Abstract
Living eukaryotic systems evolve delicate cellular mechanisms for responding to various environmental signals. Among them, epigenetic machinery (DNA methylation, histone modifications, microRNAs, etc.) is the hub in transducing external stimuli into transcriptional response. Emerging evidence reveals the concept that epigenetic signatures are essential for the proper maintenance of cellular metabolism. On the other hand, the metabolite, a main environmental input, can also influence the processing of epigenetic memory. Here, we summarize the recent research progress in the epigenetic regulation of cellular metabolism and discuss how the dysfunction of epigenetic machineries influences the development of metabolic disorders such as diabetes and obesity; then, we focus on discussing the notion that manipulating metabolites, the fuel of cell metabolism, can function as a strategy for interfering epigenetic machinery and its related disease progression as well.
Keywords: epigenetics, DNA methylation, histone modifications, microRNA, cellular metabolism, metabolites
Epigenetics
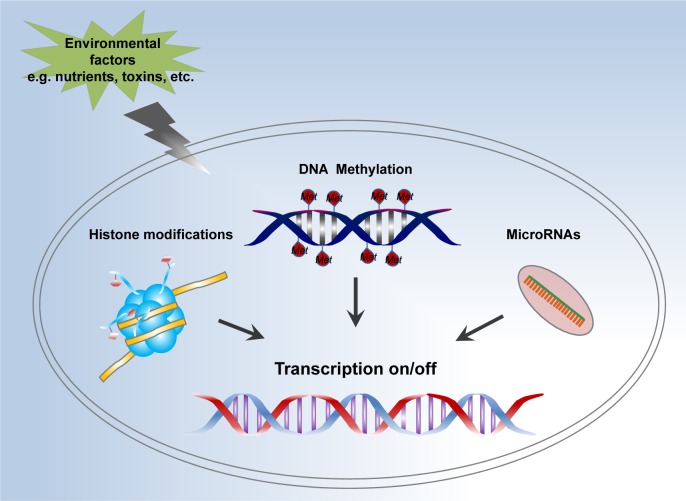
In biology, epigenetics is characterized by regulations of gene expression without alteration of the underlying DNA sequence.1,2 In epigenetic manifestations, external or environmental factors (including nutrition, stress, toxins, and medical history) play an important role in regulating the expression of certain genes.3 Hence, epigenetic investigations aim to understand how external factors regulate gene expression, and even phenotypic traits. Actually, the term epigenetics, with the prefix “epi” taken from the Greek language (meaning outside of, or around), emerged in the 1990s. Until 2008, the formulated definition of epigenetic traits was present as a stably heritable phenotype without changes in DNA sequences at a Cold Spring Harbor meeting.4 During the past 15 years, plenty of cases and mechanisms are discovered and studied in the field of epigenetics. Among them, three major mechanisms are categorized by biologists to instruct epigenetic regulations (Fig. 1): DNA methylation, histone posttranslational modifications (PTMs), and noncoding RNAs.2
Figure 1.
Mechanisms of epigenetic modifications. Cell transduces the environmental changes (eg, nutrients, toxins, etc.) into epigenetic modifications including DNA methylation, histone PTMs, and noncoding RNAs (especially the microRNAs), which eventually turn certain genes on or off and regulate the transcription process without changing DNA sequence.
DNA methylation
DNA methylation is among the best studied and characterized epigenetic modifications. A methyl group is added to the C-5 position of a cytosine adjacent to a guanine residue (CpG dinucleotides), which normally leads to gene suppression.2 In mammals, the CpGs are predominately methylated. However, the CpG islands, which locate in the promoter region of housekeeping and developmental regulator genes with dense CG distribution, are largely resistant to DNA methylation. During the progression of diseases, CpGs of some key genes are reported to be abnormally hyper- or hypomethylated, which further result in transcriptional misregulation.5 It is well demonstrated that in mammalian cells, DNA methylation is performed by DNA methyltransferases DNMT1, DNMT3A, and DNMT3B. Among them, DNMT1 propagates DNA methylation patterns during DNA replication, while 3A and 3B are involved in establishing de novo patterns of DNA methylation during development and cell fate determination.6,7 DNA can also be demethylated through several enzymatic reactions. For instance, the 10 to 11 translocation (TET) proteins can mediate the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), finally leading to DNA demethylation.8
Histone PTMs
Chromatin is a macromolecular structure that consists of DNA, protein, and RNA. DNA wraps around histone octamer to form nucleosomes, the fundamental unit of chromatin. Each histone octamer involved in the organization of nucleosomes contains two copies of four histones (H2A, H2B, H3, and H4). The precise structure of a nucleosome has been illustrated, and the functions of these protein–DNA or protein–protein interactions are suggested to be important for many cellular processes.9,10 Since chromatin is usually packed with nucleosomes, which is believed to be a challenge for transcription accessibility, chromatin should be remodeled to allow the access of transcription machinery to DNA.11 In the past few decades, compelling evidences suggest that histone modifications are able to change chromatin structure and are associated with both transcriptional activation and repression.12 Therefore, the chromatin features are essential for the molecular biologist to analyze the transcriptional regulations and cellular status.13–17
In general, histone modifications mainly include acetylation, methylation, phosphorylation, and ubiquitination. Recently, some novel modifications are identified by tandem mass spectrometry (MS/MS), such as histone lysine GlcNAcylation, butyrylation, malonylation, and crotonylation.18,19 Among them, acetylation is one of the most widely studied epigenetic modifications that commonly occurs on the basic amino acids (lysine and arginine). Basically, acetylation can neutralize the positive charge of basic residues and reduce the affinity between histones and DNA for gene activation.19 This process is catalyzed by histone acetyltransferases (HATs). HATs are traditionally divided into two different classes (type A and B) based on their subcellular localization. Type A HATs locate in the nucleus and are involved in the regulation of gene expression through acetylation of nucleosomal histones. Type B HATs locate in the cytoplasm and are responsible for acetylating newly synthesized histones prior to their assembly into nucleosomes.20–22 The acetylation of histones can also be removed by histone deacetyltransferases (HDACs). HDACs are classified into four classes (I, II, III, and IV) according to their functions and DNA sequence similarity. Classes I, II, and IV are considered as “classical” HDACs whose activities are zinc dependent and inhibited by trichostatin A (TSA). Whereas class III enzymes, known as sirtuins, are NAD+ dependent and not affected by TSA.23–25 Histones can also be modified by other acyl moieties, eg, crotonyl, but the mechanisms and the functional consequences of acetylation versus crotonylation are still unclear. Sabari et al26 indicated that p300, which belongs to the type A HATs, processes both crotonyltransferase and acetyltransferase activities, and the induced transcription by crotonylation is greater than acetylation. These results provide a new clue to clarify the specificities of different histone acylations in regulating metabolic-related gene expressions. Methylation is another widely studied histone modification, which is catalyzed by histone methyltransferases.27 Methylation can influence gene expression in both positive and negative ways. For example, trimethylation of lysine 4 on H3 (H3K4me3) is strongly associated with gene activation, while trimethylation of lysine 27 on H3 (H3K27me3) is reported to be associated with gene silencing.28,29 Histone demethylation is performed by two classes of histone demethylases: lysine-specific demethylase (LSD) family proteins (LSD1 and LSD2) and JmjC domaincontaining histone demethylase (JHDM).27 The histone PTMs are not independent of each other, some crosstalks exist among them. Recent studies of Li et al indicated that a Serine-responsive SAM-containing Metabolic Enzyme complex, which consists of pyruvate kinase, serine metabolic enzymes, and SAM synthetases, can interact with a H3K4 methyltransferase Set1 and mediate the crosstalk between H3T11 phosphorylation (H3Tp11) and H3K4me3.30
MicroRNAs
MicroRNAs (miRNAs), the short noncoding RNAs (approximately 22 nucleotides in length), are able to regulate target gene expression via multiple molecular pathways. Normally, miRNAs can interfere with gene expression through their complementary binding to the target mRNAs, which either inhibit translation or cause mRNA degradation.31,32 Currently, miRNAs are identified in almost all metazoan genomes and even in some viruses.33 Bioinformatic analysis of known miRNAs suggests that they can regulate the large majority of mRNAs and that a single miRNA can also target multiple mRNAs. Recent findings implicate that miRNAs participate in either regulating expression of certain genes or RNA degradation in important cellular processes, such as apoptosis, cell proliferation, differentiation, and metabolism.32,34 Furthermore, enzymes involved in epigenetic modifications can also be governed by miRNAs. It is reported that some miRNAs can directly inhibit the activities of enzymes, which are related to histone acetylation, thereby considered as key regulators during epigenetic cascade. For example, miR-34a can directly inhibit the activities of SIRT1 to regulate cholesterol homeostasis (discussed in “miRNAs and cellular metabolism” section).35,36
Epigenetic Regulation on Metabolism
The prevalence of type 2 diabetes (T2D), obesity (OB), and other metabolic diseases has caused wide public attention during past two decades. Increasing studies also reveal that the epigenetic regulation can modulate metabolic dysfunctions in many aspects, which further broadens our understandings about metabolic disease progressions. Here, we discuss the recent research progress about how epigenetic machineries: DNA methylation, histone PTMs, and miRNA regulate the metabolisms.
DNA methylation and metabolic disorders
DNA contains the genetic information of a cell, and methylation of DNA bases can be highly influential on regulating gene activity, including some important genes that are involved in metabolic pathways.37–39 Over the past decade, evidence has been accumulated to define the regulatory role of DNA methylation in metabolic disorders such as diabetes and OB by taking the advantage of molecular genetic dissection and genome-wide association study (GWAS) analysis.40
The balance of blood glucose is very important for human health, the pancreatic β cells in islets can secrete insulin (INS) in response to the increased blood glucose level to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to avoid hypoglycemia.41 Studies by Kuroda et al42 showed that the CpG sites in mouse Ins2 and human INS promoter are unmethylated compared to other tissues or non-islet fraction. Overall, DNA methylation that occurred at insulin promoter correlates negatively with insulin gene expression in human pancreatic islet samples. Patients with T2D show an increased level of DNA methylation and a reduction of insulin mRNA expression compared to nondiabetic donors.43 In mammals, the de novo DNA methylation process is mainly catalyzed by DNMT3A and DNMT3B.6,7 Interestingly, deletion of Dnmt3a in mouse β cell prevents the execution of metabolic switch during β-cell maturation, resulting in a loss of glucose-stimulated insulin secretion (GSIS). Mechanistically, loss of DNMT3A leads to a reduction of DNA methylation at two key metabolic switch genes, namely, hexokinase 1 (HK1) and lactate dehydrogenase A, while their mRNA expressions are upregulated. However, Ins1 and Ins2 are not affected in Dnmt3a-KO islet cells, which suggest that DNMT3A is not involved in transcriptional regulation of Ins genes.41 Thus, it is still an open question that how INS is regulated by DNA methylation.
The GWAS studies indicate that metabolic diseases (eg, diabetes and OB) are also associated with the alterations of DNA methylation at specific genomic loci such as peroxisome proliferator-activated receptor γ (PPARγ) and glucose transporter 4 (GLUT4).44 PPARγ is a key regulator of adipocyte differentiation and required for inducing the expression of adipocyte-specific genes.45 GLUT4 is a critical regulator in glucose uptake, whose alterations are related to insulin resistance (IR) and OB. It is reported that the promoter of both PPARγ2 and GLUT4 are differentially methylated during the differentiation of 3T3-L1 cells.46,47 With the treatment of DNA methylation inhibitor (5′-aza-cytideine), the mRNA expressions of both PPARγ and GLUT4 are increased.48 These studies provide direct evidence that DNA methylation can regulate metabolic gene expression. Thus, the manipulation of DNA methylation provides an ideal target for understanding the metabolic disease progression and the potential therapy.
PTMs and T2D
Histone modifications can regulate gene expression by modifying the structure of chromatins as described. Recently, an increasing number of evidence is accumulated to elucidate the roles of histone PTMs in metabolic diseases, especially in T2D.49,50
For diabetic patients, metabolic memory means inflammation and vascular complications after controlling the blood glucose level, which presents major challenges to treatment.51 Recent findings suggest that epigenetic mechanisms may be responsible for metabolic memory. Compared with normal cells, trimethylation on H3 lysine 9 (H3K9me3) was significantly reduced in vascular smooth muscle cells (VSMCs) derived from the db/db mice (diabetic mouse, the typical mouse model of T2D). Similarly, the protein level of SUV39H1 (a methyltransferase, which catalyzed H3K9me3 methylation) was also decreased in db/db VSMC. Overexpression of SUV39H1 in db/db VSMC partially rescued this diabetic phenotype. These studies indicate a protective role of H3K9me3 and SUV39H1 in metabolic memory in cases of diabetes.52 Vecellio et al analyzed cardiac mesenchymal cells (CMSCs) from volunteers with normal blood glucose level (ND-CMSC) and type 2 diabetic patients (D-CMSC). Compared with control cells, acetylation on histone H3 lysine 9 (H3K9Ac) and lysine 14 (H3K14Ac) was decreased, as well as phosphorylation at histone H3 serine 10 (H3S10P), whereas the H3K9me3 and H3K27me3 were significantly increased. Remarkably, modification patterns of H3K9Ac and H3K14Ac were restored by treating with HAT activator, pentadecylidenemalonate 1b (SPV-106),53 and the diabetes-associated decreased histone H3 acetylation could also be restored by treating with HDAC inhibitors (valproic acid).54,55 These studies provide a novel way for reversing the misregulated epigenetic memory of diabetic patients in cardiac tissue.
MiRNAs and cellular metabolism
MiRNAs are found to be responsible for regulating various biological processes and viewed as an emerging and exciting topic of biomedical research. In recent years, it has been increasingly reported that miRNAs are associated with disease development.56,57 However, the underlining molecular mechanisms are still ambiguous. Here, in order to get a comprehensive understanding of the associations between miRNAs and metabolic disease progression, we collect and summarize the latest progress in miRNA studies (see Table 1).
Table 1.
Involvement of miRNAs in cellular metabolic pathways.
| miRNAs | TARGETS | FUNCTIONS | REFERENCES |
|---|---|---|---|
| miR-9 | SIRT1 | Insulin secretion | 35 |
| miR-10b | ABCA1/ABCG1 | Cholesterol transport | 91 |
| miR-14 | Reaper | Fat metabolism | 92 |
| miR-24 | SOX6, BHLHE22 | Insulin biosynthesis | 93 |
| miR-26 | ADAM17, SOX6, BHLHE23 | Adipogenesis | 94 |
| miR-27 | ANGPTL3, GPAM | Lipid metabolism | 95 |
| miR-29b | BCKD complex | Amino acid metabolism | 31 |
| miR-33 | ABCA1, ABCG1, CPT1A, SREBF1 | Cholesterol transport and cholesterol homeostasis | 96, 97 |
| miR-34a | SIRT1, NAMPT | Cholesterol and lipid homeostasis | 60 |
| miR-96 | Granuphilin, Noc2 | Insulin secretion | 36 |
| miR-103 | CAV-1 | Insulin signaling and glucose homeostasis | 98 |
| miR-107 | CAV-1 | Insulin signaling and glucose homeostasis | 98 |
| miR-122 | HMGCR, G6PC, AMPKα1 | Triglyceride metabolism; cholesterol biosynthesis | 99 |
| miR-124a | SNAP25, Noc2, Rab3A | Insulin secretion | 36 |
| miR-130 | PPARγ | Adipogenesis | 100 |
| miR-143 | ERK5 | Preadipocyte differentiation | 101 |
| miR-148 | SOX6, BHLHE22 | Insulin biosynthesis | 93 |
| miR-182 | SOX6, BHLHE23 | Insulin biosynthesis | 102 |
| miR-208 | MED13 | Insulin signaling and glucose homeostasis | 103 |
| miR-223 | GLUT4 | Glucose metabolism | 59 |
| miR-335 | PPARγ, AP2, FAS | Lipid metabolism | 104 |
| miR-370 | CPT1A | Insulin secretion | 105 |
| miR-375 | PDK, Myotrophin | Insulin secretion | 106 |
| miR-378/378* | FABP4, FAS, SCD1, Resistin | Preadipocyte differentiation | 107 |
Rottiers and Naar58 showed that miRNAs are essential for glucose homeostasis. For example, miR-223 was upregulated in T2D patients through quantitative analysis of miRNA expression. Overexpression of miR-223 via adenovirusmediated delivery significantly elevated the glucose uptake in rat cardiomyocytes. For the target genes, miR-223 did not affect the expression of myocyte enhancer factor 2C (MEF2C) and insulin-like growth factor 1 (IGF1) receptor, which were predicted to be related to glucose metabolism. Further studies also indicated that miR-223-induced glucose uptake was independent of insulin signaling pathway and AMPK activity, both of which play essential roles during glucose homeostasis. Interestingly, the protein expression level of GLUT4 was 1.3-fold upregulated. Consistently, introduction of a synthetic miR-223 inhibitor decreased GLUT4 expression. Moreover, the induced glucose uptake by overexpression of miR-223 can be restored by knocking down of GLUT4 via siRNA technology. Therefore, miR-223 regulates glucose metabolism by affecting GLUT4 expression in cardiomyocytes.59 In obese and aging mice, miR-34a was elevated with the decreased level of NAD+. miR-34a directly targets SIRT1, NAMPT (a rate-limiting enzyme for NAD+ biosynthesis), and represses the expression of both genes. In addition, the hepatic overexpression of miR-34a reduces the levels of NAMPT, SIRT1, and NAD+, while it increases the acetylation levels of SIRT1-targeted transcriptional regulators, including peroxisome proliferator-activated receptor γ co-activator 1a (PGC1a), sterol regulatory element-binding proteins 1c (SREBP1c), farnesoid X receptor, and nuclear factor kappa B (NF-κB), finally mimicking the outcomes of OB.60 Taken together, these findings reveal the regulatory roles of miRNA in cellular metabolism and suggest that miRNA may be putative therapeutic targets for metabolic diseases.
Metabolites Modulate Epigenetics
Chromatin-dependent gene modifications interact with the cellular metabolism reciprocally. Metabolic-related genes can be regulated by epigenetic modifications, and many metabolites of cell metabolism can serve as essential cofactors for chromatin-modifying enzymes and control the transcription or translation processes. Therefore, the fluctuation of local metabolites’ concentrations is considered as novel signaling cues for regulating gene expressions.61 Here, we take several metabolites from glycolysis and tricarboxylic acid (TCA) cycle as examples to discuss the regulatory role on epigenetics. