Abstract
Previous studies have shown that microRNA-186 (miR-186) is overexpressed in various human cancers and is associated with the regulation of the carcinogenic processes. However, the underlying mechanisms of this microRNA in melanoma remain largely unknown. In the present study, the overexpression of miR-186 was identified in melanoma tissues and melanoma cells compared to the expression of miR-186 in the matched tumor adjacent tissues and normal human epidermal melanocytes. Overexpression of miR-186 promoted the proliferation and anchorage-independent growth of melanoma cells, whereas inhibition of miR-186 reduced this effect. Bioinformatics analysis also revealed cylindromatosis (CYLD), a putative tumor suppressor, to be a potential target of miR-186. Luciferase reporter assays showed that miR-186 directly targeted the 3′-untranslated regions of CYLD messenger RNA. Additional experiments showed that overexpression of miR-186 promoted the proliferation of melanoma cells, which was consistent with the inhibitory effects induced by knockdown of CYLD. In summary, the present study indicated that miRNA-186 plays a crucial role in melanoma growth and its oncogenic effect is mediated chiefly through the direct suppression of CYLD expression.
Keywords: miR-186, melanoma, CYLD, cell proliferation
Introduction
Human melanoma is one of the most aggressive and frequently diagnosed cancers in humans (1). For numerous years, the incidence of melanoma has continually increased, despite advancements in the understanding of the initiation and progress of melanoma (2). Therefore, the identification of important molecules in the progression of melanoma is urgently required, in order to develop new preventive and diagnostic strategies targeting these markers (3,4).
An increasing number of studies have documented that microRNAs (miRNAs) play essential roles in multiple cancers, which lead to messenger (m)RNA degradation by targeting the 3′-untranslated region of target mRNAs (5,6). Among several miRNAs regulating cell proliferation, miRNA-186 (miR-186) has been shown to be one of the important determinants of cell proliferation in various types of cancers (7–10). The results of the present study revealed that upregulation of miR-186 in melanoma is associated with development of melanoma. Additional findings that miR-186 directly targets cylindromatosis (CYLD), thereby promoting cell proliferation of human melanoma cells. In summary, the present findings suggested that miR-186 is mediated with progression of melanoma and may serve as a new target for treatment of melanoma.
Materials and methods
Clinical specimens
Skin tissues were obtained from 8 patients and histopathologically diagnosed following surgery at Guangzhou First People's Hospital, Guangzhou Medical University (Guangzhou, China). The present study was approved by the Ethics Committee of Guangzhou First People's Hospital, Guangzhou Medical University (Guangzhou, China). All samples were collected and analyzed with the written informed consent of the patients.
Cell culture
The human melanoma A375-S2, SKMEL-28, SKMEL-5, MeWO and RPMI-7951 cell lines were purchased from the National Rodent Laboratory Animal Resource (Shanghai, China). All melanoma cell lines were grown in Gibco Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) and 100 units/ml Invitrogen penicillin-streptomycin (Thermo Fisher Scientific, Inc.). Normal human epidermal melanocytes (NHEMs) from adult skin (PromoCell GmbH, Heidelberg, Germany) were maintained in serum- and phorbol myristate acetate-free melanocyte growth medium M2 (PromoCell GmbH). The cell lines were cultured in a humidified incubator at 37°C in a 5% CO2 and 95% air atmosphere for 2–3 days.
