Abstract
Gastric cancer (GC) is one of the most common types of cancer of the digestive tract. Invasion of tumor cells into surrounding tissue and metastasis are among the most significant checkpoints in tumor progression. It is known that matrix metalloproteinases (MMPs) are involved in these processes; however, knowledge of their molecular interaction networks is still limited. Investigation of these networks could provide a more comprehensive picture of the function of MMPs in tumorigenesis. Furthermore, it could be used to develop new approaches to targeted anticancer therapy. In this study, we performed microarray analysis, and 1666 genes that were aberrantly expressed in GC tissues were identified (fold change >2, P<0.05). In addition, quantitative polymerase chain reaction analysis has confirmed that MMP1, MMP3, MMP7, MMP10, MMP11 and MMP12 expression is upregulated in GC. In addition, the MMP3 expression level was negatively correlated with GC differentiation (P<0.05). By integrating the microarray information and BioGRID and STRING databases, we constructed an MMP-related molecular interaction network and observed that 18 genes (including MMPs) were highly expressed in GC tissues. The most enriched of these 18 genes in the Gene Oncology (GO) and pathway analysis were in extracellular matrix disassembly (GO biological process) and extracellular matrix-receptor interaction (KEGG pathway), which are closely correlated with cancer invasion and metastasis. Collectively, our results suggest that the MMP-related interaction network has a role in GC progression, and therefore further studies are required in order to investigate these network interactions in tumorigenesis.
Keywords: matrix metalloproteinase, gastric cancer, molecular interaction network, GO analysis, pathway analysis
Introduction
Gastric cancer (GC) is one of the most common malignancies of the digestive tract. It is particularly prevalent in China, with incidence and mortality rates above the world averages. Furthermore, according to the cancer statistics, it is the third leading cause of cancer-related mortality in China, with more than 300,000 mortalities per year (1). GC mortality remains high despite numerous efforts in the elucidation of its causes and progression due to its low rate of early detection (2). Patients are often diagnosed in the advanced stages of the disease, when it is accompanied by widespread invasion and distant metastasis of cancer cells with little possibility for radical surgery (3,4). Therefore, one of the possible ways to improve the prognosis of patients with GC is to develop new therapeutic approaches targeting its invasion and metastasis. Tumor differentiation, invasion and metastasis are controlled by multiple factors during the multistep processes of extracellular matrix (ECM) composition and degradation. In addition, new tumor blood vessels secrete various proteolytic enzymes which further add to the complex picture of tumor progression (5,6).
Matrix metalloproteinases (MMPs) are a group of zinc-dependent proteases involved in ECM and basement membrane degradation. They promote the formation of new blood vessels, and are involved in numerous processes associated with tumor cell growth, differentiation, invasion, diffusion and metastasis (7,8). Indeed, MMPs are involved in a number of complex networks of gene/protein interactions, which regulate a variety of physiological and pathophysiological processes (9). High expression of MMPs (including MMP1, MMP7 and MMP9) was reported for GC (10,11), esophageal cancer (12), lung cancer (13) and other malignant tumors (14), and increased expression of MMPs is closely associated with GC progression and poor prognosis (15).
It has been noted that gene/protein clusters tend to be jointly involved in the regulation of various biological processes or pathways (16,17). Therefore, one of the potentially most effective approaches to examining these complex interactions would be to analyze entire networks of potentially involved signaling molecules. The Biological General Repository for Interaction Datasets (BioGRID, http://thebiogrid.org/) is a gene/protein interaction database that contains a collection of conventionally as well as high-throughput obtained experimental data for several model organisms including humans (18). The STRING database (http://string-db.org/) integrates and quantifies interactions between known and predicted proteins from a number of different organisms, and is able to synchronize the selected gene cluster gene ontology (GO) and pathway enrichment analysis (19,20).
