Abstract
Cluster of differentiation (CD)44 has been implicated in cancer metastasis to bone. Clinical and experimental studies have suggested that the standard isoform of CD44 (CD44s) and the variant isoform of CD44 (CD44v) enhance metastasis. The present study examined the differential roles of CD44s and CD44v, particularly CD44v8-10, in the development of bone metastases. For this purpose, MDA-MB-231 human breast cancer cells and A549 human lung cancer cells were stably transduced with epithelial splicing regulatory protein 1 (ESRP1), which regulates the alternative splicing of several genes, including CD44. The introduction of ESRP1 induced a splicing switch from CD44s to CD44v, particularly to CD44v8-10, while the total amount of CD44 was rarely affected. However, ESRP1 did not significantly affect cell proliferation, migration, invasion or tumor sphere formation in vitro. Furthermore, ESRP1 did not cause significant differences in the development of bone metastases in a mouse model. As an alternative approach, cancer cells transduced with the CD44v8-10 gene were also established. The overexpression of CD44v8-10 in MCF-7 human breast cancer cells, which rarely express any isoform of CD44, promoted cell migration and sphere formation, whereas the overexpression of CD44v8-10 in MDA-MB-231 cells, which endogenously express high levels of CD44s, did not exert these effects. The results of the present study collectively suggest that the ability of CD44v8-10 to promote tumor aggressiveness and bone metastases is similar to that of CD44s. CD44v8-10 and CD44s may represent potential therapeutic targets for the treatment of bone metastases.
Keywords: CD44v8-10, CD44s, ESRP1, bone metastases, cancer stem cells
Introduction
Cluster of differentiation (CD)44 is a transmembrane glycoprotein that functions as an adhesion molecule for extracellular matrices, primarily hyaluronan (1,2). The expression of CD44 in cancer cells has also been implicated in cell migration, invasion and metastasis (1,2). Furthermore, CD44 has recently been recognized as a major marker for cancer stem cells (CSCs) in several types of cancer (2,3).
The CD44 gene consists of 20 exons, 10 of which (exons 1–5 and 16–20) encode the shortest and ubiquitously expressed standard isoform of CD44 (CD44s) (1,2). The other 10 exons (exons 6–15, also referred to as exons v1-v10) are variant exons inserted by alternative splicing in various combinations, thereby generating various variant isoforms of CD44 (CD44v) (1,2). The insertion of variant exons lengthens the extracellular domain and alters the binding and signaling properties of CD44. The expression of CD44v is restricted to a few epithelial tissues, including several types of cancer (2). Although extensive research has been conducted, the association between the expression of CD44v and tumor aggressiveness remains controversial (1,2,4–6).
CD44v8-10, also known as CD44R1 and CD44E, is one of the isoforms of CD44v, and contains the variant exons 13–15 (v8-v10) (1,2). Clinical studies have demonstrated that CD44v8-10 is expressed in various human epithelial cancers, including breast, lung, colon, bladder and gastric cancer, and its expression is correlated with metastasis (7–10). Furthermore, recent studies suggest that the expression of CD44v8-10 is associated with CSC-like phenotypes (11,12).
The bone is one of the most frequent organs to be affected by metastatic cancer (13). Bone metastases cause devastating bone pain and skeletal-related events, including pathological fractures, spinal compression and hypercalcemia, thereby severely deteriorating the quality of life of patients (13). However, the mechanisms underlying bone metastases have not yet been fully elucidated.
Clinical studies have suggested that a positive correlation exists between the expression of CD44 and bone metastasis (14,15). Furthermore, our recent preclinical study demonstrated that CD44, particularly CD44s, confers CSC-like properties to cancer cells and enhances their metastatic potential to bone (16). However, the roles of CD44v in the development of bone metastases have yet to be explored. Thus, the present study examined the differential roles of CD44s and CD44v, particularly CD44v8-10, in bone metastases using a well-characterized animal model.
Materials and methods
Cell cultures
The human breast cancer cell lines MDA-MB-231 and MCF-7 were obtained from the American Type Culture Collection (Manassas, VA, USA). The human lung cancer cell line A549 was obtained from the Health Science Research Resources Bank (Osaka, Japan). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 100 µg/ml kanamycin sulfate (Meiji Seika Kaisha, Ltd., Tokyo, Japan), and were maintained at 37°C in a humidified atmosphere of 5% CO2 in air.