Plasmids and transfection
To induce the ectopic expression of CYLD, CYLD open reading frames containing a 3′-UTR was amplified by polymerase chain reaction (PCR) and then cloned into pGL3 vectors (Promega Corporation, Madison, WI, USA) downstream of the the Renilla luciferase complementary DNA. miR-186 mimic, miR-186 inhibitor, miR-186-mut and negative control (NC) were purchased from GeneCopoeia, Inc. (Guangzhou, China) and transfected into melanoma cells using Invitrogen Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using Invitrogen TRIzol Reagent (Thermo Fisher Scientific, Inc.) and RT was performed using the miScript Reverse Transcription kit (Qiagen, Hilden, Germany). For the quantification of miRNA expression, qPCR was performed using the miScript Reverse Transcription and miScript SYBR Green PCR kits, according to the manufacturer's instructions (Qiagen). The relative miR-186 expression levels following normalization to the expression of U6 small nuclear RNA were calculated using 2-ΔΔCq (11). Quantitative PCR was performed using the QuantiNova SYBR Green PCR Kit (Qiagen China Co., Ltd., Shanghai, China) and a 7500 Sequence Detection system (Applied Biosystems Life Technologies, Foster City, CA, USA). The primers selected were as follows: Cyclin D1 forward, 5′-TCCTCTCCAAAATGCCAGAG-3′ and reverse, 5′-GGCGGATTGGAAATGAACTT-3′; p21 forward, 5′-CATGGGTTCTGACGGACAT-3′ and reverse, 5′-AGTCAGTTCCTTGTGGAGCC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GACTCATGACCACAGTCCATGC-3′ and reverse, 3′-AGAGGCAGGGATGATGTTCTG-5′. The PCR reaction conditions for all assays were as follows: 95°C for 30 sec, followed by 40 cycles of amplification (95°C for 5 sec, 59°C for 30 sec and 72°C for 30 sec). The expression data was normalized to the geometric mean of GAPDH to control the variability in expression levels and calculated using 2-ΔΔCq.
Methyl thiazolyl tetrazolium (MTT) and colony formation assays
The viability of the SKMEL-28 cells was measured by MTT assay. The SKMEL-28 cells (3,000 cells/well) were seeded in 96-well plates in medium containing 10% FBS. The cell cultures were stained on days 1–5. At the end of the stipulated time, 20 µl of 5 mg/ml MTT solution (Sigma-Aldrich) was added to each well and incubated for 4 h at 37°C. Following the incubation period, the culture medium was removed and 150 µl dimethyl sulfoxide (Sigma-Aldrich) was added. The absorbance at 490 nm was measured using a Multiskan microplate reader (Thermo Fisher Scientific, Inc.).
For the colony formation assay, the SKMEL-28 cells were plated in 6-well plates at a density of 1,000 cells per well, and then incubated for 10 days in medium containing 10% FBS. The colonies were stained with crystal violet (Beyotime Biotechnology Institute of Biotechnology, Inc., Shanghai, China) and counted using an Olympus BX41 microscope fitted with the cellSens software (Olympus, Center Valley, PA, USA).
Anchorage-independent growth assay
In total, 1,000 SKMEL-28 cells were resuspended in 2 ml complete medium supplemented with 0.3% agar (Sigma-Aldrich). The agar-cell mixture was plated on top of a bottom layer consisting of 1% agar in complete medium. The cells were incubated for 14 days at 37°C until colony formation occurred, and the colonies were then stained with 0.5% crystal violet prior to counting under a microscope (DM IRBE; Leica, Heidelberg, Germany), and images of the cell colonies were captured at an original magnification of ×100. Colonies >0.1 mm in diameter were counted.
Flow cytometric analysis
A total of 48 h subsequent to transfection, the SKMEL-28 cells were harvested and washed with PBS, and fixed with 70% ethanol. The fixed cells were then treated with 20 µg/ml RNaseA and 50 µg/ml propidium iodide (Sigma-Aldrich) for 30 min, and stained cells were immediately analyzed by a FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Luciferase assays
The pGL3-luciferase reporter gene plasmid pGL3-CYLD-3′-UTR (GeneCopoeia, Guangzhou, China) was cotransfected into the cells with 15 pmol of the miR-186 mimic, miR-186 control or miR-186-mut and 5 ng pRL-TK TKRenilla plasmid (Promega Corporation) using Invitrogen Lipofectamine 2000 reagent. The activity of firefly and andRenilla luciferase was assessed 48 h subsequent to transfection using the dual luciferase assay kit (Beyotime Biotechnology Institute of Biotechnology, Inc.), according to the manufacturer's instructions. The results are expressed as the ratio of of Renilla luciferase activity to firefly luciferase activity.
