Abstract
Branched-chain amino acid transaminase 1 (BCAT1) has been associated with numerous types of tumors; however, few previous studies have evaluated the expression and role of BCAT1 in hepatocellular carcinoma (HCC). In the present study, the expression of BCAT1 was detected by reverse transcription-quantitative polymerase chain reaction and immunoblotting in six HCC cell lines and 74 pairs of HCC and adjacent non-cancerous liver tissues. In addition, the correlation between the expression levels of c-Myc and BCAT1 was analyzed using immunohistochemistry. Furthermore, RNA silencing was performed using c-Myc-specific or BCAT1-specific small interfering RNA, after which wound healing and Transwell cell invasion assays were performed. Finally, the clinicopathological characteristics of BCAT1 in patients with HCC were analyzed. It was shown that the expression of BCAT1 was significantly higher in HCC tissues compared with adjacent non-tumor tissues (P<0.001), and in HCC cell lines compared within the L-02 hepatic cell line (P<0.001). In addition, immunohistochemical analyses indicated that the expression of BCAT1 was positively correlated with c-Myc (r=0.706, P<0.001). BCAT1 expression was shown to be downregulated in c-Myc-knockdown cells, and silencing of BCAT1 expression reduced the invasion and migration of HCC cells. Furthermore, a clinical analysis indicated that BCAT1 expression in HCC tissues was significantly associated with the tumor-node-metastasis stage, tumor number and tumor differentiation (all P<0.05), and that BCAT1 was able to predict the 5-year survival and disease-free survival rates of patients with HCC (both P<0.001). The results of the present study suggested that BCAT1 expression is upregulated in patients with HCC, and that BCAT1 may serve as a potential molecular target for the diagnosis and treatment of HCC.
Keywords: branched-chain amino acid transaminase 1, hepatocellular carcinoma, prognostic factor, c-Myc
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and its incidence is increasing. Furthermore, HCC is the third leading cause of cancer-associated mortality worldwide, partly due to its high recurrence rate and early metastasis (1,2). In 2000, there were 564,000 new cases and 549,000 mortalities from HCC worldwide, indicating the devastating prognosis of this tumor (3). In 2008, 746,300 new cases of HCC were diagnosed worldwide, and 695,900 HCC-related mortalities were reported. In total, >700,000 new cases are diagnosed each year throughout the world and >600,000 mortalities are attributed to HCC each year (4). At present, the majority of patients with HCC are diagnosed at the advanced stage due to lack of specific clinical manifestations, meaning that patients often miss out on the chance of receiving curative treatments, such as liver resection (5). In addition, patients with HCC often have a poor prognosis due to the aggressive nature of the malignancy, including a high recurrence rate and metastasis. Therefore, an improved understanding of the mechanisms underlying the recurrence and metastasis of HCC is required in order to identify effective prognostic and therapeutic biomarkers of HCC.
Branched-chain amino acid transaminase 1 (BCAT1), which is also known as cytosolic branched-chain aminotransferase and ECA39, is located at chromosome 12p12.1. It encodes the cytosolic form of the branched-chain amino acid transaminase enzyme, which catalyzes the reversible transamination of branched-chain α-keto acids to branched-chain L-amino acids essential for cell growth (6–10). It has previously been suggested that the aberrant expression of BCAT1, and the concomitant defect in branched-chain amino acid transamination, leads to hypervalinemia and hyperleucine-isoleucinemia, and may have an important role in the cell growth, proliferation and apoptosis of numerous tumor types (8,11–14). Furthermore, BCAT1 overexpression has been reported in non-neoplastic diseases of the liver, including chronic hepatitis C and non-alcoholic fatty liver disease (15–17). However, the expression and role of BCAT1 in HCC remains unclear.
Previous studies have reported that BCAT1 serves as an oncogenic protein that is upregulated by several signaling molecules, including c-Myc (18–20). c-Myc is an oncogene and transcription factor involved in the tumorigenesis of multiple cancers, including Burkitt's lymphoma and breast cancer, by targeting genes harboring the c-Myc-binding element (CACGTG) downstream of their transcription start site (11). Therefore, c-Myc may have an important role in the development and progression of HCC (20).
BCAT1 has previously been associated with numerous malignancies due to its role in cell proliferation, cell cycle progression, differentiation and apoptosis (8,10–14). However, little is known regarding the role of BCAT1 in HCC. To the best of our knowledge, the present study is the first to assess the association between BCAT1 and HCC. The study aimed to determine whether BCAT1 may serve as a potential prognostic and therapeutic biomarker for HCC.
Materials and methods
Cell lines
The L-02, SMMC-7721, BEL-7402, Huh-7, HepG2 and MHCC-97H cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), and cultured in a humidified 5% CO2 incubator at 37°C.
