Abstract
Gastric cancer (GC) is a prevalent cancer, which remains incurable, and therefore requires an alternative treatment method. Celecoxib is a nonsteroidal anti-inflammatory drug that targets cyclooxygenase-2, and exhibits anticancer effects. The present study aimed to investigate the anti-GC mechanism of celecoxib using bioinformatics methods. Gene expression datasets GSE56807 (GC tissues and normal gastric tissues) and GSE54657 (celecoxib-treated and non-treated human GC epithelial AGS cells) were downloaded from the Gene Expression Omnibus database. Two groups of differentially expressed genes (DEGs) were identified using limma package in R language. The criterion for GSE56807 was a false discovery rate of <0.05, while that for GSE54657 was P<0.01. Overlapping DEGs from the two datasets were screened out. Subsequently, pathway enrichment analysis was performed using Database for Annotation, Visualization and Integrated Discovery software (P<0.1; gene count ≥2). In addition, the protein-protein interactions (PPIs) among the overlapped DEGs were obtained based on IntAct, Database of Interacting Proteins, Biomolecular Interaction Network Database and Human Protein Reference Database. Finally, a PPI network was visualized using Cytoscape software. A total of 137 overlapped DEGs were obtained, and DEGs with opposite regulation directions in the two datasets were significantly enriched in focal adhesion and leukocyte transendothelial migration. Subsequently, a PPI network of overlapped DEGs was constructed. Comprehensively, a total of 8 key DEGs [cysteine and glycine rich protein 1 (CSRP1), thrombospondin 1 (THBS1), myosin light chain 9 (MYL9), filamin A (FLNA), actinin alpha 1 (ACTN1), vinculin (VCL), laminin subunit gamma 2 (LAMC2) and claudin 1 (CLDN1)] were upregulated in GC tissues and downregulated in celecoxib-treated cells. In conclusion, celecoxib may exhibit anti-GC effects by suppressing the expression of CSRP1, THBS1, MYL9, FLNA, ACTN1, VCL, LAMC2 and CLDN1, and inhibiting leukocyte transendothelial migration and focal adhesion. However, relevant experiments are required to confirm the conclusion of the present study.
Keywords: celecoxib, gastric cancer, anticancer mechanism, pathway enrichment analysis, protein-protein interaction
Introduction
Gastric cancer (GC) is the fifth most prevalent cancer and the third leading cause of cancer-associated mortality (1). The prognosis of GC is generally poor, since GC metastasize frequently from gastric glands to other parts of the body. The 5-year survival rate of GC is <10% (2). Therefore, study concerning the treatment of GC is of great importance.
Treatments for GC include surgery, radiation therapy and chemotherapy (3). Drugs used in GC treatment primarily consist of fluorouracil or its analog capecitabine, carmustine, mitomycin C, semustine, doxorubicin, cisplatin and taxotere (3,4). However, the results of these treatments are unsatisfactory, and GC remains incurable; therefore, requiring an alternative treatment method (2).
Celecoxib is a classic nonsteroidal anti-inflammatory drug (NSAID) targeting cyclooxygenase-2 (COX-2). Previous epidemiological studies have demonstrated that prolonged treatment with NSAIDs may reduce the risk of GC (5,6). Reportedly, celecoxib possesses anticancer effects, since it downregulates AKT serine/threonine kinase 1, glycogen synthase kinase 3β and forkhead box O1, and upregulates caspase-9 in the mitochondrial apoptotic pathway (7). In addition, celecoxib regulates cell cycle arrest, mitochondrial cytochrome C release and caspase activation in cancer cells (8). Furthermore, celecoxib suppresses the invasion of GC by affecting the expression of E-cadherin, vascular endothelial growth factor and COX-2, and interfering with nuclear factor-κB signaling, Snail signaling and microvessel density (9,10). However, the anti-GC mechanism of celecoxib remains unclear and requires further study.
The present study aimed to improve the understanding of the anti-GC mechanism of celecoxib using bioinformatics methods. Two gene expression datasets (GSE56807, GC vs. normal gastric tissues; GSE54657, celecoxib-treated vs. non-treated GC cells) were downloaded from the Gene Expression Omnibus (GEO) database. GSE56807 was uploaded by Wang et al (11), who investigated hypoxia inducible factor-1α-regulated transcription factors and regulatory signaling pathways in GC. The present study identified two sets of differentially expressed genes (DEGs) and their overlapped DEGs from the two datasets. Furthermore, pathway enrichment analysis was performed, and a protein-protein interaction (PPI) network was constructed to predict the targets of celecoxib.
