore, statements concerning the use of MGMT methylation in the early detection of lung cancer by analyzing non-malignant samples cannot be reasonably made. However, two reports (3,12) with normal tissues had greater methylation frequencies (~20%) compared with the 7.1% MGMT methylation found in bronchial or 2.4% found in lung specimens in the present study, which are similar to the 3% methylation reported by Safar et al (16).
The findings of low RASSF1A promoter methylation levels are in accordance with other reports, which describe methylation frequencies of 0.0–6.0% in normal tissues (5,13,16,37,38), 12.8% methylation in benign lung tissues (3) and 15.0–45.0% in NSCLC (3–5,7,8,13–16,37,38).
Differences in MGMT and RASSF1A methylation frequencies between the results of the present study and the results of previous studies may be due to different analysis methods. In contrast to the MethyQESD technique (36), previous studies used methods such as bisulfite-conversion, which bears the risk of incomplete conversion of unmethylated cytosines (39), leading to an overestimation of methylation. Furthermore, the use of methylation specific primers without enough discrimination capacity between methylated and unmethylated DNA can also result in false positive methylation results (39). In addition, cut off values are not mentioned (12), or set at 0% (8), or methylation is analyzed only qualitatively (3,4,6,14,15,17). Certain studies employed a nested PCR (3,6), which improves sensitivity but can lower specificity and reproducibility (40). This effect is further demonstrated by the experiments of Lee et al (38), which show that MGMT methylation differed depending on whether methylation specific PCR (qualitative analysis) or pyrosequencing (quantitative analysis) was used. Therefore, the comparability of methylation studies is limited.
In the present study, only low levels or no methylation of RASAL1 (tumor tissue, 4.8%; lung, 2.4%; bronchus, 0.0%) were observed, and consequently no association with clinicopathological parameters was found. To the best of our knowledge, only two other studies have dealt with methylation of RASAL1 in lung carcinoma: Jin et al (18) observed methylation in 2/4 lung cancer cell lines; whereas Calvisi et al (20) reported a methylation frequency of 16.7% (5/30) in primary tumors, but did not describe the characteristics of the patient population nor the histology of the tumors analyzed. This is especially important as RASAL1 shows tumor and tissue-specific expression (19) with increased expression levels in endocrine organs (19), which raises the question of whether RASAL1, in general, is a cancer relevant gene in lung adenocarcinoma.
In the present study, no methylation of PDCD4 or MTSS1 was found in any of the 3 tissue types. Promoter methylation of PDCD4 and MTSS1 genes has been previously examined in other tumors (41,42), but not yet in lung cancer. According to the current results, factors other than methylation could be involved in the regulation of the expression of MTSS1, such as binding of DNA (cytosine-5-)-methyltransferase 3β to the MTSS1 5′ region (43,44), and PDCD4, such as transcription growth factor β (22), zinc finger protein 148 and histone modifications (44).
In contrast to the other analyzed genes, TUSC3 showed frequent methylation in all three tissue types: Bronchus, 67.9% (19/28); lung, 31.0% (13/42); and tumor, 59.5%, (25/42). Thus, TUSC3 promoter methylation could be an early event during bronchial tumor development, and the detection of TUSC3 methylation could be beneficial for the early detection of lung cancer. Regarding prognostic aspects, the present study shows that patients with TUSC3 methylation in tumor tissues lived significantly longer compared with patients without this epigenetic modification (Fig. 1). In addition, patients that were diagnosed with lung cancer when they were older than 60 years of age lived significantly longer (P=0.034) compared with patients diagnosed when they were younger than 60 years of age, and showed slightly more methylation in bronchus and tumor tissues, although without statistical significance. This finding could be due to an association between longer survival and TUSC3 methylation. Tumor methylation was significantly associated with R0-resection and smaller tumor size as T-stadiums were subdivided into a and b. Although longer survival is an evident result, the potential causality between TUSC3 methylation and longer survival remains to be elucidated.