In this study we examined the complex networks associated with MMPs and their role in GC. In order to create an interaction network of aberrantly expressed MMPs potentially associated with GC tumorigenesis, we integrated high-throughput gene chip expression data with the interaction data obtained from the BioGRID and STRING databases.
Materials and methods
Patients and samples
Ten patients (six males and four females) with GC, as confirmed by a pathologist, were included in this study. They were admitted to the gastrointestinal surgery department of the First Affiliated Hospital of Jilin University, Changchun, China, between May 2013 and May 2014. The median patient age was 57.6 years (range, 48 to 71 years). None of the patients received any preoperative radiotherapy, chemotherapy or other cancer treatment. Tumor and corresponding normal tissue (more than 5 cm from the tumor) were obtained during surgery and immediately snap-frozen in liquid nitrogen. The clinicopathological characteristics of patients and their tumors are presented in Table I. The study was approved by the ethics committee of the College of Basic Medical Sciences at Jilin University and written informed consent was obtained from all patients.
Table I.
Clinicopathological features of gastric cancer patients included in the study.
| Clinicopathological feature | Number of cases | P-value |
|---|---|---|
| Age (years) | 0.7937 | |
| <60 | 5 | |
| ≥60 | 5 | |
| Gender | 0.4172 | |
| Male | 5 | |
| Female | 5 | |
| Smoker | ||
| No | 8 | |
| Yes | 2 | |
| Differentiation status | 0.0336a | |
| Well or moderate | 6 | |
| Poor | 4 |
P<0.05.
RNA extraction and microarray analysis
Total RNA was extracted from 10 gastric tumors and corresponding normal tissue using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA concentration and purity were examined by ultraviolet light tube viscometer (NanoDrop Technologies, Wilmington, DE, USA) and the RNA integrity was examined by formamide denaturing gel electrophoresis. Only RNAs with an optical density (OD) 260/280 ratio of 1.8–2.0 and good integrity were used in the array analysis. The RNA concentration was adjusted to 1 µg/µl for reverse transcription. Next, biotin labeling cRNA synthesis, purification and fragmentation were performed using an IVT labeling kit following the instructions. Following hybridization (using a Eukaryotic Hybridization Control kit as well as a Hybridization, Wash and Stain kit), washing and staining (using the Hybridization, Wash and Stain kit), Affymetrix GeneChip Human Exon 1.0 ST chips were scanned with the GeneChip® operating software (GCOS) in GeneChip® Scanner 3000 7G, and differentially expressed genes were selected according to their expression in tumor tissue relative to normal gastric tissue (fold change ≥2, and P≤0.05). For the differentially expressed genes, data cluster analysis was performed using the R program (21). The reagents and instruments used in the experimental process were all from Affymetrix (Affymetrix, Inc., Santa Clara, CA, USA).
Quantitative polymerase chain reaction (qPCR) analysis
To further examine the mRNA expression of several MMPs including MMP1, MMP3, MMP7, MMP10, MMP11 and MMP12 in GC tissues and corresponding normal tissue, qPCR analysis was performed. Total RNA was reverse transcribed to cDNA with a cDNA synthesis kit according to the manufacturer's instructions (Takara, Dalian, China). Next, 2 µl cDNA was used for qPCR analysis using SYBR® Premix Ex Taq™ (Takara, Dalian, China) and an Applied Biosystems 7300 Fast Real-Time PCR system (stage 1: 95°C, 30 sec, degeneration; stage 2: 95°C, 5 sec; 60°C, 31 sec; 40 cycles). β-actin expression was used as an endogenous control. The primers were designed using Primer Premier 5 software (PREMIER Biosoft, Palo Alto, CA, USA) and are presented in Table II. The results of qPCR were expressed as 2−ΔΔCt (ΔCt = Ct target - Ct β-actin, ΔΔCt = ΔCt tumor - ΔCt normal). Statistical analysis was performed with SPSS software version 17.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Table II.