Patients and specimens
A total of 74 HCC and matched normal adjacent samples (>2 cm distance from the margin of the resection) were obtained from pathologically confirmed HCC patients who had undergone surgical resection at the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) between October 2005 and September 2008. None of the patients had received any pre-operative chemotherapy or radiotherapy, and patients with evidence of concomitant extrahepatic disease were excluded from the analysis. HCC stage was classified according to the seventh edition of the tumor-node-metastasis (TNM) classification criteria of the International Union Against Cancer (21). The present study included 56 males and 18 females with a median age of 52 years (range, 33–75 years). All HCC tissues and matched pericarcinous liver tissues were immediately snap-frozen in liquid nitrogen following surgery and stored at −80°C until use. Hepatitis B surface antigen (HBsAg) and α-fetoprotein (AFP) levels were obtained from the results of laboratory tests, capsule formation was observed during surgery and Edmonson-Steiner grade (22) was evaluated by an experienced pathologist. All information was recorded for each case. All patients provided informed consent prior to surgery, and all protocols were performed in accordance with the 1975 Declaration of Helsinki. The present study was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HCC cell lines and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. In order to avoid DNA contamination, the extracted RNA was treated with RNase-free DNase I (Invitrogen; Thermo Fisher Scientific, Inc.) and quantified by spectrophotometry. Subsequently, cDNA was synthesized using the RevertAid Premium First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). qPCR was performed using the Applied Biosystems 7500 Real-Time PCR system (Thermo Fisher Scientific, Inc.) and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; Takara Bio, Inc., Otsu, Japan). The primer sequences were as follows: BCAT1 forward, 5′-CCAAAGCCCTGCTCTTTGTA-3′ and reverse, 5′-TGGAGGAGTTGCCAGTTCTT-3′; and β-actin (internal control) forward, 5′-GGGAAATCGTGCGTGACAT-3′ and reverse, 5′-CTGGAAGGTGGACAGCGAG-3′. The reaction conditions for the PCR program were as following: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 64 sec. Melting curve analyses were performed to confirm the specificity of the PCR product. Relative mRNA expression levels were determined using the 2−ΔΔCq method (23). Reactions were performed in triplicate.
Immunoblotting
Total protein was extracted from HCC cell lines and tissues using radioimmunoprecipitation buffer (catalog no. WB009; HEART Biological Technology Co. Ltd., Xi'an, China). Protein concentration was determined using the bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.). Denatured protein samples (25 µg) were separated by 10% polyacrylamide gel electrophoresis, and then electrophoretically transferred onto polyvinylidene fluoride membranes (Merck Millipore). The membranes were blocked with 5% skimmed milk in Tris-buffered saline with Tween (TBST) at 37°C for 2 h, and subsequently incubated with the primary antibodies overnight at 4°C; the membranes were incubated with rabbit anti-BCAT1 polyclonal antibody (cat. no. ab197941; 1:1,000 dilution; Abcam, Cambridge, UK), mouse anti-c-Myc monoclonal antibody (cat. no. ab32; 1:1,000 dilution; Abcam) and mouse anti-β-actin monoclonal antibody (cat. no. ab6276; 1:10,000 dilution; Abcam). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (cat. no. ab6721; 1:3,000 dilution; Abcam) and rabbit anti-mouse (cat. no. ab6728; 1:3,000 dilution; Abcam) secondary antibodies at 37°C for 2 h, followed by washing three times with TBST. Protein bands were detected using the Western Blotting Luminol Reagent (cat. no. sc-2048, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The reactions were detected using the HyGLO™ Chemiluminescent HRP Detection kit (Denville Scientific, Inc., Holliston, MA, USA). The band density was measured using Image Lab 4.0 (Bio-Rad Laboratories Inc., Hercules, CA, USA) imaging software. Expression levels of the protein are assessed by a densitometric ratio of the targeted protein to the β-actin housekeeping protein.
Immunohistochemical staining
Immunohistochemical staining was performed using paraformaldehyde-fixed, paraffin-embedded tissue sections, which were prepared according to a method described previously (14). The tissue sections were incubated with rabbit anti-BCAT1 polyclonal antibody (cat. no. ab197941; 1:50 dilution; Abcam) and mouse anti-C-Myc monoclonal antibody (cat. no. ab32; 1:200 dilution; Abcam) overnight at 4°C, followed by incubation with biotinylated goat anti-rabbit (cat. no. SV0002; Wuhan Boster Biological Technology, Ltd., Wuhan, China) and rabbit anti-mouse (cat. no. SV0001; Wuhan Boster Biological Technology, Ltd.) secondary antibodies at 37°C for 1 h. Each slide was colored with DAB (Sigma-Aldrich, St. Louis, MO, USA) in a dark room, then all the sections were rinsed with running water and counterstained with hematoxylin (cat. no. ST047; HEART Biological Technology Co. Ltd.). Subsequently, the tissue sections were assessed by light microscopy and evaluated blindly and independently by two experienced pathologists. To evaluate the association between the expression of BCAT1 and c-Myc, a semi-quantitative scoring system based on the staining intensity and the percentage of positive liver cells was applied. Immunostaining intensity was evaluated as one of the following four grades: 0, negative; 1 weak; 2, moderate; and 3, strong. The percentage of positive liver cells was categorized into one of the following groups: 0, 0%; 1, 1–10%; 2, 11–50%; 3, 51–80%; and 4, >80%. The immunostaining intensity and average percentage of positive cells were evaluated for 10 independent high magnification fields. The final weighted expression score (0–12) was obtained by multiplying the staining intensity with the percentage of positive cells. The total expression scores for BCAT1 and c-Myc were listed as continuous variables for the correlation analyses. In order to evaluate the effect of BCAT1 protein expression on overall survival, the weighted expression scores of BCAT1 protein were divided into high and low scores using the median expression score as the cutoff point.