Western blotting
Protein lysates were prepared using radioimmunoprecipitation assay buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) and equal quantities of protein were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Beyotime Biotechnology Institute of Biotechnology, Inc.). The gels were transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) for 2 h at 4°C, run at a current of 125 mA and blocked for 2 h. The membrane was incubated overnight with the following rabbit antibodies anti-CYLD (#8462), anti-cyclin D1 (#2978) and anti-p21 antibodies (#2947; all primary antibodies dilution, 1:1,000; Cell Signaling Technology, Inc.) and washed with Tris-buffered saline and Tween-20 (TBST; Beyotime Biotechnology Institute of Biotechnology, Inc.) three times, for 5 min each time. For control sample loading, the blotting membranes were stripped and re-probed with an anti-β-actin antibody (dilution, 1:5,000; A5316; Sigma-Aldrich). Subsequent to washing with TBST and incubation with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich) for 2 h at room temperature, immunocomplexes were visualized using chemiluminescence (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA), according to the manufacturer's instructions.
Statistical analysis
All statistical analyses, with the exception of microarray data, were performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Student's t-test was used to evaluate the significance of the differences between two groups of data in all the relevant experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-186 expression was upregulated in melanoma tissues and melanoma cell lines
To investigate the role of miR-186 expression in melanoma development, the present study evaluated the expression levels of miR-186 in melanoma tissues and melanoma cells by RT-qPCR.
The current results demonstrated that the expression levels of miR-186 in the melanoma tissues were consistently increased compared with the matched normal tissues adjacent to the tumor. Additionally, all 5 tested melanoma cell lines, consisting of the A375-S2, SKMEL-28, SKMEL-5, MeWO and RPMI-7951 cell lines, demonstrated significantly increased miR-186 levels compared with the levels in NHEMs. These results revealed that miR-186 played an important role in melanoma. Overall, these results suggest that miR-186 expression is significantly increased in melanoma and may act as a prognostic marker for patients with melanoma.
miR-186 promoted melanoma cell proliferation
In order to explore the effects of miR-186 on the growth of melanoma cells, the SKMEL-28 cells were transfected with miR-186 mimics, miR-186 inhibitor or the respective controls. The miR-186 precursor was found to upregulate miR-186 expression (Fig. 2A). The results of the MTT assays showed that miR-186 significantly promoted the proliferation of SKMEL-28 cells (Fig. 2B), and this was additionally confirmed by a colony formation assay (Fig. 2C). By contrast, the cell growth rates and colony numbers of the SKMEL-28 cells transfected with miR-186-in were significantly inhibited compared with the growth rates and colony numbers of cells transfected with the NC (Fig. 2B and C). In addition, miR-186-overexpressing SKMEL-28 cells had a significantly lower percentage of cells in the G1/G0 phase and increased percentage of cells in the S phase compared with the NC-transfected cells (Fig. 2D). miR-186-in demonstrated the opposite effects to miR-186. These results revealed that miR-186 promoted melanoma tumorigenicity in vitro.
Figure 2.