In certain aspects, the present study contrasts with previous studies, which found an association between TUSC3 methylation and a poorer prognosis or advanced tumor stage in other tumor types (31,33). Furthermore, the first evidence of a loss of TUSC3 expression was found in metastatic prostate cancer (30), which implies that the loss of TUSC3 expression may be associated with progressed disease in this tumor type. Contrarily, TUSC3 can be reasonably assumed to have a non-oncogenic function due to a defect in TUSC3 that was previously described to cause non-syndromic autosomal mental retardation without tumor formation in affected patients (45).
Finally, the methylation of TUSC3 may potentially occur due to collateral damage along with other gene methylation events in the 8p22 chromosome region (46). Consequently, its loss could be without direct consequence for tumorigenesis. Previous studies showed that TUSC3 methylation was not beneficial for tumor growth in one cell culture experiment (32), whereas in another experiment, it was (31).
In conclusion, the present study identified little or no promoter methylation of MGMT, RASSF1A, RASAL1, PDCD4 and MTSS1 in bronchial, lung and lung cancer tissues, but the relatively frequent methylation of TUSC3 in the same tissues. The fact that TUSC3 methylation was found to be associated with a longer survival time contradicts the hypothesis that TUSC3 has a tumor suppressor function and underlines that TUSC3 methylation has a prognostic value in lung cancer patients. In addition, the methylation of TUSC3, particularly in combination with other markers, may be useful for the early detection of lung cancer, as TUSC3 was frequently observed in the tumor and benign bronchus and lung tissues of lung cancer patients, but not in pooled blood DNA of healthy individuals. Additional studies are required to clarify the functional role of TUSC3 methylation in lung cancer. Prospective studies are recommended to be undertaken to further evaluate TUSC3 methylation as a prognostic biomarker and its usefulness for the early detection of disease in lung cancer patients.
Acknowledgements
The present study was supported by the Wilhelm-Sander-Foundation (grant no., 2000.127.1). The authors would like to thank Mrs Irene Schardt, Mrs Beate Reil, Mrs Sigi Appel and Mrs Jutta Förster for excellent technical assistance and Dr Corinna Lang-Schwarz for tissue sampling.
References
- 1.Adamietz IA, Niederle N. Lung cancer. Onkologe. 2010;16:615–628. doi: 10.1007/s00761-010-1853-3. (In German) [DOI] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Licchesi JD, Westra WH, Hooker CM, Herman JG. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res. 2008;14:2570–2578. doi: 10.1158/1078-0432.CCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 4.Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Feng Q, Hawes SE, Stern JE, Wiens L, Lu H, Dong ZM, Jordan CD, Kiviat NB, Vesselle H. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali AH, Kondo K, Namura T, Senba Y, Takizawa H, Nakagawa Y, Toba H, Kenzaki K, Sakiyama S, Tangoku A. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol Carcinog. 2011;50:89–99. doi: 10.1002/mc.20697. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Kwon YM, Kim JS, Lee H, Park JH, Shim YM, Han J, Park J, Kim DH. Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J Clin Oncol. 2004;22:2363–2370. doi: 10.1200/JCO.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 8.Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, Carvalho AL, Califano JA, Goodman SN, Westra WH, Hoque MO, Sidransky D. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res. 2011;17:4494–4503. doi: 10.1158/1078-0432.CCR-10-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, Feser WJ, Baron AE, Franklin WA, Brock MV, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res. 2012;18:3387–3395. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Shen Y, Liu B, Tong Y. Promoter methylation of BRMS1 correlates with smoking history and poor survival in non-small cell lung cancer patients. Lung Cancer. 2011;74:305–309. doi: 10.1016/j.lungcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Lan Q, Shen M, Jin J, Mumford J, Ren D, Keohavong P. Aberrant gene promoter methylation in sputum from individuals exposed to smoky coal emissions. Anticancer Res. 2008;28:2061–2066. [PMC free article] [PubMed] [Google Scholar]
- 12.Brabender J, Usadel H, Metzger R, Schneider PM, Park J, Salonga D, TsaoWei DD, Groshen S, Lord RV, Takebe N, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: Associations with clinical outcome. Clin Cancer Res. 2003;9:223–227. [PubMed] [Google Scholar]
- 13.DeJong WK, Verpooten GF, Kramer H, Louwagie J, Groen HJ. Promoter methylation primarily occurs in tumor cells of patients with non-small cell lung cancer. Anticancer Res. 2009;29:363–369. [PubMed] [Google Scholar]
- 14.Lin Q, Geng J, Ma K, Yu J, Sun J, Shen Z, Bao G, Chen Y, Zhang H, He Y, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135:1675–1684. doi: 10.1007/s00432-009-0614-4. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama R, Sugio K, Yoshino I, Maehara Y, Gazdar AF. Hypermethylation of FHIT as a prognostic marker in nonsmall cell lung carcinoma. Cancer. 2004;100:1472–1477. doi: 10.1002/cncr.20144. [DOI] [PubMed] [Google Scholar]
- 16.Safar AM, Spencer H, III, Su X, Coffey M, Cooney CA, Ratnasinghe LD, Hutchins LF, Fan CY. Methylation profiling of archived non-small cell lung cancer: A promising prognostic system. Clin Cancer Res. 2005;11:4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 17.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, ZochbauerMuller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Wang X, Ying J, Wong AH, Cui Y, Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, et al. Epigenetic silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proc Natl Acad Sci USA. 2007;104:12353–12358. doi: 10.1073/pnas.0700153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohta M, Seto M, Ijichi H, Miyabayashi K, Kudo Y, Mohri D, Asaoka Y, Tada M, Tanaka Y, Ikenoue T, et al. Decreased expression of the RAS-GTPase activating protein RASAL1 is associated with colorectal tumor progression. Gastroenterology. 2009;136:206–216. doi: 10.1053/j.gastro.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 20.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seto M, Ohta M, Ikenoue T, Sugimoto T, Asaoka Y, Tada M, Mohri D, Kudo Y, Ijichi H, Tateishi K, et al. Reduced expression of RAS protein activator like-1 in gastric cancer. Int J Cancer. 2011;128:1293–1302. doi: 10.1002/ijc.25459. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-betal-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, Ozaki I, Petersen I. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–646. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 24.Woenckhaus M, KleinHitpass L, Grepmeier U, Merk J, Pfeifer M, Wild P, Bettstetter M, Wuensch P, Blaszyk H, Hartmann A, et al. Smoking and cancer-related gene expression in bronchial epithelium an non-small-cell lung cancers. J Pathol. 2006;210:192–204. doi: 10.1002/path.2039. [DOI] [PubMed] [Google Scholar]
- 25.Dawson JC, Timpson P, Kalna G, Machesky LM. Mtss1 regulates epidermal growth factor signaling in head and neck squamous carcinoma cells. Oncogene. 2012;31:1781–1793. doi: 10.1038/onc.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K, Wang G, Ding H, Chen Y, Yu G, Wang J. Downregulation of metastasis suppressor 1 (MTSS1) is associated with nodal metastasis and poor outcome in Chinese patients with gastric cancer. BMC Cancer. 2010;10:428. doi: 10.1186/1471-2407-10-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie F, Ye L, Chen J, Wu N, Zhang Z, Yang Y, Zhang L, Jiang WG. The impact of metastasis suppressor-1, MTSS1, on oesophageal squamous cell carcinoma and its clinical significance. J Transl Med. 2011;9:95. doi: 10.1186/1479-5876-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, Sikandar SS, Gümüs ZH, Lipkin SM. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012;2:540–553. doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam DC, Girard L, Suen WS, Chung L, Tin VP, Lam WK, Minna JD, Wong MP. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol. 2006;1:932–942. doi: 10.1016/S1556-0864(15)31624-5. [DOI] [PubMed] [Google Scholar]
- 30.Bova GS, MacGrogan D, Levy A, Pin SS, Bookstein R, Isaacs WB. Physical mapping of chromosome 8p22 markers and their homozygous deletion in a metastatic prostate cancer. Genomics. 1996;35:46–54. doi: 10.1006/geno.1996.0321. [DOI] [PubMed] [Google Scholar]
- 31.Pils D, Horak P, Vanhara P, Anees M, Petz M, Alfanz A, Gugerell A, Wittinger M, Gleiss A, Auner V, et al. Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer. 2013;119:946–954. doi: 10.1002/cncr.27850. [DOI] [PubMed] [Google Scholar]
- 32.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 33.Guervòs MA, Marcos CA, Hermsen M, Nuño AS, Suárez C, Llorente JL. Deletions of N33, STK11 and TP53 are involved in the development of lymph node metastasis in larynx and pharynx carcinomas. Cell Oncol. 2007;29:327–334. doi: 10.1155/2007/635962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholz C, Nimmrich I, Burger M, Becker E, Dörken B, Ludwig WD, Maier S. Distinction of acute lymphoblastic leukemia from acute myeloid leukemia through microarray-based DNA methylation analysis. Ann Hematol. 2005;84:236–244. doi: 10.1007/s00277-004-0969-1. [DOI] [PubMed] [Google Scholar]
- 35.Zemlyakova VV, Zhevlova AI, Zborovskaya IB, Strelnikov VV, Laktionov KK, Zaletaev DV, Nemtsova MV. Methylation profile of several tumor suppressor genes in non-small-cell lung cancer. Mol Biol. 2003;37:836–840. doi: 10.1023/B:MBIL.0000008351.36435.d6. [DOI] [PubMed] [Google Scholar]
- 36.Bettstetter M, Dechant S, Ruemmele P, Vogel C, Kurz K, Morak M, Keller G, HolinskiFeder E, Hofstaedter F, Dietmaier W. MethyQESD, a robust and fast method for quantitative methylation analyses in HNPCC diagnostics using formalin-fixed and paraffin-embedded tissue samples. Lab Invest. 2008;88:1367–1375. doi: 10.1038/labinvest.2008.100. [DOI] [PubMed] [Google Scholar]
- 37.Kubo T, Yamamoto H, Ichimura K, Jida M, Hayashi T, Otani H, Tsukuda K, Sano Y, Kiura K, Toyooka S. DNA methylation in small lung adenocarcinoma with bronchioloalveolar carcinoma components. Lung Cancer. 2009;65:328–332. doi: 10.1016/j.lungcan.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Lee SM, Lee WK, Kim DS, Park JY. Quantitative promoter hypermethylation analysis of RASSF1A in lung cancer: Comparison with methylation-specific PCR technique and clinical significance. Mol Med Rep. 2012;5:239–244. doi: 10.3892/mmr.2011.608. [DOI] [PubMed] [Google Scholar]
- 39.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive melting analysis after real time-methylation specific PCR (SMART-MSP): High-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grote HJ, Schmiemann V, Kazimirek M, Böcking A. Quantitative methylation-specific PCR for the diagnosis of lung cancer. Pathologe. 2007;28:377–383. doi: 10.1007/s00292-007-0929-3. (In German) [DOI] [PubMed] [Google Scholar]
- 41.Cao Z, Yoon JH, Nam SW, Lee JY, Park WS. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. J Cancer Res Clin Oncol. 2012;138:611–619. doi: 10.1007/s00432-011-1140-8. [DOI] [PubMed] [Google Scholar]
- 42.Utikal J, Gratchev A, MullerMolinet I, Oerther S, Kzhyshkowska J, Arens N, Grobholz R, Kannookadan S, Goerdt S. The expression of metastasis suppressor MIM/MTSS1 is regulated by DNA methylation. Int J Cancer. 2006;119:2287–2293. doi: 10.1002/ijc.22106. [DOI] [PubMed] [Google Scholar]
- 43.Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X, Zhao Z, Kong KL, Dong S, Song Y, et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene. 2012;31:2298–2308. doi: 10.1038/onc.2011.411. [DOI] [PubMed] [Google Scholar]
- 44.Leupold JH, Asangani IA, Mudduluru G, Allgayer H. Promoter cloning and characterization of the human programmed cell death protein 4 (pdcd4) gene: Evidence for ZBP-89 and Sp-binding motifs as essential Pdcd4 regulators. Biosci Rep. 2012;32:281–297. doi: 10.1042/BSR20110045. [DOI] [PubMed] [Google Scholar]
- 45.Garshasbi M, Hadavi V, Habibi H, Kahrizi K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH, Kuss AW. A defekt in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy A, Dang UC, Bookstein R. High-density screen of human tumor cell lines for homozygous deletions of loci on chromosome arm 8p. Genes Chromosomes Cancer. 1999;24:42–47. doi: 10.1002/(SICI)1098-2264(199901)24:1<42::AID-GCC6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]