Small interfering RNA (siRNA) transfection
siRNAs targeting c-Myc (cat. no. sc-29226) and BCAT1 (cat. no. sc-77222), as well as control siRNA (cat. no. sc-37007), were purchased from Santa Cruz Biotechnology, Inc. MHCC-97H tumor cells were seeded at a density of 2×105 cells per well into six-well plates and cultured overnight in a humidified 5% CO2 incubator at 37°C. Subsequently, the cells were transfected with 100 nM of the BCAT1, c-Myc or control siRNA using Lipofectamine RNAi MAX Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Further experiments were performed after 48 h of transfection.
Transwell invasion assay
Matrigel was diluted in serum-free DMEM (1:3) and added to the upper chamber of a 24-well Transwell plate. HCC cells were trypsinized and counted manually under a light microscope. A cell suspension of 5×104 cells/ml in serum-free medium was prepared and 100 µl of the suspension was loaded into the upper chamber. The lower chambers were filled with 10% FBS in DMEM. Invasion was halted in a 37°C incubator (5% CO2) after ~24 h by removing the non-migrated cells from the upper chamber using a cotton swab. The HCC cells that had migrated through the membrane were stained with 0.05% crystal violet after fixing with 4% paraformaldehyde, and were counted under a microscope. At least five fields were randomly selected for counting the mean number of invaded cells in each membrane using ImageJ v1.48 software (NIH, Bethesda, MD, USA). At least three experimental replicates were performed.
Wound healing assay
MHCC97H cells transfected with BCAT1 or control siRNA were seeded at a concentration of 5×105 per well onto 6-well plates and cultured to full confluency. Scratch wounds were made across the surface of the plates using a 10-µl pipette tip and the suspension cells were removed using phosphate-buffered saline. Cells were cultured in serum-free DMEM medium in a humidified 5% CO2 incubator at 37°C for 48 h, after which images of the plates were captured using a phase-contrast microscope. At least five replicate experiments were performed.
Follow-up
Follow-up of the patients in the present study was performed on December 31, 2013. The duration was defined as the interval between the date of surgery and the date of mortality or last follow-up. The follow-up time ranged from 6–78 months and the median time was 58.5 months. All patients received follow-up visits once every 1–3 months in the first year and every 3–6 months thereafter. The follow-up protocol included a physical examination, measurement of serum AFP levels, a chest X-ray and abdominal ultrasonography. Computed tomography, magnetic resonance imaging or positron emission tomography was performed to assess the occurrence of tumor recurrence. During the follow-up period, 59 patients (79.7%) were shown to have intrahepatic tumor recurrence and 11 patients (14.9%) had developed distant tumor metastases.
Statistical analysis
Statistical analyses were performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The Spearman's rank correlation coefficient was applied to evaluate the association between ordinal data, and the χ2 test or Fisher's exact test was performed for comparisons of categorical data. The expression levels between groups were compared using the Mann-Whitney U test. Overall survival and disease-free survival rates, and mortalities associated with tumor recurrence or metastasis, were analyzed using the Kaplan-Meier method, and differences between curves were assessed using the log-rank test. Independent prognostic factors were assessed by the Cox proportional hazards stepwise regression model. Data are presented as the mean ± standard error of the mean. P-values were two-sided. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of BCAT1 in HCC tissues and cells
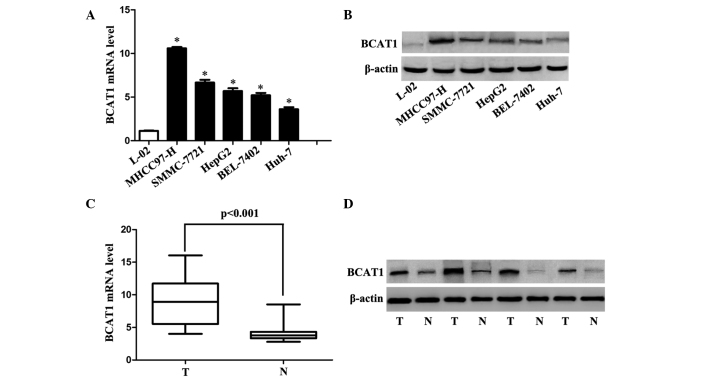
The expression levels of BCAT1 in cell lines and tissues were determined using RT-qPCR and western blotting. The expression levels of BCAT1 were significantly lower in the L-02 cells compared with the HCC cell lines (all P<0.001; Fig. 1A and B). Similarly, BCAT1 expression levels were significantly higher in HCC tissues compared with adjacent non-cancerous liver tissues (P<0.001; Fig. 1C and D).
Figure 1.
