Abstract
The long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) has been identified as an oncogene in numerous diseases, and aberrant lncRNA PVT1 expression has been associated with the development of cancer. However, the underlying mechanism by which lncRNA PVT1 affects cell invasion in esophageal cancer has been not demonstrated. In the current study, the expression of lncRNA PVT1 was found to be increased in esophageal cancer specimens (n=77) by reverse transcription-quantitative polymerase chain reaction, and was correlated with tumor stage (P=0.009) and metastasis (P<0.001). In vitro, by using transwell assay, upregulation of lncRNA PVT1 promoted the invasion of TE-1 esophageal cancer cells; while downregulation of lncRNA PVT1 inhibited Eca-109 cell invasion. In addition, western blot analysis indicated that upregulation of lncRNA PVT1 may induce epithelial-to-mesenchymal transition (EMT) by regulating the expression levels of EMT markers (E-cadherin, N-cadherin and vimentin). In conclusion, lncRNA PVT1 is able to regulate the invasion of esophageal cancer cells by inducing EMT.
Keywords: lncRNA PVT1, invasion, epithelial-mesenchymal transition, esophageal cancer
Introduction
Esophageal cancer is currently the sixth leading cause of cancer-associated mortality and the eighth most diagnosed type of cancer in China (1), where the incidence of esophageal cancer is markedly increasing, most likely due to dietary habits (2). Although great efforts have been made towards the improvement of diagnosis and therapy, the prognosis of patients with esophageal cancer remains poor (3). Common features of esophageal cancer are metastasis and relapse, which are responsible for the low 5-year survival rates of ~14% (4). Thus, to investigate the mechanism of cancer cell invasion may provide new insight into cancer progression and aid in developing improved therapies for esophageal cancer.
Long non-coding RNAs (lncRNAs) are >200 nucleotides in length and possess no protein-coding capacity (5). lncRNAs have been reported to function in multiple biological processes associated with cancer progression, such as proliferation (6), apoptosis (7) and invasion (8) of cancer cells. The gene lncRNA plasmacytoma variant translocation 1 (PVT1), which is located at 8q24.21, plays an oncogenic role in various types of human cancer, including pancreatic cancer (9), lung cancer (10) and gastric cancer (11). However, lncRNA PVT1 expression in esophageal cancer and the underlying mechanisms of its effects remain unclear.
The epithelial-to-mesenchymal transition (EMT) has been confirmed to be important in cell invasion in different cancer types (12,13). EMT is closely associated with the transformation and infiltration of tumor cells. Much research on the association between lncRNAs and tumor development has focused on the expression of EMT-associated proteins, such as the epithelial marker E-cadherin, and the mesenchymal markers N-cadherin and vimentin (14). The breakdown of tight junctions is associated with the loss of epithelial markers and acquisition of mesenchymal makers (15–17).
The current study was performed with the aim of assessing the expression of lncRNA PVT1 in esophageal cancer, and exploring the potential mechanism of lncRNA PVT1 in cell invasion. In addition, the effect of abnormal expression of lncRNA PVT1 in the regulation of EMT marker expression was investigated.
Materials and methods
Clinical samples
A total of 77 patients with esophageal cancer who had undergone routine surgery at Huai'an First People's Hospital (Huai'an, China) between May 2008 and November 2013 were enrolled. Esophageal cancer tissues and the adjacent tissues were obtained from the 77 patients; all the samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. Each patient provided written informed consent, and the study was approved by the Ethics Committee of Huai'an First People's Hospital.
Cell culture
Esophageal cancer cell lines (Kyse140, TE-1, EC18, Eca-109 and HKESC1) and a normal esophageal cell line (NEEC) were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultured in Invitrogen RPMI-1640 medium with 10% fetal bovine serum (FBS) and 100 U/ml penicillin (all from Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) at 37°C in an atmosphere of 5% CO2.
Plasmid construction and cell transfection
The sequence of lncRNA PVT1 was synthesized by Genewiz, Inc. (Suzhou, China). The plasmid was subcloned into a lentiviral vector (pLV-GFP; Addgene, Inc., Cambridge, MA, USA) and then co-transfected into HEK-293T cells (SiXin Bio Company, Shanghai, China) with Lentiviral Packaging Mix (Genewiz, Inc., Suzhou, China). TE-1 cells were transfected with lentivirus to produce cells with high expression of lncRNA PVT1 (LV-lncRNA), and underwent selection with G418. Cells transfected with LV-vector constituted the control group.
RNA interference
Small interfering RNA (siRNA; 50 nM) specifically targeting lncRNA PVT1 and a corresponding scrambled siRNA negative control (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) were transfected into Eca-109 cells in 6-well plates using Invitrogen Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
Isolation of total RNA and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the collected tissues using Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc.), and mRNA was reverse transcribed into cDNA. lncRNA PVT1 expression was quantified using a LightCycler Brilliant SYBR Green RT-qPCR kit (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer's protocol, with the following primers: Forward, TTGGCACATACAGCCATCAT; and reverse, GCAGTAAAAGGGGAACACCA. β-actin was used for normalization, with the following primers: Forward, AGCGAGCATCCCCCAAAGTT; and reverse, GGGCACGAAGGCTCATCATT. PCR was then performed on ABI PRISM 7500 System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR conditions were as follows: 94°C for 2 min, followed by 94°C for 30 sec, 60°C for 30 sec and 72°C for 1 min for 30 cycles, and 72°C for 10 min. Every independent experiment was performed three times. The 2−∆∆Cq method (18) was used to evaluate the results of RT-qPCR in all the experiments.
Wound healing assay
The transfected cells were plated into 6-well plates and cultured with RPMI-1640 medium. After 24 h, the transfected cells were wounded with a pipette tip. Serum-free RPMI-1640 medium was added, and wound closure was observed for 48 h.
Transwell assay
A transwell invasion assay was performed with BioCoat Matrigel (BD Biosciences, San Jose, CA, USA) and invasion chambers (Merck Millipore, Eschborn, Germany) with an 8-µm pore size. The transfected cells were treated with trypsin/ethylenediaminetetraacetic acid solution, washed once with serum-containing RPMI-1640 medium. Cells (1×105) in 0.2 ml serum-free RPMI-1640 medium were seeded on a Transwell apparatus. RPMI-1640 containing 10% FBS (600 µl) was added to the lower chamber. According to the manufacturer's instructions, an invasion assay was conducted following the same procedure, with the exception that the filters of the transwell chambers were coated with 45 µg Matrigel. Following incubation of the cells for 24 h at 37°C in a 5% CO2 incubator, cells on the top surface of the insert were removed by wiping with a cotton swab. Cells that invaded to the bottom surface of the insert were fixed in the 100% precooled methanol for 10 min, stained in 0.5% crystal violet for 30 min, then rinsed in phosphate-buffered saline (PBS) and subjected to microscopic inspection. The values for invasion were obtained by counting three fields per membrane and represented the average of three independent experiments.
Western blot analysis
Total proteins were collected from the established cells and quantified using a bicinchoninic acid Protein Assay Kit (Beyotime, Haimen, China). Proteins were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, blocked in 5% dry milk at 37°C temperature for 1 h, and immunostained with antibodies at 4°C overnight. Antibodies against vimentin (rabbit monoclonal anti-human vimentin; 1:100; ab76601) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; rabbit monoclonal anti-human GAPDH; 1:5,000; ab70699), N-cadherin (rabbit polyclonal anti-human N-cadherin; 1:5,000; ab18203) and E-cadherin (rabbit polyclonal anti-human E-cadherin; 1:1,000; ab15148) were purchased from Abcam (Cambridge, MA, USA). Next, the membranes were washed four times with PBS containing 0.1% Tween 20 (PBS-T; Sigma-Aldrich, St. Louis, MO, USA), and the secondary antibody (goat polyclonal anti-rabbit antibody; 1:1,000; ab6721; Abcam) was then added in PBS-T for 1 h at 37°C. The membranes were next washed three times for 15 min with PBS-T. The staining was revealed using a Pierce enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.), and exposed in a Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories, Inc., Hercules, CA). The integrated density of the band was quantified by ImageJ version 1.48u software (https://imagej.nih.gov/ij/).
Statistical methods
Statistical analysis was performed using STATA 11 software (StataCorp LP, College Station, TX, USA), and presented with GraphPad Prism version 4.0 (GraphPad Software, San Diego, CA, USA). The results obtained from experiment in vitro assays are presented as the mean ± standard error of the mean from five separate experiments in triplicates per experiment, and the data was analyzed by Wilcoxon rank-sum (Mann-Whitney) test. P<0.05 was considered to indicate statistically significant differences.
Results
lncRNA PVT1 is increased in human esophageal cancer tissues
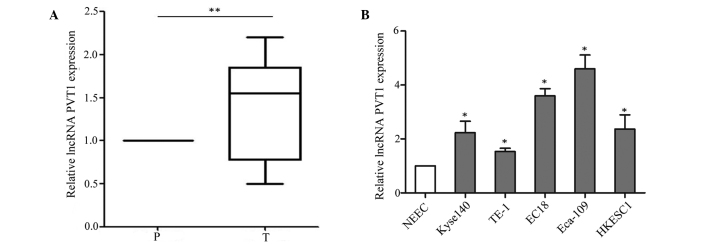
First, the expression of lncRNA PVT1 was analyzed in esophageal cancer samples (n=77) and adjacent tissues by RT-qPCR. lncRNA PVT1 expression was significantly higher in esophageal cancer tissues than adjacent normal tissues (P=0.002) (Fig. 1A). Its expression showed no association with gender, age, histological type or tumor size; however, there were significant associations with tumor stage (P=0.009) and metastasis (P<0.001) (Table I). The aberrant expression level of lncRNA PVT1 in the cancer tissues suggested that lncRNA PVT1 may play an important role in the development of esophageal cancer. Furthermore, the expression of lncRNA PVT1 was assessed in esophageal cancer cell lines (Kyse140, TE-1, EC18, Eca-109 and HKESC1) and a normal esophageal cell line (NEEC). The RT-qPCR assay indicated that lncRNA PVT1 was higher in esophageal cancer cell lines (Kyse140, TE-1, EC18, Eca-109 and HKESC1) than the normal esophageal cell line (NEEC) (P=0.016) (Fig. 1B). Based on this expression pattern, the TE-1 and Eca-109 cell lines were selected to verify the effect of lncRNA PVT1.
Figure 1.
lncRNA PVT1 is increased in esophageal cancer tissues and cell lines. (A) Expression levels of lncRNA PVT1 in human esophageal cancer tissues and corresponding adjacent tissues relative to β-actin were determined by RT-qPCR. (n=77; **P=0.002). (B) The expression levels of lncRNA PVT1 in esophageal cancer cell lines (Kyse140, TE-1, EC18, Eca-109 and HKESC1) and a normal esophageal cell line (NEEC) relative to β-actin were detected by RT-qPCR. Data are presented as the mean ± standard error of the mean. *P<0.05 for Kyse140, TE-1, EC18, Eca-109 and HKESC1 vs NEEC. lncRNA PVT1, long non-coding RNA plasmacytoma variant translocation 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table I.
Expression level of lncRNA PVT1 in esophageal cancer tissues.
| lncRNA PVT1 expression, n | ||||
|---|---|---|---|---|
| Factor | Samples, n | High (≥median) | Low (<median) | P-value |
| Total | 77 | 39 | 38 | |
| Gender | 0.554 | |||
| Male | 33 | 18 | 15 | |
| Female | 44 | 21 | 23 | |
| Age (years) | 0.420 | |||
| <60 | 36 | 20 | 16 | |
| ≥60 | 41 | 19 | 22 | |
| Histological types | 0.424 | |||
| Squamous cell carcinoma | 38 | 21 | 17 | |
| Adenocarcinoma | 39 | 18 | 21 | |
| Tumor stage | 0.009a | |||
| I–II | 35 | 12 | 23 | |
| III–IV | 42 | 27 | 15 | |
| Metastasis | <0.001a | |||
| No | 47 | 16 | 31 | |
| Yes | 30 | 23 | 7 | |
Statistically significant (P<0.05). lncRNA PVT1, long non-coding RNA plasmacytoma variant translocation 1.
lncRNA PVT1 regulates the invasion of cell lines in vitro
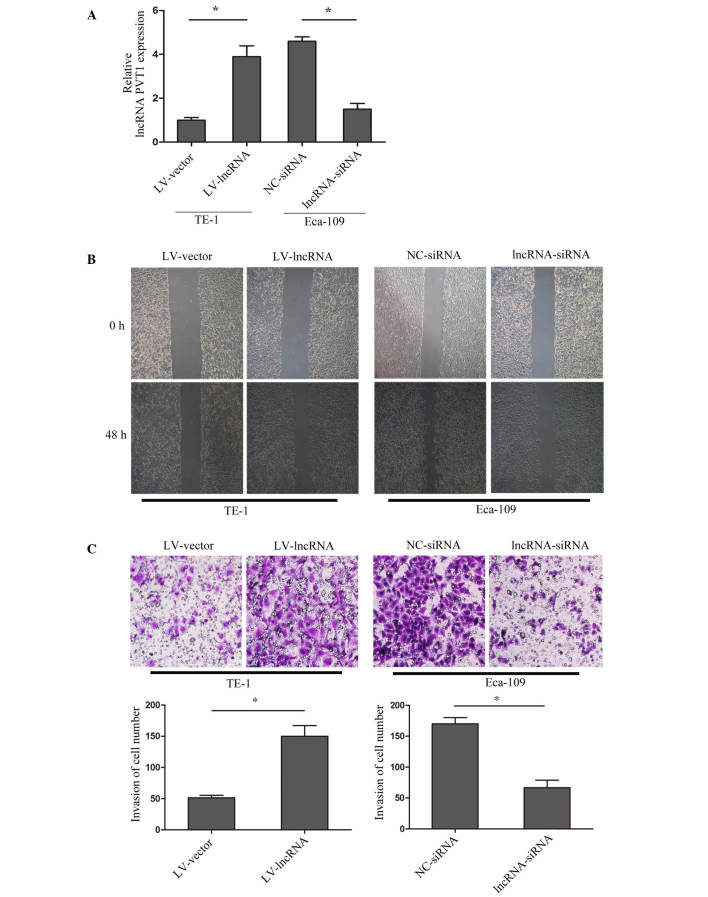
To explore the mechanism of lncRNA PVT1 and cell invasion in esophageal cancer, lncRNA PVT1 expression was upregulated in TE-1 cells by transfection with lentivirus (named LV-lncRNA), and downregulated in Eca-10 cells by siRNA interference (named lncRNA-siRNA). The transfection efficiency was validated by RT-qPCR (Fig. 2A). A wound healing assay indicated that upregulation of lncRNA PVT1 promoted the healing of cells, while suppressed lncRNA PVT1 inhibited healing (P=0.009) (Fig. 2B). Furthermore, a matrigel invasion assay showed that upregulated lncRNA PVT1 promoted cell invasion, while suppressed lncRNA PVT1 inhibited cell invasion (P=0.0031) (Fig. 2C). The results suggested that lncRNA PVT1 acts to regulate esophageal cancer cell invasion.
Figure 2.
lncRNA PVT1 regulated cells invasion. (A) lncRNA PVT1 expression was upregulated in TE-1 cells by transfecting with lentivirus containing lncRNA PVT1 (LV-lncRNA), with vector used as a control (named LV-vector); while downregulated lncRNA PVT1 expression in Eca-10 cells was achieved by transfecting with small interfering RNA (lncRNA-siRNA), with negative control siRNA used as a control (named NC-siRNA). The result was validated by reverse transcription-quantitative polymerase chain reaction. (B) Images were captured at 0 h and 48 h post-wounding and are shown at ×200 magnification. (C) A transwell assay was performed to assess the effect of lncRNA PVT1 on cell migration and invasion. Representative images of invasive cells at the bottom of the membrane stained with crystal violet are shown. All experiments were performed in triplicate and presented as the mean ± standard error of the mean. *P<0.05. Every independent experiment was performed three times. lncRNA PVT1, long non-coding RNA plasmacytoma variant translocation 1.
lncRNA PVT1 regulates EMT marker expression
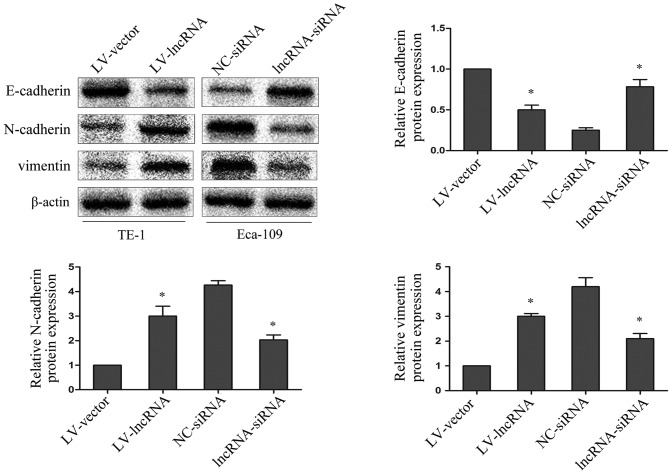
To explore whether abnormal lncRNA PVT1 expression levels may be involved in the EMT process, the epithelial marker E-cadherin, and the mesenchymal markers N-cadherin and vimentin were investigated by western blotting. Upregulated lncRNA PVT1 in TE-1 cells resulted in decreased E-cadherin protein expression and increased N-cadherin and vimentin protein expression; while suppressed lncRNA PVT1 expression in Eca-109 cells resulted in increased E-cadherin protein expression and decreased N-cadherin and vimentin protein expression (P=0.0019) (Fig. 3). This indicates that lncRNA PVT1 contributes to the regulation of EMT marker expression in esophageal cancer cell lines.
Figure 3.
lncRNA plasmacytoma variant translocation 1 may regulate the expression levels of epithelial-to-mesenchymal transition markers. E-cadherin, N-cadherin and vimentin protein expression levels in esophageal cancer cells were analyzed by western blotting. β-actin was used as a loading control. Average values of integrated optical density were assessed by analyzing five fields per slide. Data are presented as the mean ± standard error of the mean. *P<0.05 for LV-lncRNA vs. LV-vector and lncRNA-siRNA vs. NC-siRNA. lncRNA, long non-coding RNA; NC, negative control; siRNA, small interfering RNA; LV, lentivirus.
Discussion
lncRNAs have been reported to have critical regulatory roles in cancer biology (19–21). Specifically for esophageal cancer, it has already been reported that lncRNAs contribute to proliferation and invasion (22–24). The present study sought to provide evidence that upregulation of lncRNA PVT1 may induce EMT to promote cell invasion in esophageal cancer. The results indicated that the expression of lncRNA PVT1 was increased in esophageal cancer tissues (n=77) compared to corresponding adjacent tissues, and was associated with tumor stage and metastasis. Furthermore, in vitro, upregulation of lncRNA PVT1 promoted the invasion of esophageal cancer cell lines, and downregulation of lncRNA PVT1 inhibited the cell invasion.
The major causes of mortality from cancer are complications arising from cancer cell invasion. Decreased E-cadherin and elevated N-cadherin and vimentin expression are hallmarks of EMT, which is a key element in cancer invasion (25). EMT has been identified to be associated with tumor invasiveness, metastasis and prognosis (26,27). Numerous studies have established functional associations between lncRNAs and key effectors of EMT occurring in the context of carcinogenesis (28–30). In addition to cancer progression, EMT contributes to chronic epithelial injury (31), leading to tissue fibrosis and organ failure (32,33). In the current study, it was also discovered that abnormal expression of lncRNA PVT1 could regulate markers of EMT. Upregulated lncRNA PVT1 in TE-1 cells resulted in decreased E-cadherin expression and increased N-cadherin and vimentin expression relative to the control cells, while suppressed lncRNA PVT1 expression in Eca-109 cells resulted in increased E-cadherin expression and decreased N-cadherin and vimentin expression. These data suggest that lncRNA PVT1 promotes esophageal cancer invasion by inducing EMT.
Taken together, the findings of the present study indicate that overexpression of lncRNA PVT1 is associated with tumor stage and metastasis in esophageal cancer. In addition, upregulation of lncRNA PVT1 promotes cell invasion by inducing EMT in vitro. In the future, the pathway linking lncRNA PVT1 and EMT may be exploited in a therapeutic approach for the treatment of esophageal cancer.
Acknowledgements
This research was partly supported by Dr Wei Li for language revision.
References
- 1.Shigaki H, Baba Y, Watanabe M, Murata A, Ishimoto T, Iwatsuki M, Iwagami S, Nosho K, Baba H. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19:2451–2459. doi: 10.1158/1078-0432.CCR-12-3559. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Tong Y, Yang C, Gan Y, Sun H, Bi H, Cao S, Yin X, Lu Z. Consumption of hot beverages and foods and the risk of esophageal cancer: A meta-analysis of observational studies. BMC Cancer. 2015;15:449. doi: 10.1186/s12885-015-1185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao Y, Xue Y, Zhang L, Feng X, Liu W, Zhang G. Higher heat shock factor 1 expression in tumor stroma predicts poor prognosis in esophageal squamous cell carcinoma patients. J Transl Med. 2015;13:338. doi: 10.1186/s12967-015-0703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koshy M, Esiashvilli N, Landry JC, Thomas CR, Jr, Matthews RH. Multiple management modalities in esophageal cancer: Combined modality management approaches. Oncologist. 2004;9:147–159. doi: 10.1634/theoncologist.9-2-147. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, Wang L, Lian Y, Wang K, Shu Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L, Han F, Huang T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143–149. [PubMed] [Google Scholar]
- 10.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 11.Ding J, Li D, Gong M, Wang J, Huang X, Wu T, Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625–1630. doi: 10.2147/OTT.S68854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY, Li HC, Wu XN. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5:1639–1642. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Ruan B, You N, Huang Q, Liu W, Dang Z, Xu W, Zhou T, Ji R, Cao Y, et al. Downregulation of miR-200a induces EMT phenotypes and CSC-like signatures through targeting the β-catenin pathway in hepatic oval cells. PLoS One. 2013;8:e79409. doi: 10.1371/journal.pone.0079409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. doi: 10.1186/1471-2407-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong H, Xie L, Tang C, Chen S, Liu Q, Zhang Q, Zheng W, Zheng Z, Zhang H. Snail1 correlates with patient outcomes in E-cadherin-preserved gastroesophageal junction adenocarcinoma. Clin Transl Oncol. 2014;16:783–791. doi: 10.1007/s12094-013-1149-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Li H, Feng J, Cui X, Huang W, Li Y, Su F, Liu Q, Zhu J, Lv X, et al. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS One. 2013;8:e83083. doi: 10.1371/journal.pone.0083083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao YX, Cao Q, Yang Y, Mao R, Xiao L, Zhang H, Zhao HR, Wen H. Expression and prognostic significance of Golgiglycoprotein73 (GP73) with epithelial-mesenchymal transition (EMT) related molecules in hepatocellular carcinoma (HCC) Diagn Pathol. 2013;8:197. doi: 10.1186/1746-1596-8-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388. [PubMed] [Google Scholar]
- 20.Wang CM, Wu QQ, Li SQ, Chen FJ, Tuo L, Xie HW, Tong YS, Ji L, Zhou GZ, Cao G, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591–597. doi: 10.1007/s10620-013-2956-7. [DOI] [PubMed] [Google Scholar]
- 21.Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Zheng J, Deng J, You Y, Wu H, Li N, Lu J, Zhou Y. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714–1726. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li SQ, Wang CM, Tong YS, Tuo L, Wu M, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743–7754. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 24.Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H, Fang X. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2838–2849. [PMC free article] [PubMed] [Google Scholar]
- 25.Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 cluster regulates TGF-β-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 26.Guo S, Xu X, Tang Y, Zhang C, Li J, Ouyang Y, Ju J, Bie P, Wang H. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett. 2014;344:40–46. doi: 10.1016/j.canlet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Yamada S, Fuchs BC, Fujii T, Shimoyama Y, Sugimoto H, Nomoto S, Takeda S, Tanabe KK, Kodera Y, Nakao A. Epithelial-to-mesenchymal transition predicts prognosis of pancreatic cancer. Surgery. 2013;154:946–954. doi: 10.1016/j.surg.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7 doi: 10.18632/oncotarget.9883. (Epub ahead of print) doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, Ding L, Jia Y, Zhang H, Li Q, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol. 2016 doi: 10.1007/s13277-016-4892-6. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, Liu Z, Li W, Li Y, Li G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol Biosyst. 2016;12:952–962. doi: 10.1039/C5MB00685F. [DOI] [PubMed] [Google Scholar]
- 31.Vitalone MJ, Naesens M, Sigdel T, Li L, Hseih S, Sarwal MM. The dual role of epithelial-to-mesenchymal transition in chronic allograft injury in pediatric renal transplantation. Transplantation. 2011;92:787–795. doi: 10.1097/TP.0b013e31822d092c. [DOI] [PubMed] [Google Scholar]
- 32.López-Novoa JM, Nieto MA. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1:303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mucsi I, Rosivall L. Epithelial-mesenchymal transition in renal tubular cells in the pathogenesis of progressive tubulo-interstitial fibrosis. Acta Physiol Hung. 2007;94:117–131. doi: 10.1556/APhysiol.94.2007.1-2.11. [DOI] [PubMed] [Google Scholar]