Abstract
The human brain has enormously complex cellular diversity and connectivities fundamental to our neural functions, yet difficulties in interrogating individual neurons has impeded understanding of the underlying transcriptional landscape. We developed a scalable approach to sequence and quantify RNA molecules in isolated neuronal nuclei from post-mortem brain, generating 3,227 sets of single neuron data from six distinct regions of the cerebral cortex. Using an iterative clustering and classification approach, we identified 16 neuronal subtypes that were further annotated on the basis of known markers and cortical cytoarchitecture. These data demonstrate a robust and scalable method for identifying and categorizing single nuclear transcriptomes, revealing shared genes sufficient to distinguish novel and orthologous neuronal subtypes as well as regional identity within the human brain.
Main Text
While significant progress has been achieved in mice (1–3), comprehensive classification of adult human brain neurons on the basis of their single-cell transcriptomes has yet to be realized. Examination of individual neuronal gene expression profiles for functional patterns could provide unbiased insights into subtypes from defined neuroanatomical regions, which are missed by gross anatomical studies that report limited transcriptomic differences across the neocortex (4–7). Previous analyses of single adult human neurons has been dependent on methods compatible with freshly isolated neurosurgical tissues (8), which can be difficult to obtain, with limited regional sampling and depth. By contrast, post-mortem tissues provide a vastly more accessible source of both normal and diseased brain, wherein challenges to interrogating single neuronal genomes can be overcome using single nuclei (9, 10) combined with RNA sequencing. Here, we report the development of a scalable pipeline from post-mortem brain through nuclear transcriptome analyses that identifies both known and novel neuronal subtypes across the cerebral cortex in humans.
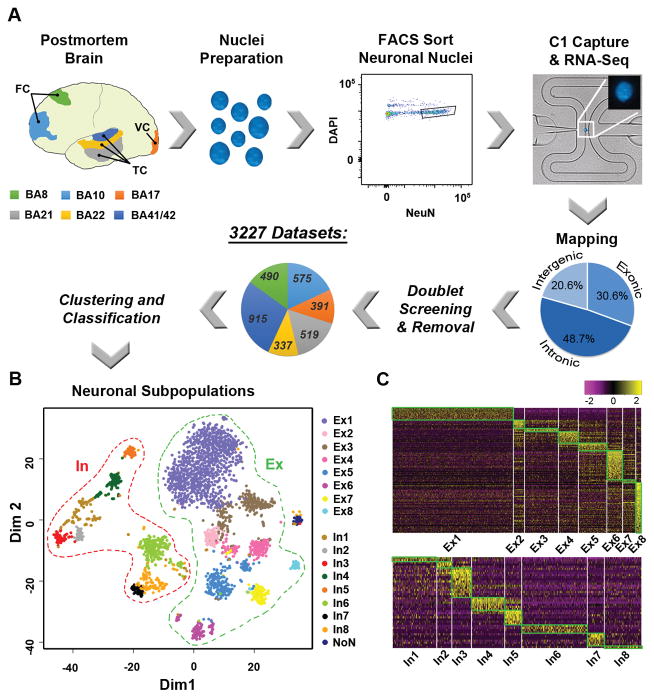
With the goal of defining transcriptomic profiles of single neurons, neuronal nuclear antigen (NeuN) was used (9) to isolate neuronal nuclei (Fig. S1) from the post-mortem brain of a normal, 51-year old female (Fig. 1A). We focused on six classically defined Brodmann Areas (BAs) with well-documented anatomical and electrophysiological properties that were derived from a single cortical hemisphere, since inter-hemispheric and inter-individual transcriptome differences were reported to be minimal (4–7). Isolation of nuclei was used to reduce transcriptomic contamination from other cells or degradation encountered with whole-neuron dissociation or laser caption micro-dissection (Fig. S2). Furthermore, sequencing of RNA from single nuclei on a limited scale has found gene expression values comparable to that of the whole cell (11, 12). Therefore, we developed and implemented a highly-scalable, single nucleus RNA sequencing (SNS) pipeline (13) (Fig. 1A, Fig. S1, Fig. S3–S8) that has broad applicability for post-mortem brains derived from multiple brain banks/repositories (Fig. S4F).
Fig. 1.
Single nucleus RNA sequencing (SNS) identified 16 neuronal subtypes over 6 neocortical regions. A. Overview of SNS pipeline. Post-mortem tissue from Brodmann Areas (BA) 8, 10, 17, 21, 22, and 41/42 were dissociated to single nuclei for NeuN+ and DAPI+ sorting and capture on C1 chips. Resultant libraries were sequenced, mapped to the reference genome (pie chart showing averaged proportions) and screened for doublet removal before clustering and classification. BA proportions are shown. FC = Frontal Cortex; TC = Temporal Cortex; VC = Visual Cortex. B. Neuronal subtypes (excitatory (Ex) and inhibitory (In)) shown by multidimensional plotting using 10-fold or greater differentially expressed genes (Table S3); NoN (no nomenclature), low expression outlier cluster. C. Heatmap showing unique marker gene expression (Table S5).
Using this pipeline, we processed 86 Fluidigm C1 chips and sequenced 4,488 single nuclei to an average depth of 8.34M reads (Table S1, Fig. S5). Genomic mapping rates revealed a high proportion of reads that corresponded to intronic sequences (Fig. 1A, Fig. S5A). The low percentage of intergenic reads argues against possible genomic contamination. Instead, the intronic reads likely captured an abundance of nascent RNA transcripts present in the nuclei. Intronic reads can be used to predict de novo expression (14), as well as whole cell gene transcription levels (15). Additionally, our single nuclei expression data inclusive of intronic reads accurately predicted cellular identity (Fig. S7), thereby providing initial validation for our SNS pipeline.
After quality filtering, including removal of doublets misclassified as single nuclei (13) (Fig. 1A, Fig. S6), we achieved 3,227 data sets across the six cortical regions (Fig. 1A, Table S2). To identify neuronal subtypes, we developed a clustering and classification strategy that was capable of resolving 17 clusters (13) (Fig. S8A) on the basis of differential expression of neuronally annotated marker genes (Tables S3–S4, Fig. S8B). These clusters showed distinct subgroup aggregation (Fig. 1B, Fig. S9A) and unique gene expression profiles associated with neuronal ontologies (Fig. 1C, Fig. S9B, Tables S5–S6). With the exception of a single cluster (NoN, n=44) deriving from one C1 chip having reduced mapping rates, 16 of these clusters were generated independent of detectable batch effects (Table S2, Fig. S10). Differential expression of inhibitory markers associated with GABAergic interneurons (Table S3) distinguished potential inhibitory (In) from excitatory (Ex) neuronal subtypes (Fig. 1B), consistent with mutually exclusive positivity of associated marker genes using a fraction of positive thresholding method (2) (Fig. 2A). As such, our dataset first differentiated two major classifications within the cerebral cortex: 972 inhibitory neurons that generally encompass interneurons and 2,253 excitatory neurons that generally encompass pyramidal or projection neurons (16). Furthermore, each subgroup within these classifications showed distinct contributions from each brain region (Fig. 2A, Table S7), likely reflecting varied proportions of these neuronal subtypes across BAs, with most variability present in the visual cortex (BA17) that is known to have distinct cytoarchitecture and gene expression profiles (7, 17).