Primer sequences used in quantitative polymerase chain reaction analysis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| β-actin | 5′-CTGGAACGGTGAAGGTGACA-3′ | 5′-AAGGGACTTCCTGTAACAATGCA-3′ |
| MMP1 | 5′-AAAATTACACGCCAGATTTGCC-3′ | 5′-GGTGTGACATTACTCCAGAGTTG-3′ |
| MMP3 | 5′-AGTCTTCCAATCCTACTGTTGCT-3′ | 5′-TCCCCGTCACCTCCAATCC-3′ |
| MMP7 | 5′-GAGTGAGCTACAGTGGGAACA-3′ | 5′-CTATGACGCGGGAGTTTAACAT-3′ |
| MMP10 | 5′-TGCTCTGCCTATCCTCTGAGT-3′ | 5′-TCACATCCTTTTCGAGGTTGTAG-3′ |
| MMP11 | 5′-CCGCAACCGACAGAAGAGG-3′ | 5′-ATCGCTCCATACCTTTAGGGC-3′ |
| MMP12 | 5′-GATCCAAAGGCCGTAATGTTCC-3′ | 5′-TGAATGCCACGTATGTCATCAG-3′ |
Protein-protein interaction (PPI) network analysis
Based on our results of which genes were differentially expressed in gastric tumors compared with normal gastric tissue, we selected the online tools BioGRID and STRING to explore the potential PPIs. The filter settings were as follows: the STRING software species source was limited to Homo sapiens, CI 0.400; the BioGRID software species source was limited to Homo sapiens, and the retrieval channel was set to gene. The network interactions from the STRING software and genes involved in the GO and pathway analysis results were obtained synchronously.
Results
Identification of differentially expressed genes in GC
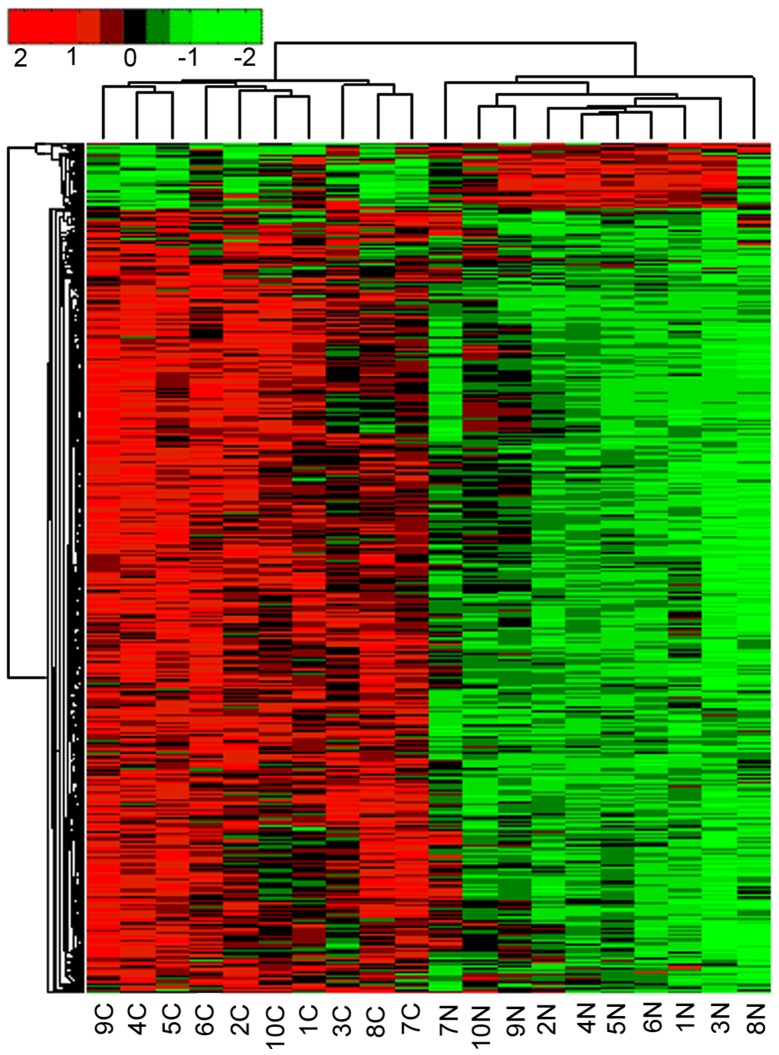
In the present study we examined the mRNA expression in GC and corresponding normal tissue samples by microarray analysis. As a result of this analysis, a total of 1666 differentially expressed genes were detected, including 1540 genes with high expression and 126 genes with decreased expression in GC tissue (data not shown). R software was employed to analyze the difference in gene clustering results, and the results are presented in Fig. 1.
Figure 1.
Clustering analysis of differentially expressed genes in gastric cancer. The vertical axis represents 1666 differential genes and the horizontal axis shows the tissue samples. N indicates control tissues and C indicates cancer tissues. The red color represents upregulated genes and the green color represents downregulated genes. Genes with fold change ≥2 and P≤0.05 in gastric cancer vs. normal tissues were screened for the clustering.
As a result of this high-throughput screening, a significant phenomenon was observed. Six MMP family members (MMP1, MMP3, MMP7, MMP10, MMP11 and MMP12) were highly expressed in GC tissue. To further confirm these findings, qPCR was performed and the obtained results were in accordance with those obtained by microarray analysis. Of the analyzed MMPs, the highest expression difference was observed for MMP3 (Fig. 2).
Figure 2.
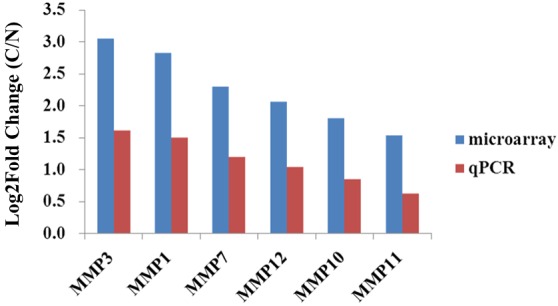
Differential expression of matrix metalloproteinases in gastric cancer tissues. The vertical axis represents the log2-transformed fold changes (C/N) of gene expression (P<0.05) in microarray and quantitative polymerase chain reaction analysis.
Association between MMP mRNA expression and clinicopathological features of GC patients
Next, MMP3 mRNA expression was examined in relation to the clinicopathological features of GC patients. For this purpose the GC tissues with MMP3 expression above the median value were defined as high MMP3 expression (n=5), and tumor samples with MMP3 expression below the median value were defined as low MMP3 expression (n=5). According to this analysis MMP3 mRNA expression was negatively correlated with the tumor differentiation status (P<0.05, Table I). In addition, no association was observed between MMP3 mRNA expression and the patients' age, gender or smoking status. The mRNA expression of other MMPs differentially expressed in gastric caner demonstrated no significant association with the patients' age, gender, smoking or differentiation status.
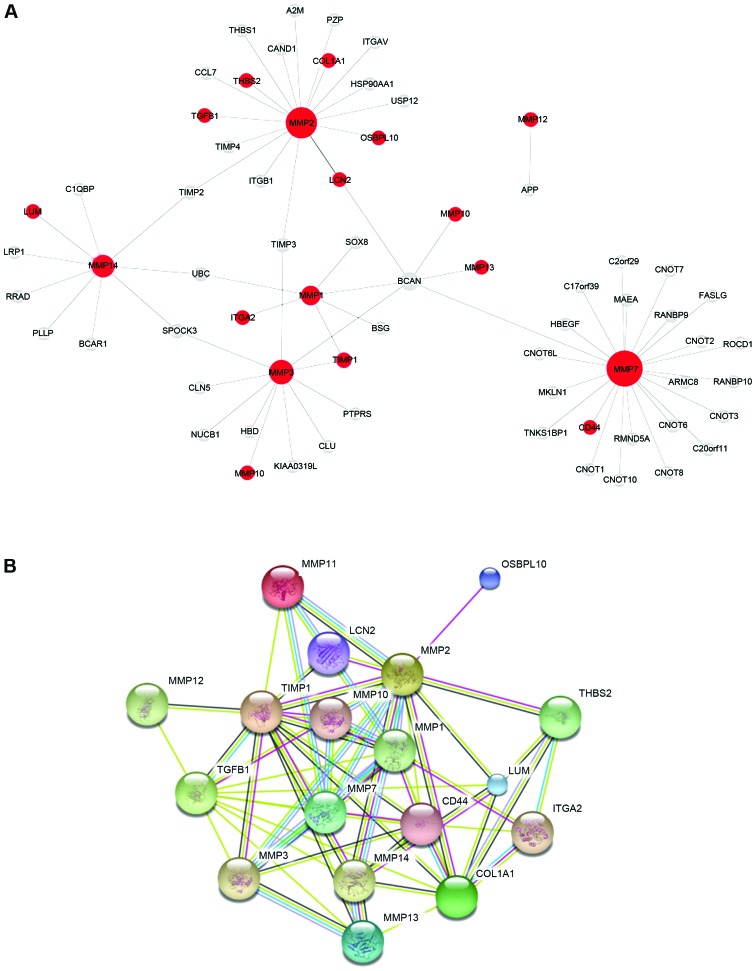
Construction and analysis of MMP-related PPI network
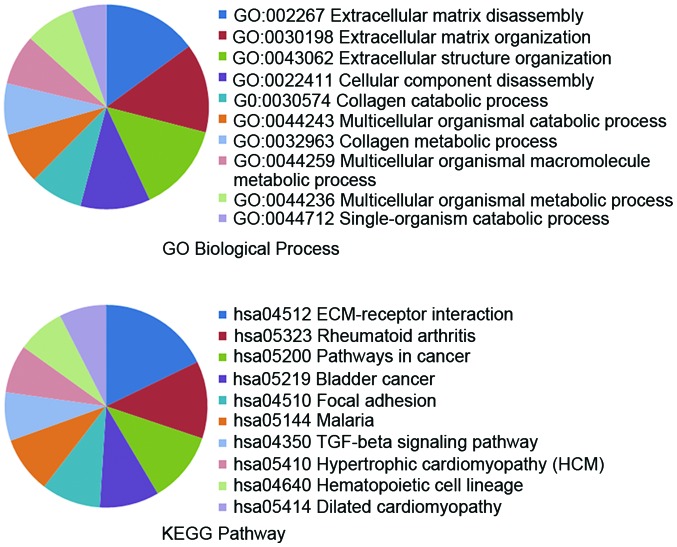
Next, according to the results of the microarray analysis, we examined the BioGRID molecular database to obtain the MMP PPI network in GC, and 18 genes were detected (Fig. 3A). When these genes were examined in the STRING database they were observed to form a significant functional network (Fig. 3B). This complex network is known to be involved in several essential biological processes and pathways (Fig. 4), including ECM disassembly and the ECM-receptor interaction pathway, both closely related to cancer invasion and metastasis (22,23).
Figure 3.
Protein-protein interaction (PPI) network of matrix metalloproteinases (MMPs) in gastric cancer tissues. (A) MMP PPI network from the BioGRID database; the 18 red circles represent differentially expressed (upregulated) genes in gastric cancer tissues (P<0.05). (B) MMP PPI network of the 18 differentially expressed genes from the STRING database.
Figure 4.
Gene ontology (GO) and pathway analysis of 18 differentially expressed genes in the protein-protein interaction (PPI) network. The STRING database revealed the GO and pathway terms of 18 genes in the PPI network. Here we collected biological process terms in GO analysis, as well as the KEGG pathway terms.
Discussion
Matrix metalloproteinases have numerous functions at the cellular and tissue level, in physiological and pathophysiological states, and form complex interaction networks depending upon the processes in which they are involved (24). Over the past decades much has been learned about their role and interactions with other molecules; however, studies performed before the rise of the high throughput molecular techniques have often been limited to only a few interactions that could be examined simultaneously by conventional methods. In this study, we have performed microarray analysis in ten samples of gastric tumors and corresponding normal tissue. Among the most differentially expressed genes several MMPs have been identified, and therefore we have decided to further examine their role in the broader context of their interactions in GC. In our study, the MMP-related PPI network revealed a set of 18 genes, including 9 MMP family members, that were upregulated in GC tissues, with MMP3 exhibiting the most significant upregulation. Enrichment analysis of the 18 differentially expressed genes in the STRING database focused on biological processes that are associated with the functions of the ECM and the ECM-receptor pathway, both of which are associated with cell migration and metastasis. This is in accordance with previous studies in which the role of MMPs in tumorigenesis was examined. Indeed, several studies have reported upregulation of MMP members in GC as well as in other types of cancers, including hepatocellular carcinoma, colorectal cancer, breast cancer and prostate cancer.
In our study the most upregulated MMP was MMP3. Previous studies have reported its role in cancer. For example, in nasopharyngeal carcinoma, MMP3 and MMP9 are upregulated, and co-expression of these two molecules increased cell invasive properties (25). Another study reported that Cyclophilin A (CypA) promotes the metastatic properties of hepatocellular cancer at least in part through the upregulation of MMP3 (26). MMP3 and other MMP members have also been linked with glioma angiogenesis, tumor remodeling and infiltration (27). However, our study is the first to identify an aberrantly expressed MMP-related PPI network in GC tissues.
One of the issues that we encountered in our study was the limited number of cases and scarce clinicopathological information, including the data on distant and lymph node metastasis. Nevertheless, there were a number of notable findings. For instance, in our study MMP3 expression was negatively correlated with the differentiation status of GC tissues, which offers a new perspective to further explore the role of MMPs in GC tumorigenesis.
The gene expression dataset and online database tools provided us with comprehensive information concerning gene expression and regulation in GC. In a previous study by Feng et al (28), PPI networks in GC were constructed based on microarray data and the data from the STRING database, and as a result, STAT1 and EGR2 were proposed as new potential biomarkers of GC metastasis. In another study, researchers identified GC-related genes using an algorithm and the PPI network, and protein interaction information was also retrieved from STRING (29). Studies on PPI networks in other malignancies also demonstrated the same trend. Zhang et al (30) analyzed ChIP-Seq data in prostate cancer, and constructed a PPI network with BioGRID and cytoscape software. As a result, they identified several critical genes that provided a direction for future research into prostate cancer, and this research provided us with a novel perspective on GC.
In conclusion, in the present study we examined aberrantly expressed MMPs and the related PPI network in GC tissues using the data provided by microarray and the PPI database. As a result, MMP3 emerged as one of the most unregulated MMPs, and in our study its expression was negatively correlated with the differentiation status of GC. Our study was performed using a small set of samples and therefore our results could be considered as preliminary findings; however, they have laid the foundation for further studies which could be performed using a larger set of samples.
References
- 1.He YT, Hou J, Chen ZF, Qiao CY, Song GH, Meng FS, Jin HX, Chen C. Trends in incidence of esophageal and gastric cardia cancer in high-risk areas in China. Eur J Cancer Prev. 2008;17:71–76. doi: 10.1097/CEJ.0b013e3282b6fd97. [DOI] [PubMed] [Google Scholar]
- 2.Worm K, Weaver DG, Green RC, Saeui CT, Dulay DM, Barker WM, Cassel JA, Stabley GJ, DeHaven RN, LaBuda CJ, et al. Discovery of N-(3-(morpholinomethyl)-phenyl)-amides as potent and selective CB2 agonists. Bioorg Med Chem Lett. 2009;19:5004–5008. doi: 10.1016/j.bmcl.2009.07.057. [DOI] [PubMed] [Google Scholar]
- 3.Jin X, Zhu Z, Shi Y. Metastasis mechanism and gene/protein expression in gastric cancer with distant organs metastasis. Bull Cancer. 2014 doi: 10.1684/bdc.2013.1882. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Wang Y, Dong S, Ge C, Xiao Y, Li R, Ma X, Xue Y, Zhang Q, Lv J, et al. Association of CXCR1 and 2 expressions with gastric cancer metastasis in ex vivo and tumor cell invasion in vitro. Cytokine. 2014;69:6–13. doi: 10.1016/j.cyto.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Steinestel K, Brüderlein S, Lennerz JK, Steinestel J, Kraft K, Pröpper C, Meineke V, Möller P. Expression and Y435-phosphorylation of Abelson interactor 1 (Abi1) promotes tumour cell adhesion, extracellular matrix degradation and invasion by colorectal carcinoma cells. Mol Cancer. 2014;13:145. doi: 10.1186/1476-4598-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P, Wang X, Hu CH, Hu TH. Bioinformatics analysis with graph-based clustering to detect gastric cancer-related pathways. Genet Mol Res. 2012;11:3497–3504. doi: 10.4238/2012.September.26.5. [DOI] [PubMed] [Google Scholar]
- 7.Hansmann F, Herder V, Kalkuhl A, Haist V, Zhang N, Schaudien D, Deschl U, Baumgärtner W, Ulrich R. Matrix metalloproteinase-12 deficiency ameliorates the clinical course and demyelination in Theiler's murine encephalomyelitis. Acta Neuropathol. 2012;124:127–142. doi: 10.1007/s00401-012-0942-3. [DOI] [PubMed] [Google Scholar]
- 8.Griner SE, Joshi JP, Nahta R. Growth differentiation factor 15 stimulates rapamycin-sensitive ovarian cancer cell growth and invasion. Biochem Pharmacol. 2013;85:46–58. doi: 10.1016/j.bcp.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theocharis AD, Gialeli C, Bouris P, Giannopoulou E, Skandalis SS, Aletras AJ, Iozzo RV, Karamanos NK. Cell-matrix interactions: focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J. 2014;281:5023–5042. doi: 10.1111/febs.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long ZW, Wang JL, Wang YN. Matrix metalloproteinase-7 mRNA and protein expression in gastric carcinoma: a meta-analysis. Tumour Biol. 2014;35:11415–11426. doi: 10.1007/s13277-014-2441-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang BG, Du T, Zang MD, Chang Q, Fan ZY, Li JF, Yu BQ, Su LP, Li C, Yan C, et al. Androgen receptor promotes gastric cancer cell migration and invasion via AKT-phosphorylation dependent upregulation of matrix metalloproteinase 9. Oncotarget. 2014;5:10584–10595. doi: 10.18632/oncotarget.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Ren Y, Yu X, Qian F, Bian BS, Xiao HL, Wang WG, Xu SL, Yang J, Cui W, et al. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod Pathol. 2014;27:775–783. doi: 10.1038/modpathol.2013.189. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Morales JM, Dorantes-Heredia R, Arrieta O, Chávez-Tapia NC, Motola-Kuba D. Neutrophil gelatinase-associated lipocalin (NGAL) and matrix metalloproteinase-9 (MMP-9) prognostic value in lung adenocarcinoma. Tumour Biol. 2015;36:3601–3610. doi: 10.1007/s13277-014-2997-3. [DOI] [PubMed] [Google Scholar]
- 14.Heinke J, Kerber M, Rahner S, Mnich L, Lassmann S, Helbing T, Werner M, Patterson C, Bode C, Moser M. Bone morphogenetic protein modulator BMPER is highly expressed in malignant tumors and controls invasive cell behavior. Oncogene. 2012;31:2919–2930. doi: 10.1038/onc.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Chen LJ, Zhou HC, Yang RB, Lu Y, Xia YL, Werner M, Patterson C, Bode C, Moser M. Prognostic value of matrix metalloproteinase-9 in gastric cancer: a meta-analysis. Hepatogastroenterology. 2014;61:518–524. [PubMed] [Google Scholar]
- 16.Ibrahim ZM, Ngom A. The relative vertex clustering value - a new criterion for the fast discovery of functional modules in protein interaction networks. BMC Bioinformatics. 2015;16(Suppl 4):S3. doi: 10.1186/1471-2105-16-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anwar S, Dikhit MR, Singh KP, Kar RK, Zaidi A, Sahoo GC, Roy AK, Nozaki T, Das P, Ali V. Interaction between Nbp35 and Cfd1 proteins of cytosolic Fe-S cluster assembly reveals a stable complex formation in Entamoeba histolytica. PLoS One. 2014;9:e108971. doi: 10.1371/journal.pone.0108971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gene Ontology Consortium, corp-author. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D448. doi: 10.1093/nar/gkm883. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu A, Wang C, Sun S. Screening candidate genes associated with bladder cancer using DNA microarray. Mol Med Rep. 2014;10:3087–3091. doi: 10.3892/mmr.2014.2667. [DOI] [PubMed] [Google Scholar]
- 22.Kim SO, Kim MR. [6]-gingerol prevents disassembly of cell junctions and activities of MMPs in invasive human pancreas cancer cells through ERK/NF-kappa B/snail signal transduction pathway. Evid Based Complement Alternat Med. 2013;2013:761852. doi: 10.1155/2013/761852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, Fabry B, O'Neill GM. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PLoS One. 2012;7:e35058. doi: 10.1371/journal.pone.0035058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki I, Noro T, Tsutsui N, Yamanouchi E, Kuroda H, Nakano M, Yokomori H, Inagaki Y. Fibrogenesis and carcinogenesis in nonalcoholic steatohepatitis (NASH): involvement of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinase (TIMPs) Cancers. 2014;6:1220–1255. doi: 10.3390/cancers6031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorentini C, Bodei S, Bedussi F, Fragni M, Bonini SA, Simeone C, Zani D, Berruti A, Missale C, Memo M, et al. GPNMB/OA protein increases the invasiveness of human metastatic prostate cancer cell lines DU145 and PC3 through MMP-2 and MMP-9 activity. Exp Cell Res. 2014;323:100–111. doi: 10.1016/j.yexcr.2014.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Dai C, Zhu H, Chen S, Wu Y, Li Q, Zeng X, Wang W, Zuo J, Zhou M, et al. Cyclophilin A promotes human hepatocellular carcinoma cell metastasis via regulation of MMP3 and MMP9. Mol Cell Biochem. 2011;357:387–395. doi: 10.1007/s11010-011-0909-z. [DOI] [PubMed] [Google Scholar]
- 27.Campbell NE, Kellenberger L, Greenaway J, Moorehead RA, Linnerth-Petrik NM, Petrik J. Extracellular matrix proteins and tumor angiogenesis. J Oncol. 2010;2010:586905. doi: 10.1155/2010/586905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng D, Ye X, Zhu Z, Wei Z, Cai Q, Wang Y. Comparative transcriptome analysis between metastatic and non-metastatic gastric cancer reveals potential biomarkers. Mol Med Rep. 2015;11:386–392. doi: 10.3892/mmr.2014.2709. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y, Shu Y, Shi Y, Li LP, Yuan F, Ren H. Identifying gastric cancer related genes using the shortest path algorithm and protein-protein interaction network. Biomed Res Int. 2014;2014:371397. doi: 10.1155/2014/371397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Huang Z, Zhu Z, Liu J, Zheng X, Zhang Y. Network analysis of ChIP-Seq data reveals key genes in prostate cancer. Eur J Med Res. 2014;19:47. doi: 10.1186/s40001-014-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]