Materials and methods
Microarray data
Two gene expression datasets, GSE56807 (11) and GSE54657 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54657), were downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo/) (12). Dataset GSE56807 consisted of 10 samples, which included 5 pairs of GC and normal gastric tissues. Its corresponding platform is GPL5175 [HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [transcript (gene) version]. Dataset GSE54657 consisted of 6 samples, which included 3 celecoxib-treated human GC epithelial AGS cell line samples and 3 non-treated AGS cell line samples. The celecoxib-treated AGS samples were harvested following incubation with 20 µM celecoxib for 24 h in triplicate. The corresponding platform for GSE54657 is GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version].
Data preprocessing and DEGs screening
The downloaded raw gene expression data were preprocessed based on R language (13). Data in different chips were normalized using the Robust Multichip Averaging algorithm (14). Subsequently, limma version 3.22.1 software (www.bioconductor.org/packages/release/bioc/html/limma.html) (15) in R language was applied to identify DEGs. For DEGs between GC tissues and normal gastric tissues (GSE56807), the P-value was adjusted using the Benjamini-Hochberg method (16). Adjusted P<0.05, also known as false discovery rate (FDR), was set as the criterion for DEG screening. For DEGs between celecoxib-treated AGS samples and non-treated AGS samples (GSE54657), P<0.01 was set as the cut-off criterion. DEGs shared by the two DEG groups were defined as overlapped DEGs, which exhibited the same or opposite regulation directions in the two datasets.
Hierarchical clustering analysis
In order to determine the sample-specificity of the overlapped DEGs, bidirectional hierarchical clustering analysis (BHCA) (17) was performed using pheatmap version 1.0.8 package (https://cran.r-project.org/web/packages/pheatmap/) in R language. DEGs with similar expression patterns were clustered.
Pathway enrichment analysis
Pathway enrichment analysis of the overlapped DEGs was performed using Database for Annotation, Visualization and Integrated Discovery software version 6.7 (david.abcc.ncifcrf.gov/) (18) based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (19). P<0.1 and gene count ≥2 were set as the cut-off criteria.
Construction of a PPI network