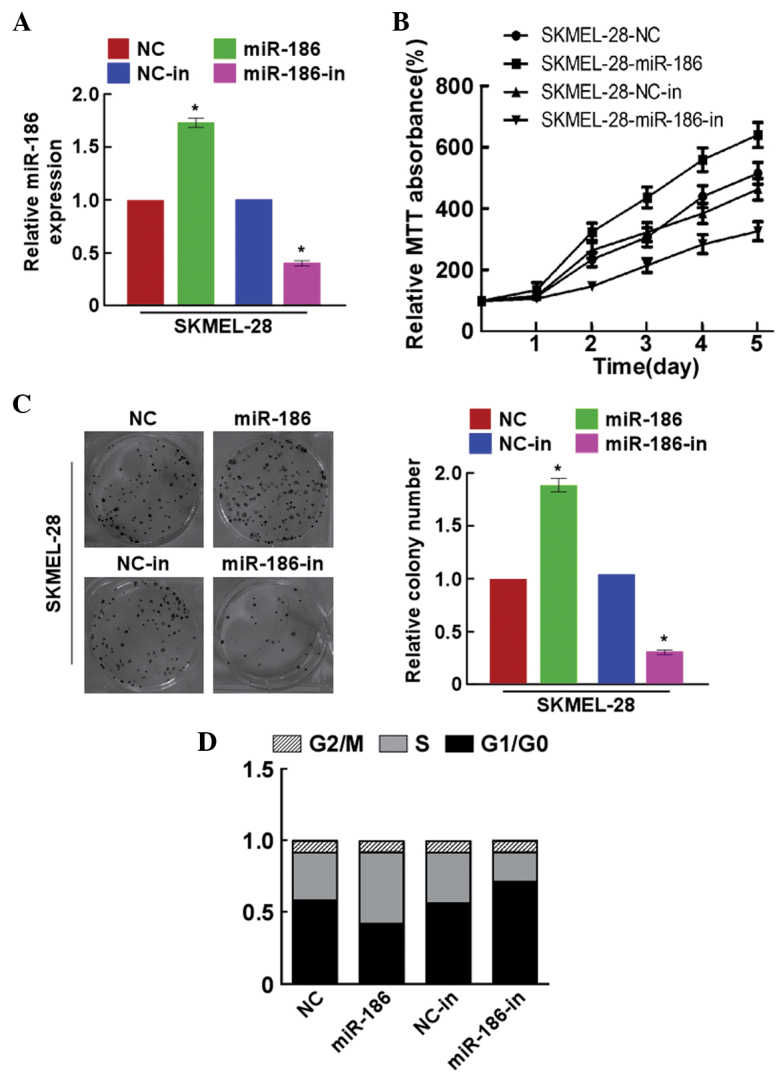
miR-186 upregulation promoted melanoma cell proliferation. (A) Validation of miR-186 expression levels by polymerase chain reaction analysis subsequent to transfection. (B) MTT assays revealed that upregulation of miR-186 promoted the growth of SKMEL-28 cells and miR-186-in inhibited the growth of SKMEL-28 cells. (C) Representative micrographs (left) and quantification (right) of crystal violet staining of the cell colonies. (D) Cell cycle distribution in SKMEL-28 cells with miR-186, miR-186-in or miR-NC transfection. *P<0.05 versus control. miR-186, microRNA-186; in, inhibitor; NC, negative control; PCR, polymerase chain reaction; MTT, methyl thiazolyl tetrazolium.
miR-186 directly targets CYLD by binding to its 3′-UTR and alters the levels of proteins associated with cell proliferation and cell cycle in melanoma cells
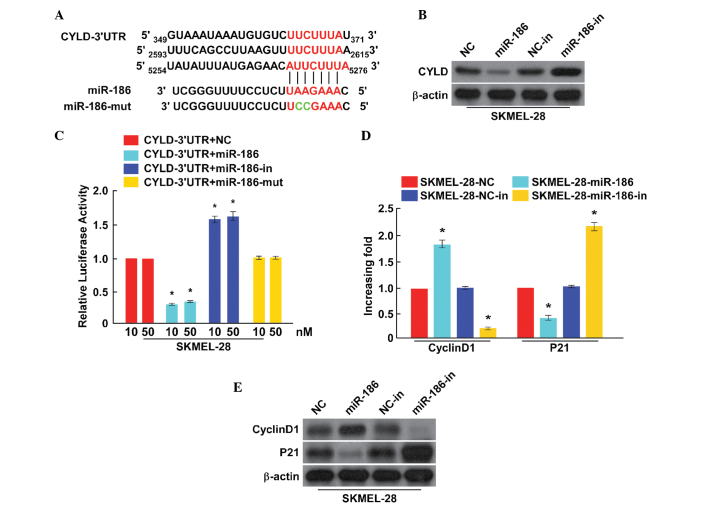
To explore the mechanism that facilitates the effects on cell proliferation and cell cycle induced by miR-186, the putative targets of miR-186 were analyzed by a bioinformatics screen that was developed using TargetScan. The present study focused on CYLD, from which originated a 3′-UTR containing a binding site of miR-186 (Fig. 3A).