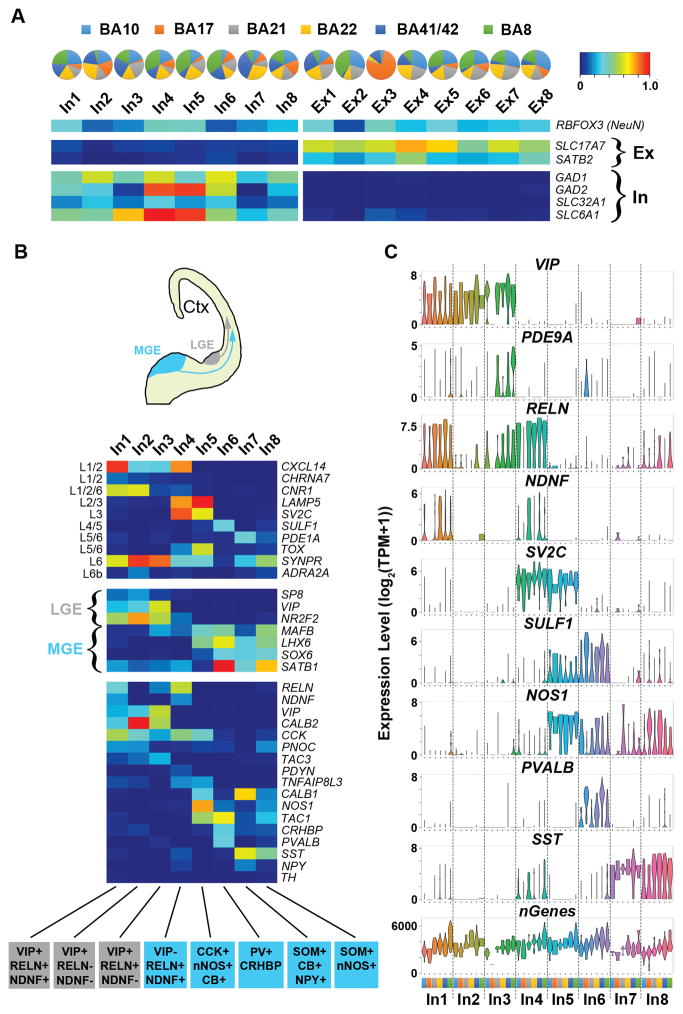
Fig. 2.
SNS reveals distinct interneuron subtypes. A. Pie charts display relative proportions of subtypes amongst BAs, and fraction of positive (FOP) heatmaps for inhibitory (In) and excitatory (Ex) marker genes. B. Diagram of subpallial origins of interneurons from either the lateral or medial ganglionic eminence (LGE, MGE) with FOP heatmaps (see A for scale) for marker genes associated with cortical layer (L) (upper panel), subpallial origin (middle panel) and interneuron classification (bottom panel). Potential interneuron subtypes are indicated below. SOM, somatostatin or SST; NPY, neuropeptide Y; CB, calbindin-D-28k or CALB1; VIP, vasoactive intestinal peptide; RELN, reelin; nNOS, neuronal nitric oxide synthase or NOS1; PV, parvalbumin or PVALB; CCK, cholecystokinin; NDNF, neuron-derived neurotrophic factor; CRHBP, corticotropin releasing hormone binding protein. C. Violin plots showing select marker gene expression values by BA (colors indicated in A) for each inhibitory neuron subtype. nGenes, total number of genes identified.
In order to further annotate inhibitory neuron subtypes, we examined expression of known marker genes associated with cortical layers, developmental origin, and interneuron classification (13) (Fig. 2B). On the basis of in situ human brain expression data (Fig. S11) (17), our inhibitory neuron subtypes were found to distribute spatially from the pial surface (most superficial boundary) to white matter (deepest boundary) of the neocortex, and could be grouped by the developmental origin of interneurons from subcortical regions of the medial, lateral, or caudal ganglionic eminences (MGE, LGE or CGE) (Fig. 2B) (18, 19). Furthermore, distinct profiles of interneuron classification markers revealed subtypes that parallel those identified from the mouse somatosensory cortex (3) (Fig. 2B–C, Fig. S12A). Cortical regional heterogeneity within subtypes was also observed, as evident by a layer 3 population (In4) that showed a specific absence of RELN/SST expression in BA17 (Fig. 2C, Fig. S11B, D). As such, our data distinguished inhibitory neuron subtypes having heterogeneous distributions within the neocortex.
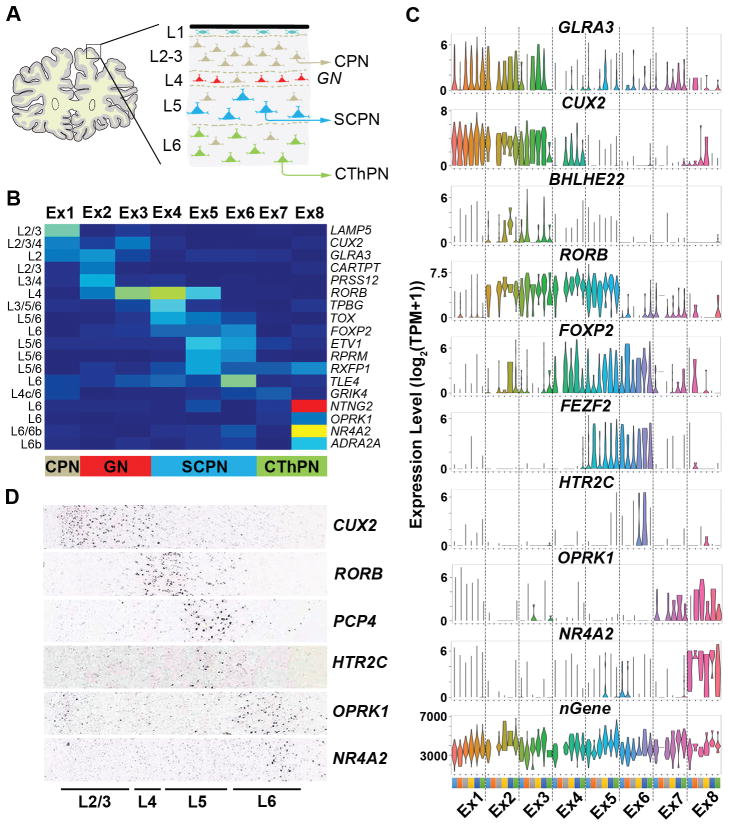
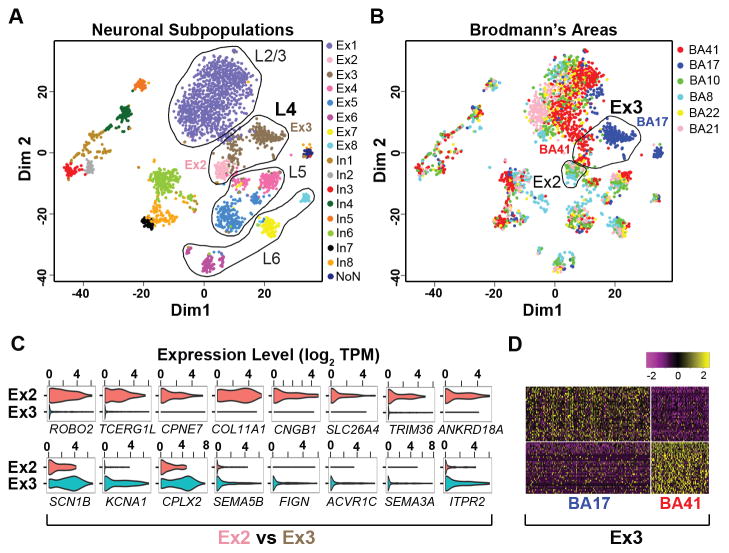
Most excitatory cortical projection or pyramidal neurons can be categorized by their layer position established during neocortical development (17) combined with their axonal projections (16) (Fig. 3A). Our excitatory neuron subgroups, which were also in high concordance with subtypes identified in mice (3) (Fig. S12B), expressed known markers associated with a superficial-to-deep cortical distribution (13) (Fig. 3B–D, Fig. S13), with more than one subtype occupying most layers. Our data set was able to resolve cortical region specificity, as seen for the BHLHE22 positive (Fig. 3C, Fig. S13A,D) layer 4 subtypes Ex2 and Ex3 (Fig. 4A), where Ex2 derived predominantly from rostral regions, BA8 and BA10, and Ex3 from caudal regions, BA17 and BA41/42 (Fig. 2A, Fig. 4B). Consistently, these subgroups showed distinct gene expression (Fig. 4C, Table S8) associated with neuronal electrophysiology and connectivity (Table S9). Furthermore, we were able to resolve intra-subtype heterogeneity, in terms of BA-specific expression patterns, which was observed in all subtypes (Fig. 4B), as for example within the Ex3 subtype between BA17 and BA41/42 regions (Fig. 4B,D; Table S10). As such, regional neurophysiological differences in cortical regions may be attributed to not only variations in the proportions of interneuron and projection neuron subtypes, but also to cell-intrinsic transcriptomic differences amongst single neurons within a subtype. Consistent with this possibility, we found genes having known variability between the visual and temporal cortices from in situ hybridization (ISH) studies (17) also had transcriptomic differences that could be attributed to subtypes defined by our data set (13) (Fig. S14A, Table S11). Therefore, our data highlight subtle yet region-defining gene expression signatures amongst specific neuronal subtypes that could not be detected from bulk analyses (Fig. S14B).
Fig. 3.
Excitatory neuronal subtypes show distinct spatial organization. A. Schematic of the prefrontal cortex showing projection neuron layers (L) and expected axonal projection destinations (layer 4 granule neurons typically receive outside inputs for distribution of signals locally). B. FOP heatmap (see Fig. 2A for scale) for layer specific marker genes showing expected cortical layer identity (L2–L6b) and excitatory neuron sub-classification. CPN = cortical projection neuron; GN = granule neuron; SCPN = subcortical projection neuron; CThPN = corticothalamic projection neuron. C. Violin plots showing selected marker gene expression values by Ex subtype and BA represented by colors (see Fig. 2A). nGenes = total number of genes identified. D. RNA ISH showing layer-specific expression of selected markers in the temporal cortex (Allen Human Brain Atlas, Table S11).
Fig. 4.
Neuronal subtypes reveal heterogeneity amongst BAs. A. Multidimensional plot showing projection neuron subtypes distributed according to their predicted cortical layer (L) identity. Layer 4 Ex2 and Ex3 subtypes are indicated. B. Clusters shown in (A) colored by BA and with BA41/42 and BA17 subpopulations of Ex3 indicated. C. Violin plots showing differentially expressed genes between Ex2 and Ex3 subtypes (Table S8). D. Heatmap showing genes differentially expressed between BA17 and BA41/42 within the Ex3 subtype (Table S10).
To further understand the extent of heterogeneity that may exist within subtypes, we identified genes varying globally (Table S12, Fig. S15A) or expressed differentially within each BA (Table S13, Fig. S15B) for each subgroup. While a subset of In and Ex subgroup-variable genes were associated with differential expression between brain regions, a large proportion were unique (Fig. S15C). Therefore, the potential exists for not only intra-regional cortical transcriptomic differences, but also further intra-subtype heterogeneity. This might reflect a technical need for increased sampling depth for further subtype resolution, yet may also indicate the potential for even more diversity within subtypes associated with a broader range of individualized neuronal activities. Consistent with these observations, proportions of subgroup variable genes were associated with neuronal subtype classification, post-synaptic function and known regional expression variability (Fig. S15C). These data support further local and regional functional heterogeneity existing amongst defined subtypes.
Our results demonstrate that post-mortem SNS can identify expected and novel neuronal subtypes that provide insight into brain function through distinct profiles of activity defining genes (Fig. S16, Table S14). Furthermore, given that only a very small subset of layer specific markers used in our analyses (CARTPT, CHRNA7, PDYN, RELN) were found to have ISH differences between individual donors (17), our subtypes can be expected to be globally representative. Indeed, our subtypes remain highly conserved in mice (3), with differences highlighting evolutionary changes in potential orthologues (Fig. S12). Our data sets reveal shared gene expression signatures that can distinguish subtypes and regional identity, supporting a transcriptional basis for well-known differences in cortical cytoarchitecture. Additional heterogeneity found within single neuronal transcriptomes may further reflect activities of complex neuronal networks that vary with function and time, as well as underlying genomic mosaicism that exists in human cortical neurons (10, 20–23). Our study thus lays the groundwork for high-throughput global human brain transcriptome mapping using nuclei derived from readily available post-mortem tissues for analyses of normal individuals, as assessed here, as well as myriad diseases of brain and mind.
Supplementary Material
Figure S1. Overview of single nuclei sampling methodology. A. Schematic of human brain at the level of BA8 showing approximate region sampled, typical tissue quantity processed for fluorescent activated cell sorting (FACS), the approximate proportion of NeuN+ nuclei obtained, the quantity of NeuN+ nuclei needed for a single C1 loading, and the average single nuclei capture rate. Expected sample scaling and minimal tissue needed for a single C1 experiment is summarized. B. Samples generated using pooled sorted NeuN+ nuclei from BA8, BA10, BA17, BA21, BA22 and BA41/42 as well as matching tissue sections were analyzed for expression of oligodendrocyte (Oligo.), astrocyte (Astro.), endothelial (Endo.) and neuronal (Neuro.) marker genes (17). Violin plots show expression values for associated nuclear (Nuc.) and tissue (Tiss.) sample groupings. C. Data sets from ~120 pooled nuclei derived from either BA21 or BA17 were used to confirm enrichment for neurons or glia in NeuN+ and NeuN− sorts, respectively. D. Histograms showing the frequency of all single NeuN+ nuclei analyzed in this study binned by level of RBFOX3 (NeuN) expression. Neuronal nuclei were distinguished on the basis of either SLC17A7 (excitatory) or GAD1 (inhibitory) marker gene expression.
Figure S2. Limited RNA recovery from laser capture microdissection (LCM) of post-mortem brain. A. Fresh-frozen BA8 cerebral cortex sections stained with hematoxylin were subjected to LCM; well-separated cells were manually outlined by software, (B) cut out of the tissue, and (C) projected into 96-well caps that were visually verified. Scale bar = 75μm. D. Bioanalyzer trace results showing total RNA yields extracted from 100 cells either hand cut or isolated using LCM from fresh frozen brain sections. Results were compared with 1000 pg control human brain reference RNA.
Figure S3. SmartSeq Plus provides more uniform transcript coverage. Exon read coverage of all transcripts from SmartSeq and SmartSeq Plus libraries prepared from RNA extracted from bulk sorted BA8 neuronal nuclei.
Figure S4. Protocol comparison and improved sampling depth. A. Scatter plots showing high Pearson correlation coefficients (r) for expression values from all protein coding genes averaged across 10 single BA8 nuclei libraries generated using SmartSeq, SmartSeq Plus (SmartPlus) and SmartSeq Plus containing PolydIdC (SmartPoly) protocols. B. Proportion of reads mapped to different gene types for bulk tissue (t) and bulk nuclei (n) datasets generated using the SmartSeq Plus protocol, as well as single nuclei (1–3) datasets generated using the indicated protocols. C. Total number of genes detected averaged over indicated number of single nuclei datasets that were generated using the different indicated protocols. Arrow indicates improved protein-coding gene detection with SmartSeq Plus compared to the standard SmartSeq protocol. D. Ratio of mapped reads that were to either ERCC or genes for a single preparation of BA8 nuclei, comparing libraries: from nuclei processed immediately after sorting (no preservation); after cryofreezing (frozen with DMSO) using a medium C1 chip; after cryofreezing (frozen with DMSO) using a small C1 chip; after cryofreezing using a small C1 chip and the SmartPoly protocol. E. Corresponding comparison of conditions using scatter plots from averaged expression values (10 single nuclei, all protein coding genes) that show high correlation between conditions. F. Comparison of SmartPoly protocol across different brains using scatter plots from averaged expression values (10 single nuclei, all protein coding genes) that show high correlation between brain samples from different repositories (Brain 1 = BA10 tissue from patient 1568, NICHD Brain and Tissue Bank; Brain 2 = BA9 tissue from PT-WZTO, Genotype-Tissue Expression (GTEx) Biobank; Brain 3 = BA9 tissue from PT-NPJ8, GTEx Biobank).
Figure S5. Mapping statistics. A. Top panel: Proportion of all reads that were: unmapped; multi-mapped; uniquely mapped to ERCC transcripts; uniquely mapped to reference genes (exon and intron regions); or uniquely mapped between genes (intergenic). Middle panel: relative proportion of reads uniquely mapped to: ERCC transcripts; intergenic regions; or reference genes. Bottom panel: the proportion of reads uniquely mapped to the genome that were associated with: intergenic regions; introns; or exons. Results are shown for all 0 capture (0C), single capture (1C) and multiple capture (2+C) nuclei libraries (top and middle panels) or single capture only (1 Nuclei, bottom panel). B. Plots showing the frequency distribution of total number of reads sequenced and ERCC Pearson correlation r values [log10(counts+1) versus log10(concentration)] for all single nuclei libraries. C. Plots showing the frequency distributions of genes detected across single nuclei libraries using different gene count cutoffs.
Figure S6. Doublet screening and filtering. A. Heatmap of expression for excitatory (SLC17A7, SATB2, CBLN2) and inhibitory (GAD1, GAD2, SLC6A1) marker genes across 30 groups of neuronal nuclei data sets generated from the first round of clustering and classification. Arrow indicates cluster CL8 showing co-expression of these marker types. B. Multidimensional plot showing the 30 identified clusters and known two capture (2C) data sets (DIM, Dimension). Clusters with high overlap of 2C data sets are indicated. C. The percentage of data sets contributing to each cluster was calculated separately for small C1 chips and medium C1 chips and compared with the proportion of 2C data sets associated with each cluster. Arrows indicate clusters showing the highest number of prospective “doublets”. D. Percentage of identified “doublets” and their association with use of medium C1 chips across successive C1 runs.
Figure S7. Single-nuclei RNA sequencing permits cell type identification. A. Cluster dendrogram of gene expression (Log2(TPM+1)) using all protein coding genes, a subset of glial marker genes or a subset of neuronal marker genes (Table S16). Analyzed samples were generated using pooled sorted neuronal nuclei (n) from BA8 section 7 (s7n) or section 9 (s9n), BA10, BA17, BA21, BA22 and BA41/42 (BA41n) as well as matching tissue (t) sections using the SmartSeq Plus protocol or original SmartSeq protocol (BA8s7n(Smart)). Brain section numbers are assigned according to the University of Maryland Brain and Tissue Bank (Brain sectioning – Protocol Method 2). For comparison, single nuclei data sets from combined excitatory (Ex) and inhibitory (In) subtypes (n = 3084) were averaged (AveSN). B. Scatter plots comparing averaged single nucleus data and averaged bulk sorted nuclei or tissue data for protein coding, neuronal or glia marker genes. C. Scatter plots comparing single nucleus data, averaged 10 single nuclei data, or averaged 100 single nuclei data from BA21, with data from matched bulk nuclei or tissue (protein coding genes). D. Representative scatter plots comparing single nucleus data sets (protein coding genes). Associated Pearson r values are indicated (B–D).
Figure S8. Overview of subtype clustering and classification. A. Sample splitting at each step of the clustering and classification strategy showing: number of nuclei at each level; genes associated with each splitting (A–W, Table S3); final clusters associated with excitatory (Ex) and Inhibitory (In) neurons. Outlier cluster (n = 44, NoN) in the inhibitory branch is indicated by a dark circle. B. GO annotations associated with differentially expressed genes (DEGs) defining cluster splitting (fold change or FC ≥ 2) within excitatory or inhibitory subgroup branches (A) (Bonferroni adjusted p values < 0.05) (Table S4). C. Proportion of genes used for each branch of excitatory or inhibitory subgroup clustering (A) that were differentially expressed either between 2 and 10-fold or greater than 10 fold.
Figure S9. Differentially expressed subtype marker genes. A. Heatmap showing expression of 10-fold or more differentially expressed genes (Table S3) used for multidimensional plotting of neuronal subtypes (Fig. 1B, Fig. 4A, Fig. S10A). B. Heatmaps showing consistency in expression (top panel, TPM calculated from exon and intron reads) and corresponding fraction of positive values (bottom panel, TPM calculated from only exon reads) for unique marker genes (Table S5) identified for each neuronal subtype.
Figure S10. Neuronal subtypes do not show batch bias. A. Multidimensional plots as shown in Fig. 1B for neuronal subgroups indicating each experimental C1 run (Table S1). Arrow indicates an outlier cluster (n = 44, NoN, Table S2) derived from the 20150122B C1 run. B. Multidimensional plot using ERCC expression values showing all clusters identified in our analyses, demonstrating that unlike the outlier cluster indicated in (A) (arrow), our neuronal subtypes were identified independently of random batch specific expression differences.
Figure S11. Confirmation of subtype identity by RNA ISH. A. Fraction of positive heatmap of inhibitory (expressing GAD1) and excitatory (expressing SLC17A7) subtypes and a subset of representative combinatorial marker genes. B. Allen Human Brain Atlas ISH data (Table S11). Cortical stains are oriented from layer 1 (L1) to layer 6 (L6). A and B. Numbered boxes represent In subtype-specific combinations (e.g. 1 = In1) with spatial orientation in the cortex indicated through RNA ISH. Box 1 or In1 represents a layer 1 VIP+CNR1+RELN+ subgroup. Box 2 or In2 represents a VIP+CNR1+RELN− layer 2/3 subgroup that co-expresses OPRK1, as confirmed by neurons co-stained for OPRK1 and GAD1 in this region (C) (indicated by * in B and C). OPRK1 also labels SLC17A7 expressing layer 6/6b Ex7/Ex8 neurons (region indicated by ** in B and C). Box 4 or In4 represents a layer 3/4 RELN+SV2C+ subgroup that co-expresses SST in BA8, BA10 and BA21, but not in BA17, BA22 or BA41/42 (Fig. 2C). Consistently, In4-associated SST expression can be found within the temporal (Temp.) cortex (BA21) but not the visual cortex (BA17) (B). D. RNAscope co-staining of RELN and SST in BA8 and BA17 showing co-positive cell distributions that are consistent with RNA-seq data. Box 6 or In6 represents a layer 4/5 subgroup co-expressing SULF1 and PVALB. Box 3 or In3 represents a VIP+ RELN+ subgroup positive for PDE9A, which is also specifically expressed in the layer 5 Ex6 subgroup and shows a consistent expression pattern with HTR2C (A). E. PDE9A-positive In3 and Ex6 expression was confirmed by RNAscope co-staining with GAD1 (double positive restricted to layers 2/3) and SLC17A7 (double positive restricted to layers 5/6). F. RNAscope counts consistent with RNA-seq data.
Figure S12. Subtype comparison with a recent mouse study on the somatosensory cortex (3). A. Violin plots showing core interneuron marker genes in In subtypes and across BA regions, indicating a similar proportion of VIP+, SOM+ (SST) and PV+ subtypes between mouse and human, with the exception of additional RELN+ subtypes likely associated with differences in sampling methods between studies. A schematic for combinatorial expression summarizes species-specific differences. Int = mouse subtypes identified (3); In = human subtypes. B. Violin plots of excitatory markers used to define mouse pyramidal subtypes showing associated expression in human Ex subtypes and across BA regions. A high concordance in the pattern of expression can be found using both human subtypes and Allen Human Brain Atlas ISH data (Table S11). Cortical images are oriented from left (pial layer) to right (white matter) and regions sampled are indicated. Associated species-specific similarities or differences in observed cortical layer identity are summarized for each marker gene, including a shift in SCPN marker THSD7A in mouse to CPN in human and an observed shift in a claustrum pyramidal (clauPyr) neuronal subtype in mouse to layer 6b in human that may reflect evolutionary changes or differences in the regions examined between these studies.
Figure S13. Confirmation of layer identity in BA8 by RNA ISH. A. RNA in situ hybridization (ISH) analyses on BA8 cortical sections showing counts of positive cells for CBLN2 (Layers 2/3/6), BHLEH22 (Layer 4) and PCP4 (Layer 5) in image fields spanning from the pial layer (upper cortex) through to the white matter (lower cortex). Violin plots are corresponding gene expression values for excitatory neuron subtypes across all BA regions. B. RNAscope technology was used to stain BA8 sections for the layer markers RELN, MFGE8, PCP4 and CBLN2. Single-brown chromogenic or fluorescent staining (top) and average counts (bottom) are shown. Insets are of representative single positively stained neurons. Positive counts were derived from over 22 vertical sections spanning from pial surface and upper cortex (top) to lower cortex. Positive cells were counted by two independent observers over two independent regions. C. Fraction of positive heatmap showing the layer 4 specific marker BHLHE22 in Ex2 and Ex3 subgroups (*) which have correspondingly low positivity of its negatively regulated target gene CDH11 (37). D. Representative RNAscope images of BHLHE22 positive cells used for counts shown in (A), and which also show the expected absence of CDH11. Inset is a representative CDH11 positive cell that is negative for BHLHE22. The proportion of BHLHE22 positive cells having an absence or presence of CDH11 expression is shown through RNAscope counts and is consistent with RNA-seq data.
Figure S14. Neuronal subtype heterogeneity between brain regions. A. Violin plots showing expression values (black boxes) for genes with known differential expression between BA17 (visual cortex) and BA21 (temporal cortex) (17). Stains are associated RNA ISH analyses of cortical sections oriented with pial layer at the top (Allen Human Brain Atlas, Table S11). Double arrows indicate regions associated with the indicated differential expression. B. Violin plots showing indicated marker gene expression values for: each inhibitory (In) and excitatory (Ex) subtype and brain region (bar colors, bottom); each brain region from combined neuronal (NeuN+) data; each brain region from the specific subgroup showing BA17/BA21 expression differences (black arrows) associated with RNA ISH staining differences shown in (A). Additional subtypes that may account for these RNA ISH differences are indicated (gray arrows).
Figure S15. Subgroup variable genes. A. Plot of average expression and dispersion (binned log(Variance/mean)) for subgroup Ex1, indicating genes that show variance (z-score cutoff = 2) across these single nuclei (Table S12). B. Multidimensional plot on Ex1 nuclei using genes identified as being differentially expressed between BA regions of this subgroup (10-fold cutoff, Table S13). Nuclei show a distribution consistent with their spatial origination (Occipital, Temporal, Frontal lobes). DIM = dimension. C. Venn diagrams showing overlap between: all subgroup-derived variable genes (A, Table S12); all differentially expressed genes between BA regions for each subgroup (B, Table S13); genes defining subgroup clustering (DEG, Table S3); genes associated with the human post-synaptic density (hPSD) (38); and genes within the top five percentile for stable expression differences across cortical parcels (DS (Cortex)) (7).
Figure S16. Subtype expression patterns of electrophysiological-relevant genes. Top panels: fraction of positive values for ion channel and neurotransmitter-related genes (see Table S14) are shown for In and Ex subtypes (minimum FOP value of 0.1 in at least one cluster). Bottom panels: Fraction of positive values for select genes are shown for Ex subtypes, demonstrating potential for unique subtype-specific electrophysiological properties.
Table S1: Summary of C1 experimental conditions and outcome
Table S2: Single nuclei library details and group/subgroup identifiers
Table S3: Differentially expressed genes (fold change indicated as ≥ 2 and ≥ 10) underlying group cluster separation (A–W, See Fig. S8a), where “left” denotes genes upregulated in the left branch and “right” denotes genes upregulated in right branch.
Table S4. GO annotations for differentially expressed genes defining subgroup classifications (Table S3)
Table S5: Unique group specific genes and associated fraction of positive values using exon only derived TPM (see Fig. S9b)
Table S6. GO annotations for unique subgroup marker genes (Table S5)
Table S7: A. Distribution of brain regions amongst classification subgroups. B. Relative distribution of brain regions amongst classification groups on the basis of normalized input contributions (values are percentage of total within the group)
Table S8: Genes differentially expressed between Ex2 and Ex3 subgroups
Table S9. GO annotations for genes differentially expressed between Ex2 and Ex3 subgroups (See Table S8)
Table S10: Genes differentially expressed between BA41/42 and BA17 brain regions within the Ex3 subgroup
Table S11. Allen Human Brain Atlas ISH citations (See Fig. 3D, Fig. S11, Fig. S12, Fig. S14)
Table S12. Variable genes identified within each subgroup
Table S13. Genes identified in each subgroup having 10-fold or more difference in expression level between at least two Brodmann Areas (BA)
Table S14. Fraction of positive values for genes associated with neurotransmitter function or ion channels (see Fig. S16)
Table S15. Description of Imaging and methods used for RNAscope Validation
Table S16: Neuronal and glia marker genes (see Fig. S7)
Summary.
Single-nucleus RNA sequencing of neurons from the adult human cerebral cortex revealed transcriptomic signatures sufficient to identify neuronal subtypes and neuroanatomical areas while also revealing transcriptomic heterogeneity.
Acknowledgments
Flow cytometry was performed both at the UCSD Human Embryonic Stem Cell Core and TSRI Flow Cytometry Core. Initial C1 runs were performed at the UCSD Stem Cell Genomics Core. The data tables accompanying this work are provided as Excel files in the supplementary materials. Clustering-and-Classification code used to identify neuronal subtypes and instructions (Readme.txt) for its operation in R are provided as supplementary files. We thank Fluidigm Inc. (M. Ray, R.C. Jones, P. Steinberg) for instrument support and technical advice in adaptation of the C1 protocol for nuclei. Sequencing data has been deposited with dbGaP (accession phs000833.v3.p1), curated by the NIH Single Cell Analysis Program – Transcriptome (SCAP-T) Project (http://www.scap-t.org) and annotated in supplementary material (Table S2). We thank G. Kennedy for help with RNAscope. Funding support was from the NIH Common Fund Single Cell Analysis Program (1U01MH098977). GEK was additionally supported by Neuroplasticity of Aging Training Grant (5T32AG000216-24).
References
- 1.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 3.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 4.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawrylycz M, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832–1844. doi: 10.1038/nn.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman DM, et al. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. Elife. 2015;4 doi: 10.7554/eLife.05116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnaswami SR, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.See supplementary text, materials and methods available at Science Online.
- 14.Graf A, et al. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc Natl Acad Sci U S A. 2014;111:4139–4144. doi: 10.1073/pnas.1321569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 16.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng H, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 20.Lodato MA, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra JW, et al. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 25.Edelstein AD, et al. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1 doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardou P, Mariette J, Escudie F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Abruzzese RV, Polejaeva I, Davis S, Ji W. Amplification of nanogram amounts of total RNA by the SMART-based PCR method for high-density oligonucleotide microarrays. Clin Chem. 2005;51:2354–2356. doi: 10.1373/clinchem.2005.056721. [DOI] [PubMed] [Google Scholar]
- 31.Baker SC, et al. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 32.Fan J, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt D, et al. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron. 2014;82:350–364. doi: 10.1016/j.neuron.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinsey GL, et al. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77:83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaerlinger B, Hickel P, Etienne N, Guesnier L, Maroteaux L. Agonist actions of dihydroergotamine at 5-HT2B and 5-HT2C receptors and their possible relevance to antimigraine efficacy. Br J Pharmacol. 2003;140:277–284. doi: 10.1038/sj.bjp.0705437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross SE, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Overview of single nuclei sampling methodology. A. Schematic of human brain at the level of BA8 showing approximate region sampled, typical tissue quantity processed for fluorescent activated cell sorting (FACS), the approximate proportion of NeuN+ nuclei obtained, the quantity of NeuN+ nuclei needed for a single C1 loading, and the average single nuclei capture rate. Expected sample scaling and minimal tissue needed for a single C1 experiment is summarized. B. Samples generated using pooled sorted NeuN+ nuclei from BA8, BA10, BA17, BA21, BA22 and BA41/42 as well as matching tissue sections were analyzed for expression of oligodendrocyte (Oligo.), astrocyte (Astro.), endothelial (Endo.) and neuronal (Neuro.) marker genes (17). Violin plots show expression values for associated nuclear (Nuc.) and tissue (Tiss.) sample groupings. C. Data sets from ~120 pooled nuclei derived from either BA21 or BA17 were used to confirm enrichment for neurons or glia in NeuN+ and NeuN− sorts, respectively. D. Histograms showing the frequency of all single NeuN+ nuclei analyzed in this study binned by level of RBFOX3 (NeuN) expression. Neuronal nuclei were distinguished on the basis of either SLC17A7 (excitatory) or GAD1 (inhibitory) marker gene expression.
Figure S2. Limited RNA recovery from laser capture microdissection (LCM) of post-mortem brain. A. Fresh-frozen BA8 cerebral cortex sections stained with hematoxylin were subjected to LCM; well-separated cells were manually outlined by software, (B) cut out of the tissue, and (C) projected into 96-well caps that were visually verified. Scale bar = 75μm. D. Bioanalyzer trace results showing total RNA yields extracted from 100 cells either hand cut or isolated using LCM from fresh frozen brain sections. Results were compared with 1000 pg control human brain reference RNA.
Figure S3. SmartSeq Plus provides more uniform transcript coverage. Exon read coverage of all transcripts from SmartSeq and SmartSeq Plus libraries prepared from RNA extracted from bulk sorted BA8 neuronal nuclei.
Figure S4. Protocol comparison and improved sampling depth. A. Scatter plots showing high Pearson correlation coefficients (r) for expression values from all protein coding genes averaged across 10 single BA8 nuclei libraries generated using SmartSeq, SmartSeq Plus (SmartPlus) and SmartSeq Plus containing PolydIdC (SmartPoly) protocols. B. Proportion of reads mapped to different gene types for bulk tissue (t) and bulk nuclei (n) datasets generated using the SmartSeq Plus protocol, as well as single nuclei (1–3) datasets generated using the indicated protocols. C. Total number of genes detected averaged over indicated number of single nuclei datasets that were generated using the different indicated protocols. Arrow indicates improved protein-coding gene detection with SmartSeq Plus compared to the standard SmartSeq protocol. D. Ratio of mapped reads that were to either ERCC or genes for a single preparation of BA8 nuclei, comparing libraries: from nuclei processed immediately after sorting (no preservation); after cryofreezing (frozen with DMSO) using a medium C1 chip; after cryofreezing (frozen with DMSO) using a small C1 chip; after cryofreezing using a small C1 chip and the SmartPoly protocol. E. Corresponding comparison of conditions using scatter plots from averaged expression values (10 single nuclei, all protein coding genes) that show high correlation between conditions. F. Comparison of SmartPoly protocol across different brains using scatter plots from averaged expression values (10 single nuclei, all protein coding genes) that show high correlation between brain samples from different repositories (Brain 1 = BA10 tissue from patient 1568, NICHD Brain and Tissue Bank; Brain 2 = BA9 tissue from PT-WZTO, Genotype-Tissue Expression (GTEx) Biobank; Brain 3 = BA9 tissue from PT-NPJ8, GTEx Biobank).
Figure S5. Mapping statistics. A. Top panel: Proportion of all reads that were: unmapped; multi-mapped; uniquely mapped to ERCC transcripts; uniquely mapped to reference genes (exon and intron regions); or uniquely mapped between genes (intergenic). Middle panel: relative proportion of reads uniquely mapped to: ERCC transcripts; intergenic regions; or reference genes. Bottom panel: the proportion of reads uniquely mapped to the genome that were associated with: intergenic regions; introns; or exons. Results are shown for all 0 capture (0C), single capture (1C) and multiple capture (2+C) nuclei libraries (top and middle panels) or single capture only (1 Nuclei, bottom panel). B. Plots showing the frequency distribution of total number of reads sequenced and ERCC Pearson correlation r values [log10(counts+1) versus log10(concentration)] for all single nuclei libraries. C. Plots showing the frequency distributions of genes detected across single nuclei libraries using different gene count cutoffs.
Figure S6. Doublet screening and filtering. A. Heatmap of expression for excitatory (SLC17A7, SATB2, CBLN2) and inhibitory (GAD1, GAD2, SLC6A1) marker genes across 30 groups of neuronal nuclei data sets generated from the first round of clustering and classification. Arrow indicates cluster CL8 showing co-expression of these marker types. B. Multidimensional plot showing the 30 identified clusters and known two capture (2C) data sets (DIM, Dimension). Clusters with high overlap of 2C data sets are indicated. C. The percentage of data sets contributing to each cluster was calculated separately for small C1 chips and medium C1 chips and compared with the proportion of 2C data sets associated with each cluster. Arrows indicate clusters showing the highest number of prospective “doublets”. D. Percentage of identified “doublets” and their association with use of medium C1 chips across successive C1 runs.
Figure S7. Single-nuclei RNA sequencing permits cell type identification. A. Cluster dendrogram of gene expression (Log2(TPM+1)) using all protein coding genes, a subset of glial marker genes or a subset of neuronal marker genes (Table S16). Analyzed samples were generated using pooled sorted neuronal nuclei (n) from BA8 section 7 (s7n) or section 9 (s9n), BA10, BA17, BA21, BA22 and BA41/42 (BA41n) as well as matching tissue (t) sections using the SmartSeq Plus protocol or original SmartSeq protocol (BA8s7n(Smart)). Brain section numbers are assigned according to the University of Maryland Brain and Tissue Bank (Brain sectioning – Protocol Method 2). For comparison, single nuclei data sets from combined excitatory (Ex) and inhibitory (In) subtypes (n = 3084) were averaged (AveSN). B. Scatter plots comparing averaged single nucleus data and averaged bulk sorted nuclei or tissue data for protein coding, neuronal or glia marker genes. C. Scatter plots comparing single nucleus data, averaged 10 single nuclei data, or averaged 100 single nuclei data from BA21, with data from matched bulk nuclei or tissue (protein coding genes). D. Representative scatter plots comparing single nucleus data sets (protein coding genes). Associated Pearson r values are indicated (B–D).
Figure S8. Overview of subtype clustering and classification. A. Sample splitting at each step of the clustering and classification strategy showing: number of nuclei at each level; genes associated with each splitting (A–W, Table S3); final clusters associated with excitatory (Ex) and Inhibitory (In) neurons. Outlier cluster (n = 44, NoN) in the inhibitory branch is indicated by a dark circle. B. GO annotations associated with differentially expressed genes (DEGs) defining cluster splitting (fold change or FC ≥ 2) within excitatory or inhibitory subgroup branches (A) (Bonferroni adjusted p values < 0.05) (Table S4). C. Proportion of genes used for each branch of excitatory or inhibitory subgroup clustering (A) that were differentially expressed either between 2 and 10-fold or greater than 10 fold.
Figure S9. Differentially expressed subtype marker genes. A. Heatmap showing expression of 10-fold or more differentially expressed genes (Table S3) used for multidimensional plotting of neuronal subtypes (Fig. 1B, Fig. 4A, Fig. S10A). B. Heatmaps showing consistency in expression (top panel, TPM calculated from exon and intron reads) and corresponding fraction of positive values (bottom panel, TPM calculated from only exon reads) for unique marker genes (Table S5) identified for each neuronal subtype.
Figure S10. Neuronal subtypes do not show batch bias. A. Multidimensional plots as shown in Fig. 1B for neuronal subgroups indicating each experimental C1 run (Table S1). Arrow indicates an outlier cluster (n = 44, NoN, Table S2) derived from the 20150122B C1 run. B. Multidimensional plot using ERCC expression values showing all clusters identified in our analyses, demonstrating that unlike the outlier cluster indicated in (A) (arrow), our neuronal subtypes were identified independently of random batch specific expression differences.
Figure S11. Confirmation of subtype identity by RNA ISH. A. Fraction of positive heatmap of inhibitory (expressing GAD1) and excitatory (expressing SLC17A7) subtypes and a subset of representative combinatorial marker genes. B. Allen Human Brain Atlas ISH data (Table S11). Cortical stains are oriented from layer 1 (L1) to layer 6 (L6). A and B. Numbered boxes represent In subtype-specific combinations (e.g. 1 = In1) with spatial orientation in the cortex indicated through RNA ISH. Box 1 or In1 represents a layer 1 VIP+CNR1+RELN+ subgroup. Box 2 or In2 represents a VIP+CNR1+RELN− layer 2/3 subgroup that co-expresses OPRK1, as confirmed by neurons co-stained for OPRK1 and GAD1 in this region (C) (indicated by * in B and C). OPRK1 also labels SLC17A7 expressing layer 6/6b Ex7/Ex8 neurons (region indicated by ** in B and C). Box 4 or In4 represents a layer 3/4 RELN+SV2C+ subgroup that co-expresses SST in BA8, BA10 and BA21, but not in BA17, BA22 or BA41/42 (Fig. 2C). Consistently, In4-associated SST expression can be found within the temporal (Temp.) cortex (BA21) but not the visual cortex (BA17) (B). D. RNAscope co-staining of RELN and SST in BA8 and BA17 showing co-positive cell distributions that are consistent with RNA-seq data. Box 6 or In6 represents a layer 4/5 subgroup co-expressing SULF1 and PVALB. Box 3 or In3 represents a VIP+ RELN+ subgroup positive for PDE9A, which is also specifically expressed in the layer 5 Ex6 subgroup and shows a consistent expression pattern with HTR2C (A). E. PDE9A-positive In3 and Ex6 expression was confirmed by RNAscope co-staining with GAD1 (double positive restricted to layers 2/3) and SLC17A7 (double positive restricted to layers 5/6). F. RNAscope counts consistent with RNA-seq data.
Figure S12. Subtype comparison with a recent mouse study on the somatosensory cortex (3). A. Violin plots showing core interneuron marker genes in In subtypes and across BA regions, indicating a similar proportion of VIP+, SOM+ (SST) and PV+ subtypes between mouse and human, with the exception of additional RELN+ subtypes likely associated with differences in sampling methods between studies. A schematic for combinatorial expression summarizes species-specific differences. Int = mouse subtypes identified (3); In = human subtypes. B. Violin plots of excitatory markers used to define mouse pyramidal subtypes showing associated expression in human Ex subtypes and across BA regions. A high concordance in the pattern of expression can be found using both human subtypes and Allen Human Brain Atlas ISH data (Table S11). Cortical images are oriented from left (pial layer) to right (white matter) and regions sampled are indicated. Associated species-specific similarities or differences in observed cortical layer identity are summarized for each marker gene, including a shift in SCPN marker THSD7A in mouse to CPN in human and an observed shift in a claustrum pyramidal (clauPyr) neuronal subtype in mouse to layer 6b in human that may reflect evolutionary changes or differences in the regions examined between these studies.
Figure S13. Confirmation of layer identity in BA8 by RNA ISH. A. RNA in situ hybridization (ISH) analyses on BA8 cortical sections showing counts of positive cells for CBLN2 (Layers 2/3/6), BHLEH22 (Layer 4) and PCP4 (Layer 5) in image fields spanning from the pial layer (upper cortex) through to the white matter (lower cortex). Violin plots are corresponding gene expression values for excitatory neuron subtypes across all BA regions. B. RNAscope technology was used to stain BA8 sections for the layer markers RELN, MFGE8, PCP4 and CBLN2. Single-brown chromogenic or fluorescent staining (top) and average counts (bottom) are shown. Insets are of representative single positively stained neurons. Positive counts were derived from over 22 vertical sections spanning from pial surface and upper cortex (top) to lower cortex. Positive cells were counted by two independent observers over two independent regions. C. Fraction of positive heatmap showing the layer 4 specific marker BHLHE22 in Ex2 and Ex3 subgroups (*) which have correspondingly low positivity of its negatively regulated target gene CDH11 (37). D. Representative RNAscope images of BHLHE22 positive cells used for counts shown in (A), and which also show the expected absence of CDH11. Inset is a representative CDH11 positive cell that is negative for BHLHE22. The proportion of BHLHE22 positive cells having an absence or presence of CDH11 expression is shown through RNAscope counts and is consistent with RNA-seq data.
Figure S14. Neuronal subtype heterogeneity between brain regions. A. Violin plots showing expression values (black boxes) for genes with known differential expression between BA17 (visual cortex) and BA21 (temporal cortex) (17). Stains are associated RNA ISH analyses of cortical sections oriented with pial layer at the top (Allen Human Brain Atlas, Table S11). Double arrows indicate regions associated with the indicated differential expression. B. Violin plots showing indicated marker gene expression values for: each inhibitory (In) and excitatory (Ex) subtype and brain region (bar colors, bottom); each brain region from combined neuronal (NeuN+) data; each brain region from the specific subgroup showing BA17/BA21 expression differences (black arrows) associated with RNA ISH staining differences shown in (A). Additional subtypes that may account for these RNA ISH differences are indicated (gray arrows).
Figure S15. Subgroup variable genes. A. Plot of average expression and dispersion (binned log(Variance/mean)) for subgroup Ex1, indicating genes that show variance (z-score cutoff = 2) across these single nuclei (Table S12). B. Multidimensional plot on Ex1 nuclei using genes identified as being differentially expressed between BA regions of this subgroup (10-fold cutoff, Table S13). Nuclei show a distribution consistent with their spatial origination (Occipital, Temporal, Frontal lobes). DIM = dimension. C. Venn diagrams showing overlap between: all subgroup-derived variable genes (A, Table S12); all differentially expressed genes between BA regions for each subgroup (B, Table S13); genes defining subgroup clustering (DEG, Table S3); genes associated with the human post-synaptic density (hPSD) (38); and genes within the top five percentile for stable expression differences across cortical parcels (DS (Cortex)) (7).
Figure S16. Subtype expression patterns of electrophysiological-relevant genes. Top panels: fraction of positive values for ion channel and neurotransmitter-related genes (see Table S14) are shown for In and Ex subtypes (minimum FOP value of 0.1 in at least one cluster). Bottom panels: Fraction of positive values for select genes are shown for Ex subtypes, demonstrating potential for unique subtype-specific electrophysiological properties.
Table S1: Summary of C1 experimental conditions and outcome
Table S2: Single nuclei library details and group/subgroup identifiers
Table S3: Differentially expressed genes (fold change indicated as ≥ 2 and ≥ 10) underlying group cluster separation (A–W, See Fig. S8a), where “left” denotes genes upregulated in the left branch and “right” denotes genes upregulated in right branch.
Table S4. GO annotations for differentially expressed genes defining subgroup classifications (Table S3)
Table S5: Unique group specific genes and associated fraction of positive values using exon only derived TPM (see Fig. S9b)
Table S6. GO annotations for unique subgroup marker genes (Table S5)
Table S7: A. Distribution of brain regions amongst classification subgroups. B. Relative distribution of brain regions amongst classification groups on the basis of normalized input contributions (values are percentage of total within the group)
Table S8: Genes differentially expressed between Ex2 and Ex3 subgroups
Table S9. GO annotations for genes differentially expressed between Ex2 and Ex3 subgroups (See Table S8)
Table S10: Genes differentially expressed between BA41/42 and BA17 brain regions within the Ex3 subgroup
Table S11. Allen Human Brain Atlas ISH citations (See Fig. 3D, Fig. S11, Fig. S12, Fig. S14)
Table S12. Variable genes identified within each subgroup
Table S13. Genes identified in each subgroup having 10-fold or more difference in expression level between at least two Brodmann Areas (BA)
Table S14. Fraction of positive values for genes associated with neurotransmitter function or ion channels (see Fig. S16)
Table S15. Description of Imaging and methods used for RNAscope Validation
Table S16: Neuronal and glia marker genes (see Fig. S7)