Based on the PPI data downloaded from IntAct (www.ebi.ac.uk/intact/) (20), Database of Interacting Proteins (dip.doe-mbi.ucla.edu/dip/Main.cgi) (21), Biomolecular Interaction Network Database (http://bind.ca) (22) and Human Protein Reference Database (http://www.hprd.org/) (23), and the studies by Rual et al (24), Stelzl et al (25) and Ramani et al (26), a PPI set was established, which investigated the PPIs among the overlapped DEGs. Subsequently, the PPI network of DEGs was visualized using Cytoscape version 3.2.0 software (www.cytoscape.org/) (27).
Results
DEG screening and BHCA
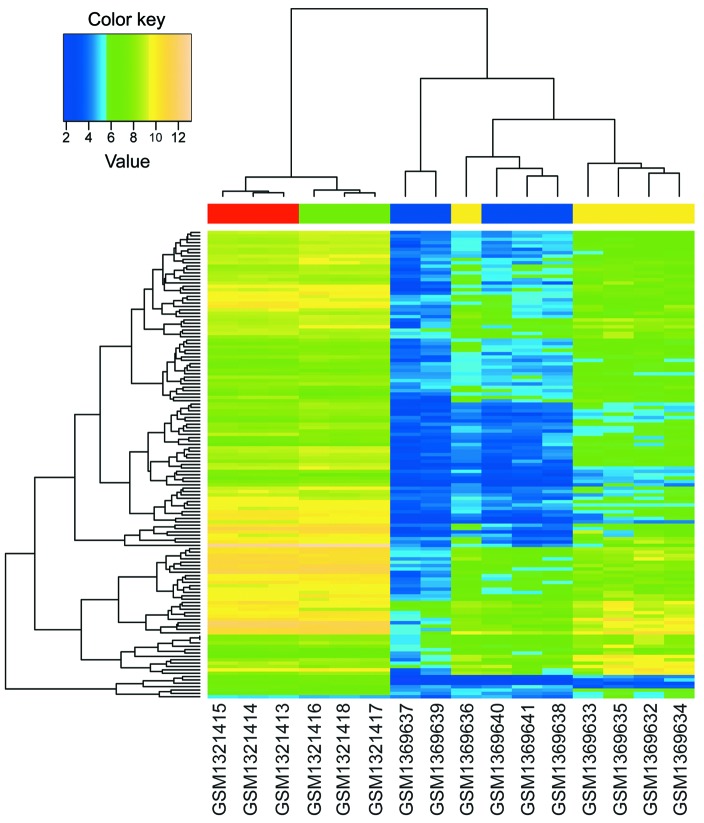
Based on the aforementioned analysis, 5190 DEGs (FDR <0.05) between GC tissues and normal gastric tissues, and 540 DEGs (P<0.01) between celecoxib-treated AGS samples and non-treated AGS samples were identified. In total, 137 overlapped DEGs were obtained by investigating the intersection of the two DEG groups. Furthermore, BHCA was performed, and the genes with similar expression patterns are shown in Fig. 1. The two datasets used in the present study were based on different platforms; therefore, gene expression levels exhibited a large difference between the two datasets. However, qualitative conclusions were obtained from Fig. 1: The overlapped DEGs could differentiate between GC and normal gastric tissues, and between celecoxib-treated and non-treated AGS samples; and celecoxib did affect the gene expression patterns of AGS cells during treatment.
Figure 1.
Bidirectional hierarchical clustering analysis of overlapped differently expressed genes. Blue terms, normal gastric tissues; yellow terms, gastric cancer samples; red terms, non-treated human gastric cancer epithelial AGS cell samples; green terms, celecoxib-treated AGS samples.
Pathway enrichment analysis
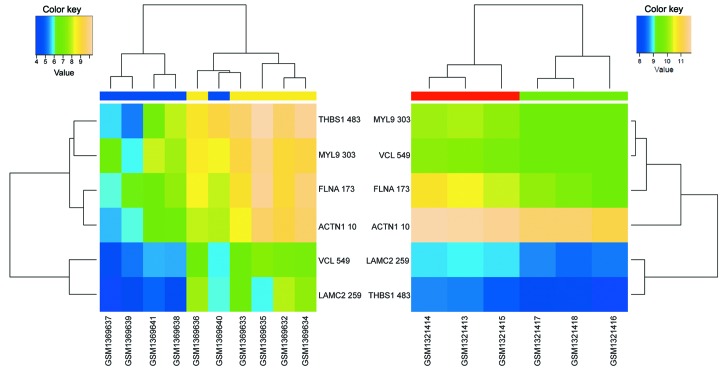
To investigate the mechanism underlying the effects of celecoxib on GC cells, KEGG pathway enrichment analysis of the 137 overlapped DEGs was performed. As shown in Table I, the overlapped DEGs were primarily enriched in four pathways (P<0.1; gene count ≥2), including lysosome, other glycan degradation, focal adhesion and leukocyte transendothelial migration pathways. Among these pathways, focal adhesion (Fig. 2) and leukocyte transendothelial migration (Fig. 3) were enriched by DEGs that exhibited opposite regulation directions in the two datasets. In fact, 6 DEGs enriched in focal adhesion [thrombospondin 1 (THBS1), myosin light chain 9 (MYL9), filamin A (FLNA), actinin alpha 1 (ACTN1), vinculin (VCL), laminin subunit gamma 2 (LAMC2)] and 4 DEGs enriched in leukocyte transendothelial migration [MYL9, VCL, ACTN1 and claudin 1 (CLDN1)] were significantly upregulated in GC tissues compared with normal gastric tissues, and significantly downregulated in celecoxib-treated AGS cells compared with non-treated AGS cells.
Table I.