Figure 3.
miR-186 suppresses CYLD expression by directly targeting the CYLD 3′-UTR and alters the levels of proteins associated with proliferation and cell cycle progression in SKMEL-28 cells. (A) Predicted miR-186c target sequence in the CYLD 3′-UTR and positions of 3 mutated nucleotides (red) in the 3′-UTR of miR-186-mut. (B) CYLD protein expression in SKMEL-28 cells transfected with miR-186 or miR-186-in was detected by western blot analysis. β-actin acted as the loading control. (C) Luciferase reporter assay of SKMEL-28 cells transfected with the pGL3-CYLD-3′-UTR reporter and miR-186, miR-186-in or miR-186-mut with increasing amounts (10 and 50 nM) of oligonucleotides. (D) Quantitative polymerase chain reaction analysis of the expression of cyclin D1 and p21 in SKMEL-28 cells. (E) Western blot analysis of the protein expression of cyclin D1 and p21 in SKMEL-28 cells. β-actin acted as the loading control. *P<0.05 versus respective control. CYLD, cylindromatosis; miR-186, microRNA-186; in, inhibitor; NC, negative control; miR-186-mut, mutated miR-186; 3′-UTR, 3′ untranslated region.
To verify the oncogenic role of miR-186, the present study investigated the expression level of CYLD protein through transfection with miR-186 mimics/inhibitors in SKMEL-28 cells. The over-expression of miR-186 was found to result in a significant reduction of CYLD protein expression in SKMEL-28 cells, while miR-186-in clearly promoted CYLD protein expression (Fig. 3B). A luciferase reporter assay was performed to determine whether miR-186 directly regulates the expression of CYLD in SKMEL-28 cells. As shown in Fig. 3C, the results indicated that a consistent and dose-dependent reduction of luciferase activity was observed in SKMEL-28 cells transfected with the miR-186 mimic, whereas the repressive effect of miR-186-in increased wild-type CYLD luciferase activity. However, overexpressing miR-186-mut had no effect on the luciferase activity of CYLD 3′-UTR. Overall, these results demonstrated that CYLD is a target of miR-186.
It was demonstrated that miR-186 promoted the growth and proliferation of melanoma and CYLD was a direct target of miR-186 (12). Previously, it has been reported that CYLD expression is associated with cell proliferation and the cell cycle. In addition, the present study observed alteration in the mRNA expression of the CYLD downstream genes cyclin D1 and p21, which are critical cell-proliferation and cell-cycle regulators. As shown in Fig. 4D, the cyclin D1 levels in SKMEL-28 cells were all significantly upregulated by miR-186, and downregulated by miR-186-in, while the expression of p21 was downregulated in cells transfected with miR-186 and upregulated in cells transfected with miR-186-in. Altogether, the present results indicated that miR-186 functionally modulates cellular proliferation and the cycle regulators cyclin D1 and p21, and are therefore relevant to cell proliferation and the cell cycle.
Discussion
Numerous studies have shown that miRNAs are a class of diverse, small, noncoding RNAs that function as critical gene regulators, and then potentially play essential roles in multiple biological processes, including cell differentiation, proliferation, angiogenesis, invasion and migration (4,13–15). It is widely recognized that miRNAs perform important roles in tumorigenesis by targeting various mRNAs. At present, the abnormal expression of several miRNAs, such as miRNA-573 (16), miRNA-106b (17), miRNA-524-5p (18) and miRNA-451a (19), has been identified in melanoma and may contribute to the development and progression of melanoma. In the present study, miR-186 was identified as a tumor-promoter miRNA in human melanoma that acts by repressing CYLD, which has been previously identified as a tumor suppressive protein, provides additional evidence of a pivotal role for miRNAs in the tumorigenesis and progression of melanoma.
Previously, miRNAs have emerged as an important regulator of various physiological and pathological processes in cancer cells (10,20,21). However, the functional role of miR-186 in the tumorigenesis of melanoma remains largely unknown. In the present study, it was found that miR-186 expression was markedly upregulated in melanoma tissues and cells. Additional experiments revealed that miR-186 overexpression dramatically promoted the proliferation of SKMEL-28 cells, accompanied by a decrease in the proportion of cells in the G1/G0 phase and an increase in the proportion of cells in the S phase, while miR-186-in showed the opposite function. These findings suggested that miR-186 was involved in the processes of the development of melanoma.
CYLD, a deubiquitination enzyme, functions as a tumor suppressor in various types of cancer (22,23). To improve the understanding of the underlying mechanisms of miR-186-induced melanoma cell growth and cell cycle progression, CYLD was identified as a potential target gene of miR-186 using a bioinformatics algorithm. The present results confirmed a vital molecular association between miR-186 and CYLD. The present experiment revealed that upregulation of miR-186 expression in SKMEL-28 cells effectively suppressed CYLD expression, while miR-186 increased the expression of the CYLD protein. Studies have indicated that CYLD interferes with cell proliferation and cell survival of cancer (23–25). The expression levels of a number of critical cell-proliferation regulators were also detected. It has also been reported that CDK proteins play critical roles in cell proliferation and cell cycle transition. Among the cell proliferation and cell cycle-associated genes, the CDK inhibitor p21 and the CDK regulator cyclin D1 were focused on in the current study. The present results showed that ectopic miR-186 expression suppressed the expression level of the cell cycle inhibitor p21, and increased the expression of the cell cycle regulator cyclin D1, consequently leading to a decrease in the proportion of SKMEL-28 cells in the G0/G1 phase and an increase the proportion of SKMEL-28 cells in the S phase.
In conclusion, the current study revealed that miR-186 expression may potentially act as a method to determine the progression of melanoma. The present results revealed that miR-186 regulated melanoma cell growth and cell cycle progression by targeting the suppression of CYLD expression. Therefore, all the results indicated that miR-186 was an oncomiR and may act as a potential therapeutic target for the treatment of melanoma.
Figure 1.
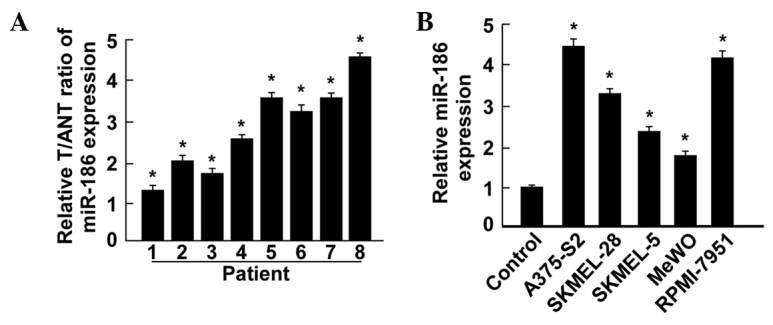
Expression of miR-186 in human melanoma tissues and cell lines. (A) Relative miR-186 mRNA expression levels in 8 paired Ts and ANTs from patients were detected by PCR analysis. (B) Quantitative PCR analysis of miR-186 expression in the control normal human epidermal melanocytes and melanoma cell lines, consisting of the A375-S2, SKMEL-28, SKMEL-5, MeWO and RPMI-7951 cell lines. Experiments were repeated at least three times. Each bar indicates the mean of three independent experiments. *P<0.05 versus control. T, primary melanoma tissue; ANT, matched adjacent non-tumor tissue; miR-186, microRNA-186; PCR, polymerase chain reaction.