Expression of BCAT1 in HCC cell lines and tissues. (A) The mRNA expression levels of BCAT1 in the immortalized L-02 normal human liver cell line and five HCC cell lines (SMMC-7721, BEL-7402, Huh-7, HepG2 and MHCC-97H) were determined using RT-qPCR (n=3; *P<0.001 vs. L-02 cells). (B) The protein expression levels of BCAT1 in the L-02 and HCC cell lines were determined using immunoblotting (n=3; P<0.001). (C) The mRNA expression levels of BCAT1 in the HCC tissues and corresponding adjacent normal tissues were determined using RT-qPCR (n=74; P<0.001). (D) The protein expression levels of BCAT1 in the HCC tissues and corresponding adjacent normal tissues were determined using immunoblotting (n=3). HCC, hepatocellular carcinoma; BCAT1, branched-chain amino acid transaminase 1; T, HCC tissues; N, non-tumorous tissues; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Association between BCAT1 expression and clinicopathological parameters
To investigate the clinical significance of BCAT1 in patients with HCC, the associations between the BCAT1 expression levels (high or low) and clinicopathological parameters, including patient gender, age, detection of HBsAg, AFP level, tumor size, tumor number, vascular invasion, cirrhosis, capsule formation, Edmondson-Steiner grade and TNM stage, were investigated. The median expression score of BCAT1 protein was used as a cutoff point to divide patients into high and low expression groups. Notably, the expression levels of BCAT1 were significantly associated with the Edmondson-Steiner grade, tumor number, vascular invasion and TNM stage (all P<0.05). However, no significant association was observed between the expression levels of BCAT1 and the patient gender, age, HBsAg, AFP level, cirrhosis and capsule formation (all P>0.05). The results are shown in Table I.
Table I.
Associations between BCAT1 expression and clinicopathologic features of patients with hepatocellular carcinoma.
| BCAT1 protein, n | ||||
|---|---|---|---|---|
| Characteristics | n | High | Low | P-value |
| Gender | 0.787 | |||
| Female | 18 | 8 | 10 | |
| Male | 56 | 29 | 27 | |
| Age, years | 1.000 | |||
| ≤45 | 17 | 9 | 8 | |
| >45 | 57 | 28 | 29 | |
| HBsAg status | 0.674 | |||
| Negative | 6 | 4 | 2 | |
| Positive | 68 | 33 | 35 | |
| Cirrhosis | 0.754 | |||
| No | 12 | 7 | 5 | |
| Yes | 62 | 30 | 32 | |
| AFP, µg/l | 0.634 | |||
| ≤400 | 29 | 13 | 16 | |
| >400 | 45 | 24 | 21 | |
| Tumor size, cm | 0.087 | |||
| ≤5 | 26 | 17 | 9 | |
| >5 | 48 | 20 | 28 | |
| Tumor number | 0.003a | |||
| Single | 47 | 17 | 30 | |
| Multiple | 27 | 20 | 7 | |
| Tumor capsule | 0.074 | |||
| Complete | 52 | 30 | 22 | |
| Incomplete | 22 | 7 | 15 | |
| Vascular invasion | 0.017a | |||
| No | 54 | 22 | 32 | |
| Yes | 20 | 15 | 5 | |
| Edmondson grade | 0.027a | |||
| I/II | 48 | 19 | 29 | |
| III/IV | 26 | 18 | 8 | |
| TNM stage | 0.017a | |||
| I+II | 45 | 17 | 28 | |
| III+IV | 29 | 20 | 9 | |
P<0.05. BCAT1, branched-chain amino acid transaminase 1; HbsAg, hepatitis B surface antigen; AFP, α-fetoprotein; TNM, tumor-node-metastasis.
High expression levels of BCAT1 are associated with a poor HCC prognosis
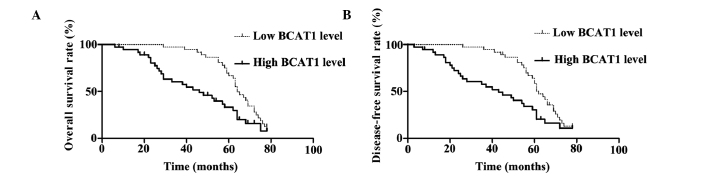
The median expression score of BCAT1 protein was used as a cutoff point to divide patients into high and low expression groups for a clinical association analysis. Univariate prognostic analyses and multivariate Cox regression models were applied to assess the association between the expression levels of BCAT1 and the overall and disease-free survival rates (Fig. 2). The patients with high BCAT1 expression levels showed significantly reduced overall and disease-free survival rates (P=0.002). The 5-year overall survival rate of the low BCAT1 expression group was 66.8%, which was significantly higher than that of the high BCAT1 expression group (33.2%) (P=0.002). In addition, the 5-year disease-free survival rate of the low BCAT1 expression group was 58.5%, which was also significantly higher compared with that of the high BCAT1 expression group (30.5%) (Fig. 2). The associations between the overall and disease-free survival rates and the clinicopathological parameters in the HCC patients were determined by a univariate analysis. The univariate analysis demonstrated that vascular infiltration, tumor number, Edmonson-Steiner classification, TNM stage and the expression level of BCAT1 were all significant prognostic factors for HCC (P<0.05). The results are shown in Table II.
Figure 2.
Kaplan-Meier survival curves according to BCAT1 expression levels in patients with hepatocellular carcinoma. (A) Overall survival rates for high and low BCAT1 protein expression groups (log-rank, P=0.002). (B) Disease-free survival rates for high and low BCAT1 protein expression groups (log-rank, P=0.002). BCAT1, branched-chain amino acid transaminase 1.
Table II.