Significantly enriched pathways in overlapped DEGs between two datasets used by the present study.
| Pathway ID | Pathway | Gene count | DEGs | RD | P-value |
|---|---|---|---|---|---|
| hsa04142 | Lysosome | 6 | NEU1, GLB1, FUCA1, CLN5, ATP6AP1, CTSD | Same | 0.002834 |
| hsa00511 | Other glycan degradation | 3 | NEU1, GLB1, FUCA1 | Same | 0.007776 |
| hsa04510 | Focal adhesion | 6 | THBS1, MYL9, FLNA, ACTN1, VCL, LAMC2 | Opposite | 0.026095 |
| hsa04670 | Leukocyte transendothelial migration | 4 | MYL9, VCL, ACTN1, CLDN1 | Opposite | 0.076932 |
Datasets, GSE56807 and GSE54657. DEGs, differently expressed genes; RD, regulation direction of the two datasets used; NEU1, neuraminidase 1; GLB1, galactosidase beta 1; FUCA1, fucosidase, alpha-L-1; CLN5, ceroid-lipofuscinosis, neuronal 5; ATP6AP1, ATPase H+ transporting accessory protein 1; CTSD, cathepsin D; THBS1, thrombospondin 1; MYL9, myosin light chain 9; FLNA, filamin A; ACTN1, actinin alpha 1; VCL, vinculin; LAMC2, laminin subunit gamma 2; CLDN1, claudin 1.
Figure 2.
Bidirectional hierarchical clustering analysis of differently expressed genes enriched in focal adhesion pathways. Blue terms, normal gastric tissues; yellow terms, gastric cancer samples; red terms, non-treated human gastric cancer epithelial AGS cell samples; green terms, celecoxib-treated AGS samples.
Figure 3.
Bidirectional hierarchical clustering analysis of differently expressed genes enriched in leukocyte transendothelial migration pathways. Blue terms, normal gastric tissues; yellow terms, gastric cancer samples; red terms, non-treated human gastric cancer epithelial AGS cell samples; green terms, celecoxib-treated AGS samples.
Construction of PPI network
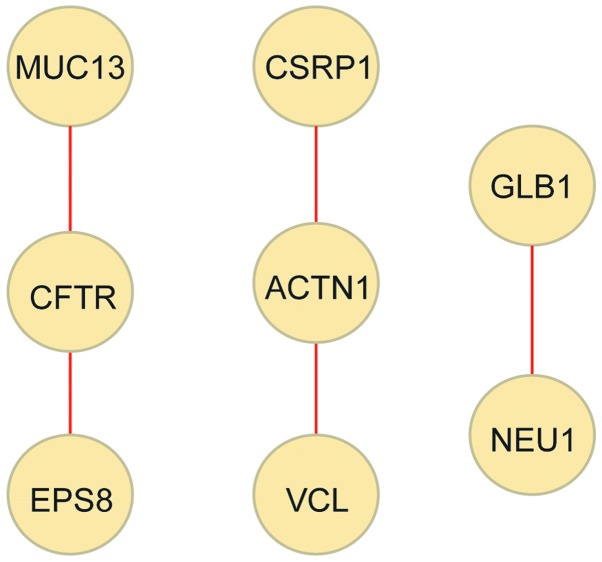
To further investigate the underlying mechanism of celecoxib in GC treatment, a PPI network of the 137 overlapped DEGs was constructed, consisting of 8 DEGs and 5 PPIs (Fig. 4). Cysteine and glycine rich protein 1 (CSRP1), VCL and ACTN1 exhibited opposite regulation directions in the two datasets. In fact, CSRP1, VCL and ACTN1 were significantly upregulated in GC tissues compared with normal gastric tissues, while significantly downregulated in celecoxib-treated AGS cells compared with non-treated AGS cells. Comprehensively, a total of 8 key DEGs (CSRP1, THBS1, MYL9, FLNA, ACTN1, VCL, LAMC2 and CLDN1) were upregulated in GC tissues and downregulated in celecoxib-treated cells.
Figure 4.
Protein-protein interaction network of the overlapped DEGs. Yellow nodes, DEGs (proteins); red lines, interactions between DEGs (proteins). DEGs, differently expressed genes; MUC13, mucin 13, cell surface associated; CSRP1, cysteine and glycine rich protein 1; GLB1, galactosidase beta 1; CFTR, cystic fibrosis transmembrane conductance regulator; ACTN1, actinin alpha 1; NEU1, neuraminidase 1; EPS8, epidermal growth factor receptor pathway substrate 8; VCL, vinculin.