Overexpression of ESRP1 and CD44v8-10
The complementary (c)DNA of human epithelial splicing regulatory protein 1 (ESRP1) in the pOTB7 vector was purchased from Dharmacon (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). Human CD44v8-10 cDNA was reverse transcribed and amplified from total RNA isolated from MDA-MB-231 cells overexpressing the ESRP1 gene. Total RNA was isolated using a High Pure RNA Isolation kit (Roche Diagnostics, Basel, Switzerland). cDNA was synthesized from 5 µg of total RNA using PrimeScript Reverse Transcriptase (Takara Bio, Inc., Shiga, Japan). The primer sequences for CD44v8-10 were 5′-CGGACACCATGGACAAGTTT-3′ (forward) and 5′-TCCAACGGTTGTTTCTTTCC-3′ (reverse). PCR was performed using PrimeSTAR GXL DNA polymerase (Takara Bio, Inc.) with a TaKaRa PCR Thermal Cycler Dice (Takara Bio, Inc.) for 30 cycles (98°C for 10 sec, 60°C for 15 sec and 68°C for 90 sec). Both cDNAs were subcloned into the pLVSIN-CMV Pur vector (Takara Bio, Inc.) and transduced into cells using a Lenti-X Lentiviral Expression System (Takara Bio, Inc.). As a control, empty vector (EV) was transduced into the cells. Colonies resistant to puromycin (0.25–1.00 µg/ml; Sigma-Aldrich; Merck Millipore) were isolated and cloned.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total cellular RNA was isolated using a High Pure RNA Isolation kit. cDNA was synthesized from 5 µg of total RNA using PrimeScript Reverse Transcriptase. Two sets of PCR primers were used to detect CD44 (8). One set (primer set 1, forward 5′-TCCCAGACGAAGACAGTCCCTGGAT-3′ and reverse 5′-CACTGGGGTGGAATGTGTCTTGGTC-3′) detected all the isoforms of CD44, while the other set (primer set 2, forward 5′-GACAGAATCCCTGCTACCAATA-3′ and reverse 5′-ATGTGTCTTGGTCTCCTGATAA-3′) specifically detected CD44v8-10 and CD44v10. The primer sequences for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were 5′-CATGGAGAAGGCTGGGGCTC-3′ (forward) and 5′-CACTGACACGTTGGCAGTGG-3′ (reverse). PCR was conducted using PCR Master Mix (Promega Corporation, Madison, WI, USA) with a TaKaRa PCR Thermal Cycler Dice for 25 cycles (at 95, 55 and 72°C for 30 sec), or for 20 cycles in the case of GAPDH. The PCR products were separated on 2% agarose gels containing ethidium bromide and visualized under UV light. The sizes of the fragments were confirmed by comparison with a 100-bp DNA ladder (Takara Bio, Inc.).
Flow cytometry
Cells were harvested by trypsinization and incubated with a phycoerythrin (PE)-conjugated anti-panCD44 antibody (1:100 dilution; 12-0441-81; eBioscience, Inc., San Diego, CA, USA) for 40 min at 4°C. Flow cytometric analysis was performed using a flow cytometer (Cytomics FC 500; Beckman Coulter, Inc., Brea, CA, USA). Data were analyzed using FlowJo software (version 7.6.5; FlowJo, LLC, Ashland, OR, USA).
Western blotting
Total protein lysates were prepared in lysis buffer [20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.4), 150 mM NaCl, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride and 0.2 mM sodium orthovanadate]. Equal amounts of proteins were separated on 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were then immunoblotted with the following antibodies: Anti-panCD44 [1:500 dilution (17)], anti-CD44v9 (1:500 dilution; LKG-M001; Cosmo Bio, Tokyo, Japan), anti-ESRP1 (1:500 dilution; HPA023720; Atlas Antibodies AB, Stockholm, Sweden) and anti-β-actin (1:1,000 dilution; A00702; GenScript, Tokyo, Japan), and visualized with a horseradish peroxidase-conjugated anti-mouse immunoglobulin (Ig)G (1:10,000 dilution; 715-035-150; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or anti-rabbit IgG antibody (1:10,000 dilution; 711-035-152; Jackson ImmunoResearch Laboratories, Inc.), followed by enhanced chemiluminescence (ECL). ECL solution was made by dissolving 0.2 mM p-coumaric acid (Sigma-Aldrich; Merck Millipore), 2.5 mM luminol (Sigma-Aldrich; Merck Millipore) and 0.02% hydrogen peroxide (Wako Pure Chemical Industries, Ltd., Osaka, Japan) in 100 mM Tris-HCl (pH 8.5). The expression of β-actin was used as a loading control.
Cell proliferation in monolayer cultures
Cells (1,000 cells/well) were plated in a 96-well plate in growth medium and cultured for 72 h. Subsequently, cell proliferation was determined using Cell Proliferation Reagent WST-1 (Roche Diagnostics). Absorbance was measured using iMark Microplate Absorbance Reader (Bio-Rad Laboratories, Inc.) at a wavelength of 450 nm.
Wound healing assay
Cells (1×105 cells/well) were plated in a 24-well plate in growth medium and incubated for 24 h. Upon confirming the formation of a complete monolayer, cells were wounded by scratching lines with a standard 200-µl plastic pipette tip. Cell migration into the wound area was observed with a phase-contrast microscope after 24 or 48 h. The percentage of the filled wound area was calculated as follows: Filled wound area (%) = (original wound area-remaining wound area)/original wound area × 100.
Cell invasion assay
Cell invasion assays were conducted using 24-well Corning BioCoat Matrigel Invasion Chambers (BD Biosciences, Franklin Lakes, NJ, USA). In brief, cells were seeded into the upper inserts (2.5×104 cells/insert) in DMEM supplemented with 1% FBS. The outer wells were filled with DMEM containing 10% FBS as a chemoattractant. After 24-h incubation, the membranes were stained with hematoxylin and mounted on slides. Invading cells were counted in five randomly selected microscopic fields. Data are expressed as the number of invaded cells/field.
Tumor sphere formation
Cells were plated in ultra-low attachment 24-well plates (Corning, Inc., Corning, NY, USA) at a density of 1,000 cells/well and cultured in DMEM/F12 (Sigma-Aldrich; Merck Millipore) supplemented with 2% MACS Supplement B27 PLUS (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), 40 ng/ml recombinant human fibroblast growth factor-2 (ProSpec, East Brunswick, NJ, USA) and 20 ng/ml recombinant human epidermal growth factor (Sigma-Aldrich; Merck Millipore). The cells were replenished with fresh medium every 3 days. After culturing for 6–12 days, the number of tumor spheres with a diameter of >100 µm was counted by light microscopy. Data are expressed as the number of tumor spheres/well.
Animal experiments
Under anesthesia with pentobarbital (0.05 mg/g body weight; Kyoritsu Seiyaku Corporation, Tokyo, Japan), MDA-MB-231 cell clones [1×105 cells/0.1 ml phosphate-buffered saline (PBS)] were injected into the left cardiac ventricle of athymic nude mice (KSN/Slc, female, 4-week-old, n=5/group; Japan SLC, Inc., Shizuoka, Japan). All mice were sacrificed 4 weeks after cell inoculation. All animal experiments were approved by the Animal Management Committee of Matsumoto Dental University (Nagano, Japan). Mice were housed at 28°C in a 12-h light:dark cycle and were fed and watered ad libitum.
Histomorphometric and immunohistochemical analyses
Histomorphometry
Paraffin sections were prepared by conventional methods and were stained with hematoxylin and eosin. Histomorphometric analysis of the tumor burden in bone was conducted as described previously (16). Data are expressed as the tumor area (mm2).
Immunohistochemistry
Paraffin sections were deparaffinized in xylene and rehydrated in a graded series of ethanol. Endogenous peroxidase activity was blocked by immersing the sections in 0.3% H2O2 in methanol for 15 min. After blocking with 3% bovine serum albumin in PBS for 30 min, sections were incubated with primary antibodies overnight at 4°C. Primary antibodies against panCD44 (17) and CD44v9 (LKG-M001; Cosmo Bio) were applied at a dilution of 1:500. Indirect immunohistochemical staining was performed using a Histofine Simple Stain MAX PO kit (Nichirei Biosciences, Inc., Tokyo, Japan). Chromogen was developed using Liquid DAB+ (Dako, Glostrup, Denmark). The slides were counterstained with hematoxylin.
Statistical analysis
Data are expressed as the mean ± standard error of the mean. Data were analyzed by analysis of variance followed by a Tukey's test for determining differences between the groups, using Mini StatMate software (version 1.1; ATMS, Tokyo, Japan). P<0.05 was considered to indicate a statistically significant difference.
Results
Effects of ESRP1 overexpression on bone metastases
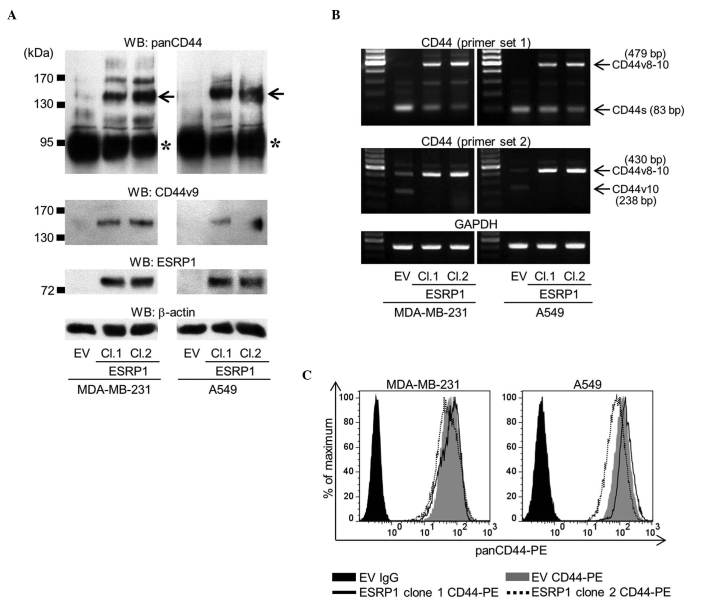
ESRP1 is an epithelial-specific splicing factor that regulates the alternative splicing of several genes, including CD44 (18–20). The overexpression of ESRP1 has been reported to cause a splicing switch from CD44s to CD44v (18–20). In order to examine the differential roles of CD44s and CD44v8-10 in bone metastases, the current study employed an approach in which the ESRP1 gene was stably transduced into MDA-MB-231 human breast cancer cells and A549 human lung cancer cells (MDA/ESRP1 and A549/ESRP1, respectively). While mock-transduced cells (MDA/EV and A549/EV) predominantly expressed CD44s, the overexpression of ESRP1 reduced the expression of CD44s, but increased that of CD44v (Fig. 1A). RT-PCR analysis confirmed that the dominant isoform of CD44v expressed in MDA/ESRP1 and A549/ESRP1 cells was CD44v8-10 (Fig. 1B). Flow cytometric analysis using an antibody that recognizes panCD44 revealed that the total amount of CD44 expressed was rarely affected by ESRP1 (Fig. 1C).
Figure 1.
Establishment of MDA-MB-231 and A549 cell clones overexpressing ESRP1. (A) The expression of panCD44, CD44v9 and ESRP1 was determined by western blot analysis (asterisk, CD44s; arrow, CD44v8-10). Two ESRP1-overexpressing clones (clones 1 and 2) of each cell line were examined in subsequent experiments, and compared with empty vector mock-transduced cells. (B) Messenger RNA expression of CD44 was determined by reverse transcription-polymerase chain reaction using two sets of primers. Primer set 1 detected all isoforms of CD44, while primer set 2 specifically detected CD44v8-10 and CD44v10. (C) Flow cytometric analysis of the total amount of CD44 protein was determined using an anti-panCD44 antibody. WB, western blot; CD, cluster of differentiation; CD44v, variant isoform of CD44; ESRP1, epithelial splicing regulatory protein 1; EV, empty vector; Cl, clone; CD44s, standard isoform of CD44; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PE, phycoerythrin; Ig, immunoglobulin.
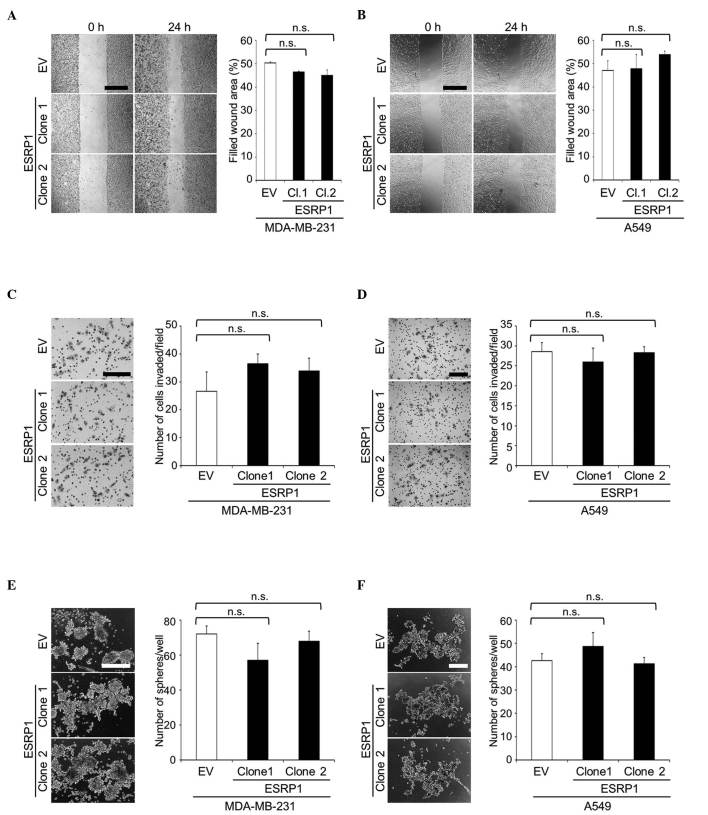
Using these clones, cellular functions were evaluated by in vitro assays. WST-1 assay revealed no significant differences in cell proliferation between mock- and ESRP1-transduced cells (data not shown). In addition, ESRP1 did not significantly affect cell migration or invasion, as determined in wound healing and matrigel invasion assays, respectively (Fig. 2A-D). Since CD44 is a widely recognized marker for CSCs (2,3), CSC-like properties were assessed by sphere formation assay. The ability of ESRP1-overexpressing cells to form tumor spheres in suspension cultures was similar to that of the control cells (Fig. 2E and F).
Figure 2.
Cell migration, cell invasion and tumor sphere formation of MDA-MB-231 and A549 cell clones overexpressing ESRP1. (A and B) Cell migration of (A) MDA-MB-231 and (B) A549 clones was determined by wound healing assay. Representative microscopic images at 0 and 24 h after wounding are shown on the left side of the panel (scale bar, 500 µm). Data are expressed as the percentage of filled wound area. (C and D) Cell invasion of (C) MDA-MB-231 and (D) A549 clones was determined by matrigel invasion assay. Representative microscopic images of membranes are shown on the left side of the panel (scale bar, 200 µm). Data are expressed as the number of cells invaded/field. (E and F) Tumor sphere formation of (E) MDA-MB-231 and (F) A549 clones in suspension cultures. Representative microscopic images are shown on the left side of the panel (scale bar, 200 µm). Data are expressed as the number of tumor spheres/well. n.s., not significant; CD, cluster of differentiation; ESRP1, epithelial splicing regulatory protein 1; EV, empty vector; Cl, clone.
The bone-metastatic potential of MDA/ESRP1 cells in vivo was examined using the intracardiac injection model. In accordance with the data obtained in vitro, ESRP1 did not cause significant differences in the development of bone metastases (Fig. 3A and B), although the expression of CD44v8-10 in MDA/ESRP1 cells was maintained at a high level in bone (Fig. 3C).
Figure 3.
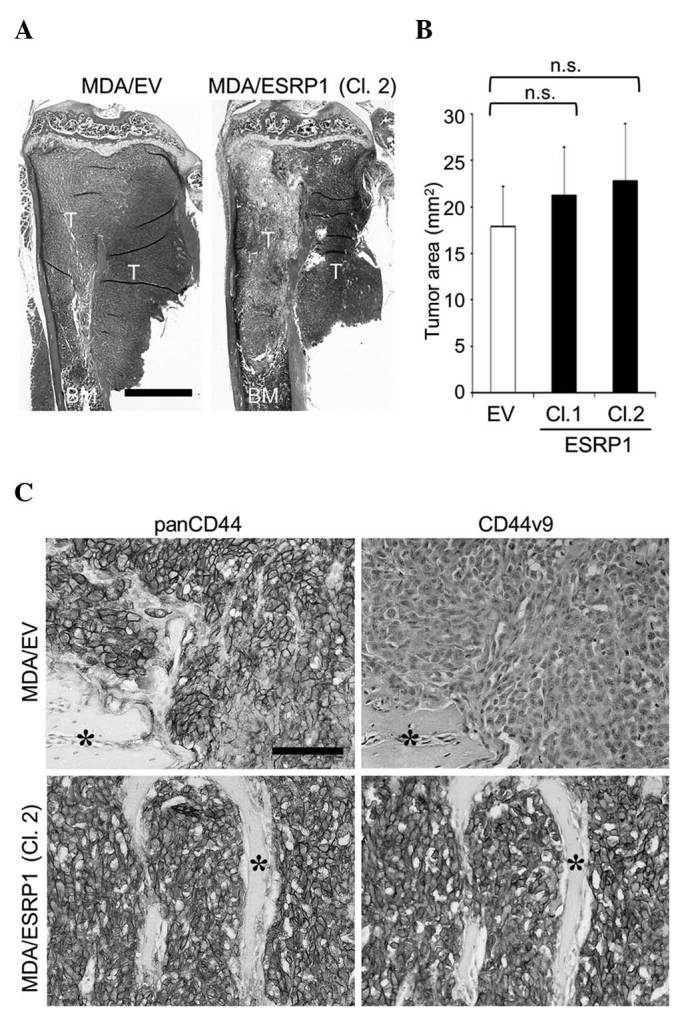
Effects of ESRP1 overexpression on the development of bone metastases of MDA-MB-231 cells. (A) Representative histological pictures of bone metastases (hematoxylin and eosin staining; scale bar, 1 mm). (B) Histomorphometric analysis of the tumor burden in bone. Data are expressed as tumor area (mm2; n=5/group). (C) Immunohistochemical detection of the expression of panCD44 and CD44v9 in cancer cells colonized in bone (asterisk, bone; scale bar, 100 µm). CD, cluster of differentiation; CD44v, variant isoform of CD44; ESRP1, epithelial splicing regulatory protein 1; EV, empty vector; Cl, clone; T, tumor; BM, bone marrow; n.s., not significant.
Effects of CD44v8-10 overexpression on bone metastases
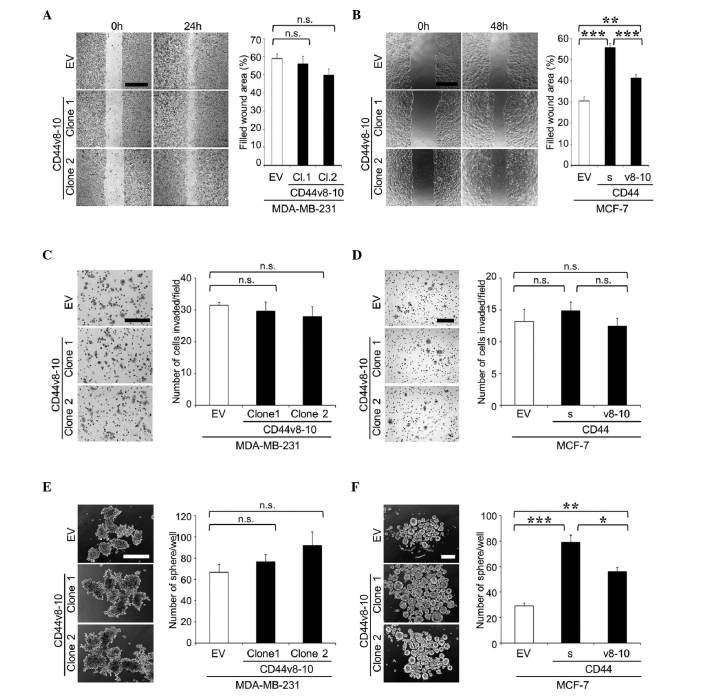
To further examine the roles of CD44v8-10, an alternative approach was undertaken in which the CD44v8-10 gene was stably transduced into MDA-MB-231 cells to generate MDA/CD44v cells (Fig. 4). MDA/CD44v cells exhibited no significant changes in cell proliferation, migration, invasion or tumor sphere formation in vitro (Fig. 5A, C and E), or in the development of bone metastases in mice (Fig. 6).
Figure 4.
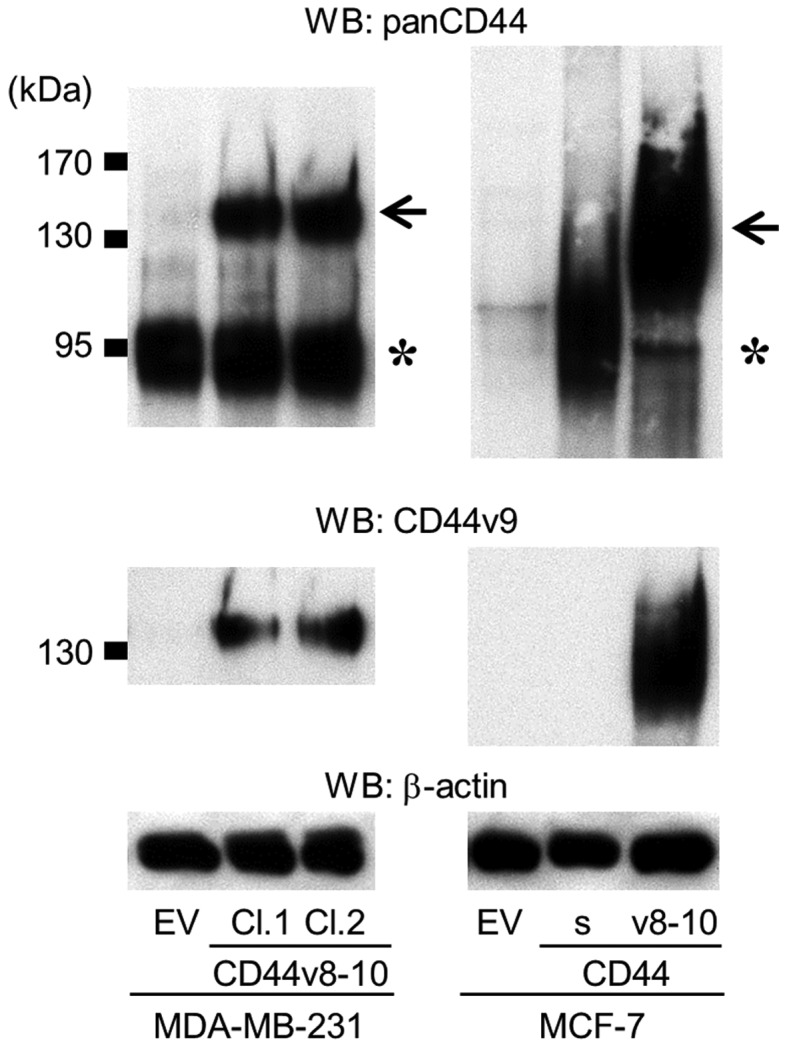
Establishment of MDA-MB-231 and MCF-7 cell clones overexpressing CD44v8-10. The expression of panCD44 and CD44v9 was determined by western blot analysis (asterisk, CD44s; arrow, CD44v8-10). MCF-7 cells overexpressing CD44s were also established. WB, western blot; CD, cluster of differentiation; CD44v, variant isoform of CD44; EV, empty vector; Cl, clone; CD44s, standard isoform of CD44.
Figure 5.
Cell migration, cell invasion and tumor sphere formation of MDA-MB-231 and MCF-7 cell clones overexpressing CD44v8-10. (A and B) Cell migration of (A) MDA-MB-231 and (B) MCF-7 clones was determined by wound healing assay. Representative microscopic images at 0 and 24 h (MDA-MB-231) or 48 h (MCF-7) after wounding are shown on the left side of the panel (scale bar, 500 µm). Data are expressed as the percentage of filled wound area. **P<0.01, ***P<0.001 vs. control. (C and D) Cell invasion of (C) MDA-MB-231 and (D) MCF-7 clones was determined by matrigel invasion assay. Representative microscopic images of membranes are shown on the left side of the panel (scale bar, 200 µm). Data are expressed as the number of cells invaded/field. (E and F) Tumor sphere formation of (E) MDA-MB-231 and (F) MCF-7 clones in suspension cultures. Representative microscopic images are shown on the left side of the panel (scale bar, 200 µm). Data are expressed as the number of tumor spheres/well. *P<0.05, **P<0.01, ***P<0.001 vs. control. n.s., not significant; CD, cluster of differentiation; CD44v, variant isoform of CD44; EV, empty vector; Cl, clone; CD44s, standard isoform of CD44.
Figure 6.
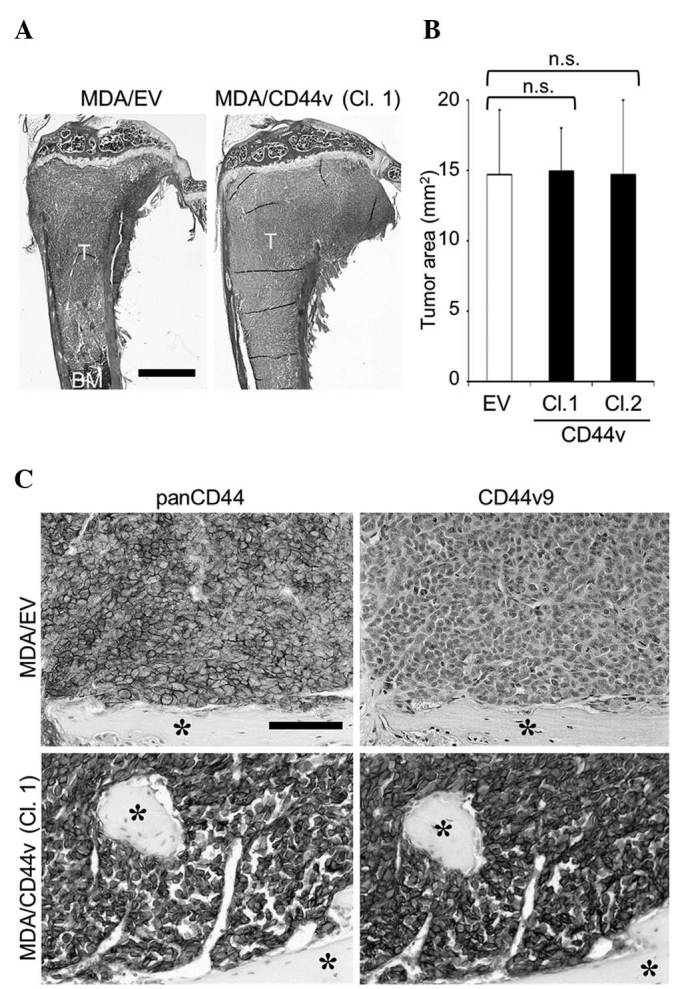
Effects of CD44v8-10 overexpression on the development of bone metastases of MDA-MB-231 cells. (A) Representative histological pictures of bone metastases (hematoxylin and eosin staining; scale bar, 1 mm). (B) Histomorphometric analysis of the tumor burden in bone. Data are expressed as tumor area (mm2; n=5/group). (C) Immunohistochemical detection of panCD44 and CD44v9 expression in cancer cells colonized in bone (asterisk, bone; scale bar, 100 µm). CD, cluster of differentiation; CD44v, variant isoform of CD44; EV, empty vector; Cl, clone; T, tumor; BM, bone marrow; n.s., not significant.
The effects of the overexpression of CD44v8-10 were similarly tested in MCF-7 human breast cancer cells (MCF-7/CD44v; Fig. 4). In contrast to MDA-MB-231 and A549 cells, MCF-7 cells expressed very low levels of any isoform of CD44 (Fig. 4). As reported previously (16), the overexpression of CD44s promoted cell migration and sphere formation in MCF-7 cells in vitro (Fig. 5B and F). MCF-7/CD44v also exhibited enhanced cell migration and sphere formation compared with the control cells (Fig. 5B and F), while neither CD44s nor CD44v8-10 stimulated cell invasion (Fig. 5D). MCF-7 cells have been demonstrated to possess low metastatic potential and rarely metastasize to bone (16). Our previous study revealed that overexpression of CD44s did not enhance the development of bone metastases (16). Similarly, in the present study, overexpression of CD44v8-10 failed to potentiate metastatic potential and developed few lesions in bone (data not shown).
Discussion
Accumulating evidence indicates that CD44 is involved in various cancer phenotypes, including enhanced cell proliferation, migration, invasion and metastasis (1,2). Consistent with these findings, the present authors recently reported that the expression of CD44 in cancer cells increases tumorigenicity, cell migration and invasion, thereby promoting cancer metastasis to bone (16). However, it currently remains unknown whether CD44s and CD44v make differential contributions to the development of bone metastases.
Similar to the overexpression of CD44s, that of CD44v8-10 in MCF-7 cells (which rarely express any isoforms of CD44) significantly increased cell migration and tumor sphere formation. These results suggest that CD44v8-10 has the potential to promote tumor progression in a similar manner to CD44s. The overexpression of ESRP1, which caused a switch in alternative splicing from CD44s to CD44v (mainly CD44v8-10) did not change the phenotypes or metastatic potential of MDA-MB-231 or A549 cells. Considering that ESRP1 did not change the total amount of CD44, the contribution of CD44v8-10 to the development of bone metastases is likely comparable to that of CD44s. However, the overexpression of CD44v8-10 did not further enhance the aggressiveness of MDA-MB-231 cells. Since MDA-MB-231 cells endogenously express high level of CD44s, the efficacy of endogenous CD44s to enhance bone metastases may have already reached its maximum level.
As discussed above, although cell migration and tumor sphere formation were stimulated in MCF-7/CD44v cells, these effects were less prominent than those observed in MCF-7/CD44s cells (Fig. 5B and F). One explanation for these results is that the expression levels of CD44s and CD44v8-10 may not be completely the same. Another possibility is that there are functional differences between CD44s and CD44v8-10. CD44s has been demonstrated to interact with its major ligand, hyaluronan, more effectively than CD44v8-10 (21). In addition, the CD44v segment is known to include binding sites for several growth factors (2). These differences may become visible under certain conditions.
Our previous study revealed that the overexpression of CD44s in MCF-7 cells failed to increase bone metastases, although cell migration and tumorigenicity were significantly enhanced (16). Similar results were obtained with MCF-7/CD44v cells, suggesting that solely CD44v8-10 is not sufficient to cause bone metastases.
Recent studies have proposed a critical contribution of cancer stem-like cells to the development of metastases (22). CD44 is one of the most well-recognized markers for CSCs (2,3). Consistent with this notion, our previous study demonstrated that CD44s confers stem cell-like phenotypes to cancer cells, thus leading to the increased formation of bone metastases (16). The present study, using MCF-7 cells, revealed that CD44v8-10 also enhanced sphere formation, suggesting that CD44v8-10, in addition to CD44s, has the ability to confer stem cell-like properties to cancer cells and may be a marker for CSCs.
It has been suggested that CD44v is predominantly expressed in epithelial cells, whereas CD44s is mainly expressed in mesenchymal cells (19). Furthermore, the induction of epithelial-mesenchymal transition (EMT) is accompanied by a shift from CD44v to CD44s (19). In contrast to these findings, Yae et al reported that the expression of the CD44 isoforms is not associated with that of EMT markers (20). In the present study, RT-PCR analysis also revealed that the overexpression of ESRP1 and CD44v8-10 did not cause consistent changes in the expression of the epithelial marker E-cadherin or the mesenchymal marker vimentin in cancer cells (data not shown). Thus, further studies are required to clarify the molecular mechanism of the association between the switch in CD44 isoform expression and EMT.
ESRP1 has been demonstrated to regulate the alternative splicing of genes other than CD44, including fibroblast growth factor receptor 2, catenin delta-1 (CTNND1) and enabled homolog (ENAH) (18). The current study confirmed by RT-PCR analysis that the overexpression of ESRP1 also changed the splicing patterns of CTNND1 and ENAH (data not shown). The involvement of the ESRP1-induced splicing switch in genes other than CD44 requires further investigation.
In conclusion, the present results collectively suggest that CD44v8-10 promotes tumor aggressiveness and bone metastases to a similar extent to CD44s, while their functional differences have yet to be elucidated in detail. Thus, CD44v8-10, in addition to CD44s, could be a potential therapeutic target for the treatment of bone metastases.
Acknowledgements
The present study was partly supported by grants from the Japan Society for the Promotion of Science (grant nos., JP21390505, JP23659883 and JP15K11093; Tokyo, Japan; Grants-in-Aid for Scientific Research) and the Naito Foundation (Tokyo, Japan).
Glossary
Abbreviations
- CD
cluster of differentiation
- CD44s
standard isoform of CD44
- CD44v
variant isoform of CD44
- CSCs
cancer stem cells
- CTNND1
catenin delta-1
- DMEM
Dulbecco's modified Eagle's medium
- ECL
enhanced chemiluminescence
- EMT
epithelial-mesenchymal transition
- ENAH
enabled homolog
- ESRP1
epithelial splicing regulatory protein 1
- EV
empty vector
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PBS
phosphate-buffered saline
- PE
phycoerythrin
- RT-PCR
reverse transcription-polymerase chain reaction
References
- 1.Ponta H, Sherman L, Herrlich PA. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 2.Zöller M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Günthert U, Hofmann M, Rudy W, Reber S, Zöller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-L. [DOI] [PubMed] [Google Scholar]
- 5.Friedrichs K, Franke F, Lisboa BW, Kügler G, Gille I, Terpe HJ, Hölzel F, Maass H, Günthert U. CD44 isoforms correlate with cellular differentiation but not with prognosis in human breast cancer. Cancer Res. 1995;55:5424–5433. [PubMed] [Google Scholar]
- 6.Seelentag WK, Günthert U, Saremaslani P, Futo E, Pfaltz M, Heitz PU, Roth J. CD44standard and variant isoform expression in human epidermal skin tumors is not correlated with tumor aggressiveness but down-regulated during proliferation and tumor de-differentiation. Int J Cancer. 1996;69:218–224. doi: 10.1002/(SICI)1097-0215(19960621)69:3<218::AID-IJC12>3.3.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Olsson E, Honeth G, Bendahl PO, Saal LH, Gruvberger-Saal S, Ringnér M, Vallon-Christersson J, Jönsson G, Holm K, Lövgren K, et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011;11:418. doi: 10.1186/1471-2407-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto I, Morisaki T, Sasaki J, Miyake H, Matsumoto M, Suga M, Ando M, Saya H. Molecular detection of cancer cells by competitive reverse transcription-polymerase chain reaction analysis of specific CD44 variant RNAs. J Natl Cancer Inst. 1998;90:307–315. doi: 10.1093/jnci/90.4.307. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi A, Urano T, Goi T, Saito M, Takeuchi K, Hirose K, Nakagawara G, Shiku H, Furukawa K. Expression of a CD44 variant containing exons 8 to 10 is a useful independent factor for the prediction of prognosis in colorectal cancer patients. J Clin Oncol. 1996;14:1122–7112. doi: 10.1200/JCO.1996.14.4.1122. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi A, Saito M, Goi T, Iida A, Takeuchi K, Hirose K, Nakagawara G, Urano T, Furukawa K, Shiku H. Expression of CD44 variant exons 8-10 in gastric cancer. Jpn J Cancer Res. 1995;86:1166–1171. doi: 10.1111/j.1349-7006.1995.tb03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, Shabbir A, So JB, Chan SL. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630–2641. doi: 10.1158/0008-5472.CAN-13-2309. [DOI] [PubMed] [Google Scholar]
- 12.Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y, Vander Griend DJ, Luo J, Kong C, Getzenberg RH, Kulkarni P. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73:4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 13.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: A fatal attraction. Nat Rev Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 15.Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, Datar RH, Cote RJ. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 16.Hiraga T, Ito S, Nakamura H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013;73:4112–4122. doi: 10.1158/0008-5472.CAN-12-3801. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura H, Kato R, Hirata A, Inoue M, Yamamoto T. Localization of CD44 (hyaluronan receptor) and hyaluronan in rat mandibular condyle. J Histochem Cytochem. 2005;53:113–120. doi: 10.1177/002215540505300113. [DOI] [PubMed] [Google Scholar]
- 18.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 21.Bartolazzi A, Jackson D, Bennett K, Aruffo A, Dickinson R, Shields J, Whittle N, Stamenkovic I. Regulation of growth and dissemination of a human lymphoma by CD44 splice variants. J Cell Sci. 1995;108:1723–1733. doi: 10.1242/jcs.108.4.1723. [DOI] [PubMed] [Google Scholar]
- 22.Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]