Univariate prognostic analysis of overall and disease-free survival rates in patients with hepatocellular carcinoma.
| Overall survival rate | Disease-free survival rate | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | n | 3-year, % | 5-year, % | P-value | 3-year, % | 5-year, % | P-value | |
| Gender | 0.891 | 0.895 | ||||||
| Female | 18 | 82.4 | 51.0 | 82.4 | 44.9 | |||
| Male | 56 | 78.2 | 50.1 | 74.6 | 44.5 | |||
| Age, years | 0.644 | 0.732 | ||||||
| ≤45 | 17 | 81.3 | 68.8 | 75.6 | 56.7 | |||
| >45 | 57 | 78.6 | 44.7 | 76.8 | 41.1 | |||
| HBsAg status | 0.079 | 0.073 | ||||||
| Negative | 6 | 50.0 | 33.3 | 50.0 | 33.3 | |||
| Positive | 68 | 81.9 | 52.0 | 78.9 | 45.7 | |||
| Cirrhosis | 0.235 | 0.318 | ||||||
| No | 12 | 66.7 | 50.0 | 66.7 | 50.0 | |||
| Yes | 62 | 81.8 | 50.4 | 78.5 | 43.5 | |||
| AFP, µg/l | 0.122 | 0.224 | ||||||
| ≤400 | 29 | 68.1 | 41.0 | 64.7 | 33.2 | |||
| >400 | 45 | 86.4 | 56.3 | 84.1 | 51.8 | |||
| Tumor size, cm | 0.764 | 0.778 | ||||||
| ≤5 | 26 | 60.1 | 47.5 | 60.4 | 47.7 | |||
| >5 | 48 | 89.4 | 52.0 | 85.2 | 47.8 | |||
| Tumor number | <0.001a | <0.001a | ||||||
| Single | 47 | 95.7 | 58.3 | 93.6 | 56.3 | |||
| Multiple | 27 | 48.4 | 36.3 | 44.9 | 23.0 | |||
| Tumor capsule | 0.739 | 0.715 | ||||||
| Complete | 52 | 76.5 | 44.0 | 72.6 | 44.3 | |||
| Incomplete | 22 | 85.7 | 66.0 | 85.7 | 45.7 | |||
| Vascular infiltration | 0.035a | 0.049a | ||||||
| No | 54 | 92.4 | 54.1 | 90.5 | 50.2 | |||
| Yes | 20 | 45.0 | 40.0 | 45.0 | 30.0 | |||
| Edmondson grade | <0.001a | <0.001a | ||||||
| I/II | 48 | 93.7 | 56.4 | 93.7 | 54.5 | |||
| III/IV | 26 | 46.3 | 37.9 | 42.6 | 25.6 | |||
| TNM Stage | 0.020a | 0.027a | ||||||
| I/II | 45 | 93.3 | 54.1 | 91.1 | 47.3 | |||
| III/IV | 29 | 48.5 | 40.7 | 45.2 | 32.9 | |||
| BCAT1 protein level | 0.004a | 0.004a | ||||||
| Low | 37 | 94.6 | 66.8 | 94.6 | 58.5 | |||
| High | 37 | 60.3 | 33.2 | 57.7 | 30.5 | |||
P<0.05. HbsAg, hepatitis B surface antigen; BCAT1, branched-chain amino acid transaminase 1; TNM, tumor-node-metastasis; AFP, α-fetoprotein.
Stratified univariate and multivariate analysis
In a multivariate analysis model, the expression levels of BCAT1 were significantly associated with the overall (HR, 2.546; 95% CI, 1.427–4.543; P=0.002) and disease-free (HR, 2.443; 95% CI, 1.392–4.290; P=0.002) survival rates (Table III). Multivariate analysis indicated that the expression level of BCAT1, Edmonson-Steiner classification and tumor number were all independent prognostic factors of HCC (Table III).
Table III.
Multivariate analysis of factors contributing to overall and disease-free survival rates in patients with hepatocellular carcinoma.
| Overall survival rate | Disease-free survival rate | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Vascular infiltration | 0.796 (0.300–2.116) | 0.648 | 0.785 (0.298–2.070) | 0.624 |
| Tumor number | 3.745 (1.162–12.064) | 0.027a | 3.337 (1.032–10.794) | 0.044a |
| TNM stage | 0.232 (0.052–1.044) | 0.057 | 0.252 (0.056–1.126) | 0.071 |
| Edmondson grade | 4.321 (1.074–17.379) | 0.039a | 4.101 (1.026–16.382) | 0.046a |
| BCAT1 protein level | 2.546 (1.427–4.543) | 0.002a | 2.443 (1.392–4.290) | 0.002a |
P<0.05. HR, hazard ratio; CI, confidence interval; BCAT1, branched-chain amino acid transaminase 1; TNM, tumor-node-metastasis.
Association between BCAT1 and c-Myc protein expression levels
To determine whether the expression level of BCAT1 was correlated with c-Myc expression in patients with HCC, immunohistochemical staining was performed. The protein expression levels of c-Myc protein level were significantly higher in the HCC tissues compared with the corresponding adjacent non-tumorous tissues (P<0.001; Fig. 3A). In addition, the correlation between BCAT1 and C-Myc protein expression levels was analyzed. Notably, there was a significant positive correlation between the protein expression levels of c-Myc and BCAT1 (r=0.706; P<0.001; Fig. 3B).
Figure 3.