Acknowledgements
This study was supported by the Department of Ophthalmology, Nansha Hospital, Guangzhou First People's Hospital, Guangzhou Medical University.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Baroudjian B, Pagès C, Lebbé C. Melanoma, from diagnosis to treatment. Rev Infirm. 2016;65:16–18. doi: 10.1016/j.revinf.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Luo C, Weber CE, Osen W, Bosserhoff AK, Eichmuller SB. The role of microRNAs in melanoma. Eur J Cell Biol. 2014;93:11–22. doi: 10.1016/j.ejcb.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Mimeault M, Batra SK. Novel biomarkers and therapeutic targets for optimizing the therapeutic management of melanomas. World J Clin Oncol. 2012;3:32–42. doi: 10.5306/wjco.v3.i3.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Cui G, Cui M, Li Y, Liang Y, Li W, Guo H, Zhao S. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol. 2014;35:8933–8937. doi: 10.1007/s13277-014-2168-6. [DOI] [PubMed] [Google Scholar]
- 8.Kim SY, Lee YH, Bae YS. MiR-186, miR-216b, miR-337-3p and miR-760 cooperatively induce cellular senescence by targeting α subunit of protein kinase CKII in human colorectal cancer cells. Biochem Biophys Res Commun. 2012;429:173–179. doi: 10.1016/j.bbrc.2012.10.117. [DOI] [PubMed] [Google Scholar]
- 9.Cai J, Wu J, Zhang H, Fang L, Huang Y, Yang Y, Zhu X, Li R, Li M. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–766. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- 10.Ries J, Vairaktaris E, Agaimy A, Kintopp R, Baran C, Neukam FW, Nkenke E. miR-186, miR-3651 and miR-494: Potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol Rep. 2014;31:1429–1436. doi: 10.3892/or.2014.2983. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Mathis BJ, Lai Y, Qu C, Janicki JS, Cui T. CYLD-mediated signaling and diseases. Curr Drug Targets. 2015;16:284–294. doi: 10.2174/1389450115666141024152421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Q, Wang T, Lu P, Zhang R, Zou J, Yuan S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J Neurosci Res. 2014;92:619–626. doi: 10.1002/jnr.23339. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Wang Z, Sun G, Wan Y, Guo J, Fu X. miR-145 inhibits migration and invasion of glioma stem cells by targeting ABCG2. Neuromolecular Med. 2014;16:517–528. doi: 10.1007/s12017-014-8305-y. [DOI] [PubMed] [Google Scholar]
- 16.Wang HF, Chen H, Ma MW, Wang JA, Tang TT, Ni LS, Yu JL, Li YZ, Bai BX. miR-573 regulates melanoma progression by targeting the melanoma cell adhesion molecule. Oncol Rep. 2013;30:520–526. doi: 10.3892/or.2013.2451. [DOI] [PubMed] [Google Scholar]
- 17.Prasad R, Katiyar SK. Down-regulation of miRNA-106b inhibits growth of melanoma cells by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget. 2014;5:10636–10649. doi: 10.18632/oncotarget.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5:9444–9459. doi: 10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babapoor S, Fleming E, Wu R, Dadras SS. A novel miR-451a isomiR, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PLoS One. 2014;9:e107502. doi: 10.1371/journal.pone.0107502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rico-Rosillo MG, Vega-Robledo GB, Oliva-Rico D. The role and importance of the microRNAs in the diagnosis and development of diseases. Rev Med Inst Mex Seguro Soc. 2014;52:302–307. (In Spanish) [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Deng LL, Shao YX, Lv HF, Deng HB, Lv FZ. Over-expressing CYLD augments antitumor activity of TRAIL by inhibiting the NF-κB survival signaling in lung cancer cells. Neoplasma. 2012;59:18–29. doi: 10.4149/neo_2012_003. [DOI] [PubMed] [Google Scholar]
- 23.Massoumi R. CYLD: A deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7:285–297. doi: 10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]
- 24.Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, et al. CYLD negatively regulates transforming growth factor-β-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blake PW, Toro JR. Update of cylindromatosis gene (CYLD) mutations in Brooke-Spiegler syndrome: Novel insights into the role of deubiquitination in cell signaling. Hum Mutat. 2009;30:1025–1036. doi: 10.1002/humu.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]