Discussion
GC is a prevalent and malignant cancer, which remains incurable. As a NSAID, celecoxib is a potentially effective chemotherapy for GC. However, the anti-GC mechanism of celecoxib remains unclear. To gain insight into the anti-GC mechanism of celecoxib, the present study systematically analyzed the gene expression data of gastric tissue and human gastric cancer epithelial AGS cells using bioinformatics methods. Consequently, a total of 137 genes were identified to be differentially expressed between GC and normal gastric tissues, and between celecoxib-treated and non-treated AGS cells. Following pathway enrichment analysis and PPI network construction, 8 key DEGs (CSRP1, THBS1, MYL9, FLNA, ACTN1, VCL, LAMC2 and CLDN1) were identified, which were upregulated in GC tissues and downregulated in celecoxib-treated cells, and enriched in focal adhesion and leukocyte transendothelial migration pathways.
Among the 8 DEGs, THBS1, MYL9, FLNA, LAMC2 and CLDN1 have been reported to participate in the development of GC, as follows: THBS1 encodes thrombospondin-1, and its expression is a prognostic factor in advanced GC (28); MYL9 encodes myosin light chain 9, which is an upregulated apoptosis-associated protein and biomarker in GC (29); FLNA encodes filamin A, which is aberrantly regulated in GC tissue and regulates migration and invasion of GC cells in vitro (30); LAMC2 encodes the γ2 chain of laminin-5, which is a major component of the basement membrane, and the expression of LAMC2 is frequently upregulated by promoter demethylation in GC (31); CLDN1 encodes claudin 1, a tight junction protein that is critical in the maintenance of epithelial integrity. The overexpression of CLDN1 has been identified in GC patients with lymph node metastasis, and is associated with decreased overall survival (32). In the present study, THBS1, MYL9, FLNA, LAMC2 and CLDN1 were aberrantly upregulated in GC tissue. This is consistent with previous studies, particularly studies concerning MYL9, LAMC2 and CLDN1 (29,31,32). In addition, these genes were significantly downregulated in celecoxib-treated cells, indicating that celecoxib may exhibit an anti-GC effect by targeting these genes.
Following PPI network construction by the present study, it was revealed that ACTN1 interacted with CSRP1 and VCL. VCL encodes vinculin, an important focal adhesion protein that is responsible for cell-matrix junction and signal transduction on the membrane. Vinculin forms a vinculin-talin-actin scaffolding complex and promotes the malignancy and invasiveness of various cancers, including breast (33), pancreatic (34) and prostate (35) cancer. Additionally, the dephosphorylation of vinculin in gastric epithelial cells results in altered cell-matrix adhesion, contractility, motility and wound repair (36). In the present study, the gene expression value of VCL was significantly upregulated in GC tissues [log2 fold change (FC)=1.59; FDR=0.0043], indicating that VCL may participate in GC progression by promoting tumor malignancy and invasiveness.
ACTN1 encodes actinin α1, which triggers the unmasking of vinculin, allowing for F-actin binding and vinculin activation. In this way, the transmission of extracellular or intracellular forces and integrin-mediated mechano-chemical signaling are enabled (37). In the present study, the gene expression value of ACTN1 was significantly upregulated in GC tissues (log2FC=2.43; FDR=0.0044), indicating that ACTN1 may participate in GC progression by activating VCL and promoting tumor malignancy and invasiveness.
CSRP1 is a member of the CSRP family, which encodes a group of proteins with LIM (Lin11, Isl-1 and Mec-3) domains. CSRPs are generally transcription regulators associated with gene regulation, cell growth and somatic differentiation (38). In the present study, CSRP1 was upregulated in GC tissues (log2FC=2.88; FDR=0.0061), and interacted with ACTN1, indicating that the upregulation of CSRP1 may promote GC progression by regulating the expression of ACTN1. It should be noted that CSRP1 (log2FC=−0.53; P=0.0002), VCL (log2FC=−0.48; P=0.0011) and ACTN1 (log2FC=−0.39; P=0.0058) were significantly downregulated in celecoxib-treated AGS cells, indicating that celecoxib may exhibit an anti-GC effect by targeting CSRP1, VCL and ACTN1.