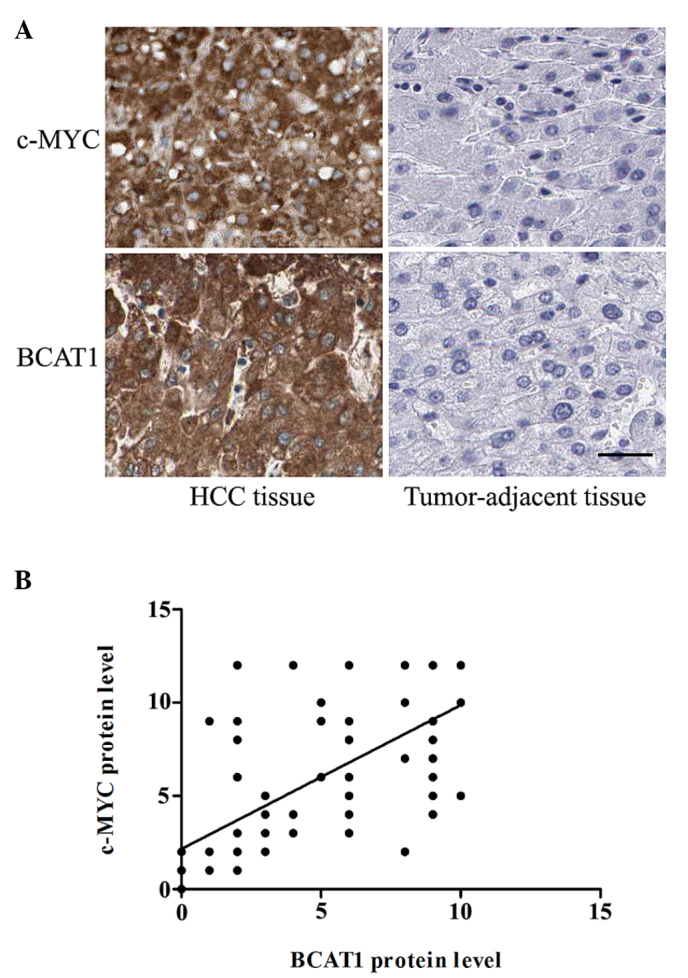
Immunohistochemical staining of BCAT1 and c-Myc in HCC and matched tumor-adjacent tissues. (A) In cases of high c-Myc protein expression, there was high BCAT1 protein expression in the same tissue section. While, in cases of low c-Myc protein expression, there was undetectable BCAT1 protein expression (scale bar, 50 µm; DAB counterstained with hematoxylin). (B) The correlation between the protein expression levels of C-Myc and BCAT1 was assessed by Spearman's rank correlation test (r=0.706, P<0.001), which showed a significant positive correlation between the levels. HCC, hepatocellular carcinoma; BCAT1, branched-chain amino acid transaminase 1.
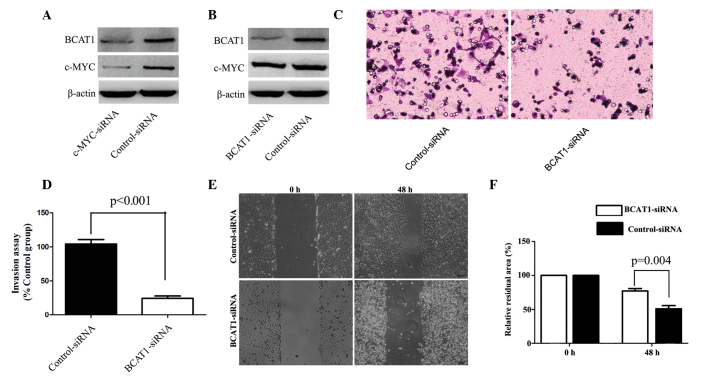
c-Myc-knockdown reduces BCAT1 expression
A previous study reported that c-Myc was able to upregulate BCAT1 expression in nasopharyngeal carcinoma (11). Therefore, to further elucidate the underlying mechanism of BCAT1 in HCC cells, MHCC-97H cells were transfected with c-Myc-specific siRNA. Silencing of c-Myc expression was shown to significantly downregulate the expression of BCAT1 in MHCC-97H cells (P=0.005; Fig. 4A). Furthermore, BCAT1-specific siRNA was used to knockdown the expression of BCAT1 in MHCC-97H cells. Compared with the control group, the silencing of BCAT1 did not significantly alter the protein expression levels of c-Myc (Fig. 4B).
Figure 4.
Effect of c-Myc- and/or BCAT1-knockdown on MHCC-97H cells. (A) The expression of c-Myc and BCAT1 in MHCC-97H cells transfected with c-Myc-specific or control siRNA was analyzed by western blotting. (B) The expression of c-Myc and BCAT1 in MHCC-97H cells transfected with BCAT1-specific or control siRNA was analyzed by western blotting. (C) Representative images of invaded MHCC-97H cells transfected with BCAT1-specific and control siRNA in the Transwell invasion assay (magnification, ×200). (D) The number of invaded MHCC-97H cells were counted under a microscope. Data are presented as the mean relative percentages of invaded cells from 5 fields. (E) Representative images showing the migration ability of MHCC-97H cells transfected with BCAT1-specific or control-siRNA, as assessed by wound healing assays (magnification, ×200). (F) Data are presented as the mean relative residual area from 5 fields. BCAT1, branched-chain amino acid transaminase 1; siRNA, small interfering RNA.
BCAT1-knockdown suppresses cell invasion and migration
To further investigate the underlying mechanism of BCAT1 in HCC, the effect of BCAT1 on MHCC-97H cell migration and invasion was investigated using BCAT1-specific and control siRNA. Compared with the control group, BCAT1-knockdown significantly repressed cell migration and invasion (Fig. 4C-F).
Discussion
At present, liver transplantation and radical liver resection are the main curative treatments for patients with HCC (24). However, the prognosis of HCC remains unsatisfactory and the overall survival rate of affected patients remains markedly low (25). Therefore, elucidation of the mechanisms underlying the pathogenesis of HCC is important in order to identify effective prognostic and therapeutic biomarkers for the disease. Increasingly, targeted therapy has exhibited encouraging outcomes for numerous tumors (26–29); thus, efforts should be made to identify novel tumor biomarkers with clinical utility for the treatment of HCC.
BCAT1, which is the cytosolic form of the enzyme BCAT, catalyzes the reversible transamination of branched-chain α-keto acids to branched-chain L-amino acids essential for cell growth (6–10). Previous studies identified positive BCAT1 expression in c-Myc-induced tumors, and demonstrated that BCAT1 was directly regulated by c-Myc through its binding to the specific DNA sequence, CACGTG. c-Myc is an oncogene and transcription factor involved in the tumorigenesis of numerous cancers, including Burkitt's lymphoma and breast cancer. Furthermore, it was reported that c-Myc upregulated BCAT1 expression, which in turn led to the proliferation, migration and invasion of nasopharyngeal carcinoma (11). Although BCAT1 has been investigated in several tumor types, including colorectal cancer (6), few previous studies have evaluated the role of BCAT1 in HCC (30). Considering that c-Myc regulates BCAT1 expression, the present study aimed to verify whether c-Myc is involved in the expression of BCAT1 in HCC tissues and cell lines. Since carcinogenesis is characterized by uncontrolled cell growth and alterations in the expression patterns of various molecules associated with the modulation of cell migration and invasion (8,11–13), the identification of these molecules is valuable for the development of effective therapeutic strategies.
In the present study, the expression levels of BCAT1 in several cell lines were initially detected, and it was demonstrated that the expression levels of BCAT1 in HCC cell lines were significantly higher compared within the L-02 immortalized normal human liver cell line. In addition, the expression levels of BCAT1 in tumor tissues derived from a relatively large population of HCC patients were determined, and it was shown that the expression levels of BCAT1 were upregulated in HCC tissue compared with tumor-adjacent tissues. Further studies demonstrated that the expression levels of BCAT1 protein were positively correlated with those of c-Myc, which indicated that c-Myc may be partially responsible for the high expression levels of BCAT1 in HCC tissues and cell lines. For better elucidation of the role and underlying mechanisms of BCAT1 in HCC cells, the effects of c-Myc- and BCAT1-knockdown on MHCC-97H cells were investigated. As was expected, c-Myc-knockdown was found to downregulate BCAT1 expression in MHCC-97H cells. Furthermore, the expression of BCAT1 was associated with the biological characteristics of HCC cells, since it was demonstrated that knockdown of BCAT1 expression repressed the migration and invasion of MHCC-97H cells. Taken together, these results support the hypothesis that BCAT1 has a critical role in the migration and invasion of HCC, and that its expression may be regulated by c-Myc. Therefore, BCAT1 may serve as a potential biomarker for the diagnosis and treatment of HCC.
In the present study, the associations between the expression levels of BCAT1 and the clinicopathological parameters and prognosis of patients with HCC were analyzed. It was demonstrated that the upregulation of BCAT1 was significantly correlated with lower overall and disease-free survival rates, and other clinicopathological parameters, including the Edmondson-Steiner grade, tumor number, vascular invasion and TNM stage.
In conclusion, the present study demonstrated that BCAT1 was upregulated in HCC tissue samples and cell lines compared with normal adjacent tissue samples and the L-02 immortalized normal human liver cell line, respectively. Furthermore, the BCAT1 expression level was positively correlated with c-Myc expression, and knockdown of c-Myc in HCC cells resulted in the downregulation of BCAT1. In addition, knockdown of BCAT1 expression was shown to repress the migration and invasion of an HCC cell line. The results of the present study suggested that BCAT1 is important for the migration and invasion of HCC and may represent a novel prognostic biomarker for the disease.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (nos. 81402039, 81272645 and 81301743).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: An update. Ann Intern Med. 2003;139:817–823. doi: 10.7326/0003-4819-139-10-200311180-00009. [DOI] [PubMed] [Google Scholar]
- 3.Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: Current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: Current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med. 2012;4:19–37. doi: 10.2147/HMER.S16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng X, Song T, Dou C, Jia Y, Liu Q. CtBP2 is an independent prognostic marker that promotes GLI1 induced epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2015;6:3752–3769. doi: 10.18632/oncotarget.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshikawa R, Yanagi H, Shen CS, Fujiwara Y, Noda M, Yagyu T, Gega M, Oshima T, Yamamura T, Okamura H, et al. ECA39 is a novel distant metastasis-related biomarker in colorectal cancer. World J Gastroenterol. 2006;12:5884–5889. doi: 10.3748/wjg.v12.i36.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuldiner O, Eden A, Ben-Yosef T, Yanuka O, Simchen G, Benvenisty N. ECA39, a conserved gene regulated by c-Myc in mice, is involved in G1/S cell cycle regulation in yeast. Proc Natl Acad Sci USA. 1996;93:7143–7148. doi: 10.1073/pnas.93.14.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eden A, Benvenisty N. Involvement of branched-chain amino acid aminotransferase (Bcat1/Eca39) in apoptosis. FEBS Lett. 1999;457:255–261. doi: 10.1016/S0014-5793(99)01054-6. [DOI] [PubMed] [Google Scholar]
- 9.Bledsoe RK, Dawson PA, Hutson SM. Cloning of the rat and human mitochondrial branched chain aminotransferases (BCATm) Biochim Biophys Acta. 1997;1339:9–13. doi: 10.1016/S0167-4838(97)00044-7. [DOI] [PubMed] [Google Scholar]
- 10.Eden A, Simchen G, Benvenisty N. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J Biol Chem. 1996;271:20242–20245. doi: 10.1074/jbc.271.34.20242. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W, Liu Z, Li Z, Zeng L, Wang L, et al. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12:53. doi: 10.1186/1476-4598-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bible E. Neuro-oncology: BCAT1 promotes cell proliferation in aggressive gliomas. Nat Rev Neurol. 2013;9:420. doi: 10.1038/nrneurol.2013.135. [DOI] [PubMed] [Google Scholar]
- 13.Zerban H, Radig S, Kopp-Schneider A, Bannasch P. Cell proliferation and cell death (apoptosis) in hepatic preneoplasia and neoplasia are closely related to phenotypic cellular diversity and instability. Carcinogenesis. 1994;15:2467–2473. doi: 10.1093/carcin/15.11.2467. [DOI] [PubMed] [Google Scholar]
- 14.Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, et al. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1281–G1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura T, Morinaga Y, Fujitani S, Takehana K, Nishitani S, Sonaka I. Oral administration of branched-chain amino acids activates the mTOR signal in cirrhotic rat liver. Hepatol Res. 2005;33:27–32. doi: 10.1016/j.hepres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Honda M, Takehana K, Sakai A, Tagata Y, Shirasaki T, Nishitani S, Muramatsu T, Yamashita T, Nakamoto Y, Mizukoshi E, et al. Malnutrition impairs interferon signaling through mTOR and FoxO pathways in patients with chronic hepatitis C. Gastroenterology. 2011;141:128–140. doi: 10.1053/j.gastro.2011.03.051. 140.e1-e2. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Yosef T, Yanuka O, Halle D, Benvenisty N. Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene. 1998;17:165–171. doi: 10.1038/sj.onc.1201939. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Yosef T, Eden A, Benvenisty N. Characterization of murine BCAT genes: Bcat1, a c-Myc target, and its homolog, Bcat2. Mamm Genome. 1998;9:595–597. doi: 10.1007/s003359900825. [DOI] [PubMed] [Google Scholar]
- 20.Peng SY, Lai PL, Hsu HC. Amplification of the c-myc gene in human hepatocellular carcinoma: Biologic significance. J Formos Med Assoc. 1993;92:866–870. [PubMed] [Google Scholar]
- 21.Liu Q, Tu K, Zhang H, Zheng X, Yao Y, Liu Q. TPX2 as a novel prognostic biomarker for hepatocellular carcinoma. Hepatol Res. 2015;45:906–18. doi: 10.1111/hepr.12428. [DOI] [PubMed] [Google Scholar]
- 22.Edmondson HA, Steiner PE. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–504. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Colombo M, Sangiovanni A. Treatment of hepatocellular carcinoma: Beyond international guidelines. Liver Int. 2015;35(Suppl 1):S129–S138. doi: 10.1111/liv.12713. [DOI] [PubMed] [Google Scholar]
- 25.Bruix J, Sherman M. American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts LR, Gores GJ. Hepatocellular carcinoma: Molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 27.Scaggiante B, Kazemi M, Pozzato G, Dapas B, Farra R, Grassi M, Zanconati F, Grassi G. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J Gastroenterol. 2014;20:1268–1288. doi: 10.3748/wjg.v20.i5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satow R, Shitashige M, Kanai Y, Takeshita F, Ojima H, Jigami T, Honda K, Kosuge T, Ochiya T, Hirohashi S, Yamada T. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16:2518–2528. doi: 10.1158/1078-0432.CCR-09-2214. [DOI] [PubMed] [Google Scholar]
- 29.Schattenberg JM, Schuchmann M, Galle PR. Cell death and hepatocarcinogenesis: Dysregulation of apoptosis signaling pathways. J Gastroenterol Hepatol. 2011;26(Suppl 1):S213–S219. doi: 10.1111/j.1440-1746.2010.06582.x. [DOI] [PubMed] [Google Scholar]
- 30.Elsemman IE, Mardinoglu A, Shoaie S, Solima TH, Nielsen J. Systems biology analysis of hepatitis C virus infection reveals the role of copy number increases in regions of chromosome 1q in hepatocellular carcinoma metabolism. Mol Biosyst. 2016;12:1496–1506. doi: 10.1039/C5MB00827A. [DOI] [PubMed] [Google Scholar]