Furthermore, THBS1, MYL9, FLNA, ACTN1, VCL and LAMC2 were identified to be significantly enriched in focal adhesion pathways by the present study, while MYL9, VCL, ACTN1, and CLDN1 were significantly enriched in leukocyte transendothelial migration pathways. Focal adhesion is essential in various important biological processes, including cell survival and apoptosis (39), and leukocyte transendothelial migration is generally activated in cancer progression, which hampers the anti-tumour responses of the host (40). In the present study, the DEGs enriched in focal adhesion and leukocyte transendothelial migration were significantly upregulated in GC tissues and downregulated in celecoxib-treated AGS cells, suggesting that these pathways were activated in GC progression, and celecoxib may exhibit an anti-GC effect by suppressing these pathways.
Overall, the present study proposes that celecoxib may exhibit an anti-GC effect by suppressing the expression of CSRP1, THBS1, MYL9, FLNA, ACTN1, VCL, LAMC2 and CLDN1, and inhibiting leukocyte transendothelial migration and focal adhesion. Although limitations exist in the present study, including the small sample size and lack of validation, the predictions proposed, based on bioinformatics analysis, provide novel directions for the understanding of the anti-GC mechanism of celecoxib. Future studies by the present authors may focus on enlarging the sample size and validating the conclusions in vitro and in vivo.
References
- 1.Fock KM. Review article: The epidemiology and prevention of gastric cancer. Aliment Pharmacol Ther. 2014;40:250–260. doi: 10.1111/apt.12814. [DOI] [PubMed] [Google Scholar]
- 2.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010:CD004064. doi: 10.1002/14651858.CD004064.pub3. [DOI] [PubMed] [Google Scholar]
- 4.Scartozzi M, Galizia E, Verdecchia L, Berardi R, Antognoli S, Chiorrini S, Cascinu S. Chemotherapy for advanced gastric cancer: Across the years for a standard of care. Expert Opin Pharmacother. 2007;8:797–808. doi: 10.1517/14656566.8.6.797. [DOI] [PubMed] [Google Scholar]
- 5.Heravi R Entezari, Hadizadeh F, Sankian M, Afshari J Tavakol, Taghdisi SM, Jafarian H, Behravan J. Novel selective Cox-2 inhibitors induce apoptosis in Caco-2 colorectal carcinoma cell line. Eur J Pharm Sci. 2011;44:479–486. doi: 10.1016/j.ejps.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Fischer SM, Hawk ET, Lubet RA. Coxibs and other nonsteroidal anti-inflammatory drugs in animal models of cancer chemoprevention. Cancer Prev Res (Phila) 2011;4:1728–1735. doi: 10.1158/1940-6207.CAPR-11-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim N, Kim CH, Ahn DW, Lee KS, Cho SJ, Park JH, Lee MK, Kim JS, Jung HC, Song IS. Anti-gastric cancer effects of celecoxib, a selective COX-2 inhibitor, through inhibition of Akt signaling. J Gastroenterol Hepatol. 2009;24:480–487. doi: 10.1111/j.1440-1746.2008.05599.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang YJ, Niu XP, Yang L, Han Z, Ma YJ. Effects of celecoxib on cycle kinetics of gastric cancer cells and protein expression of cytochrome C and caspase-9. Asian Pac J Cancer Prev. 2013;14:2343–2347. doi: 10.7314/APJCP.2013.14.4.2343. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Ran J, Tang C, Wu J, Honghua L, Xingwen L, Ning C, Qiao L. Effect of celecoxib on E-cadherin, VEGF, Microvessel density and apoptosis in gastric cancer. Cancer Biol Ther. 2007;6:269–275. doi: 10.4161/cbt.6.2.3629. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Liu M, Liu X, Huang S, Li L, Song B, Li H, Ren Q, Hu Z, Zhou Y, Qiao L. COX-2 regulates E-cadherin expression through the NF-κB/Snail signaling pathway in gastric cancer. Int J Mol Med. 2013;32:93–100. doi: 10.3892/ijmm.2013.1376. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Ni Z, Duan Z, Wang G, Li F. Altered expression of hypoxia-inducible factor-1α (HIF-1α) and its regulatory genes in gastric cancer tissues. PLoS One. 2014;9:e99835. doi: 10.1371/journal.pone.0099835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Development Core Team: R: A language and environment for statistical computing. The R Foundation for Statistical Computing. Vienna. 2012 [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [DOI] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistical Soc, Series B (Methodological) 1995:289–300. [Google Scholar]
- 17.Szekely GJ, Rizzo ML. Hierarchical clustering via joint between-within distances: Extending Ward's minimum variance method. J Classification. 2005;22:151–183. doi: 10.1007/s00357-005-0012-9. [DOI] [Google Scholar]
- 18.Huangda W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the database of interacting proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Betel D, Hogue CW. BIND: The biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 25.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 26.Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 28.Nakao T, Kurita N, Komatsu M, Yoshikawa K, Iwata T, Utsunomiya T, Shimada M. Expression of thrombospondin-1 and Ski are prognostic factors in advanced gastric cancer. Int J Clin Oncol. 2011;16:145–152. doi: 10.1007/s10147-010-0147-5. [DOI] [PubMed] [Google Scholar]
- 29.Bai Z, Ye Y, Liang B, Xu F, Zhang H, Zhang Y, Peng J, Shen D, Cui Z, Zhang Z, Wang S. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int J Oncol. 2011;38:375–383. doi: 10.3892/ijo.2010.873. [DOI] [PubMed] [Google Scholar]
- 30.Sun GG, Sheng SH, Jing SW, Hu WN. An antiproliferative gene FLNA regulates migration and invasion of gastric carcinoma cell in vitro and its clinical significance. Tumour Biol. 2014;35:2641–2648. doi: 10.1007/s13277-013-1347-1. [DOI] [PubMed] [Google Scholar]
- 31.Kwon OH, Park JL, Kim M, Kim JH, Lee HC, Kim HJ, Noh SM, Song KS, Yoo HS, Paik SG, et al. Aberrant up-regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem Biophys Res Commun. 2011;406:539–545. doi: 10.1016/j.bbrc.2011.02.082. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, Li J, Qu Y, Zhang J, Zhang L, Chen X, Liu B, Zhu Z. The expression of Claudin 1 correlates with β-catenin and is a prognostic factor of poor outcome in gastric cancer. Int J Oncol. 2014;44:1293–1301. doi: 10.3892/ijo.2014.2298. [DOI] [PubMed] [Google Scholar]
- 33.Rubashkin MG, Cassereau L, Bainer R, DuFort CC, Yui Y, Ou G, Paszek MJ, Davidson MW, Chen YY, Weaver VM. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 2014;74:4597–4611. doi: 10.1158/0008-5472.CAN-13-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Kuramitsu Y, Ueno T, Suzuki N, Yoshino S, Iizuka N, Zhang X, Akada J, Oka M, Nakamura K. Proteomic differential display identifies upregulated vinculin as a possible biomarker of pancreatic cancer. Oncol Rep. 2012;28:1845–1850. doi: 10.3892/or.2012.2004. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz C, Holz DR, Oeggerli M, Schneider S, Gonzales IM, Kiefer JM, Zellweger T, Bachmann A, Koivisto PA, Helin HJ, et al. Amplification and overexpression of vinculin are associated with increased tumour cell proliferation and progression in advanced prostate cancer. J Pathol. 2011;223:543–552. doi: 10.1002/path.2828. [DOI] [PubMed] [Google Scholar]
- 36.Moese S, Selbach M, Brinkmann V, Karlas A, Haimovich B, Backert S, Meyer TF. The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell Microbiol. 2007;9:1148–1161. doi: 10.1111/j.1462-5822.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- 37.Goldmann WH, Auernheimer V, Thievessen I, Fabry B. Vinculin, cell mechanics and tumour cell invasion. Cell Biol Int. 2013;37:397–405. doi: 10.1002/cbin.10064. [DOI] [PubMed] [Google Scholar]
- 38.Weiskirchen R, Günther K. The CRP/MLP/TLP family of LIM domain proteins: Acting by connecting. Bioessays. 2003;25:152–162. doi: 10.1002/bies.10226. [DOI] [PubMed] [Google Scholar]
- 39.Petit V, Thiery JP. Focal adhesions: Structure and dynamics. Biol Cell. 2000;92:477–494. doi: 10.1016/S0248-4900(00)01101-1. [DOI] [PubMed] [Google Scholar]
- 40.Enarsson K, Lundin BS, Johnsson E, Brezicka T, Quiding-Järbrink M. CD4+ CD25high regulatory T cells reduce T cell transendothelial migration in cancer patients. Eur J Immunol. 2007;37:282–291. doi: 10.1002/eji.200636183. [DOI] [PubMed] [Google Scholar]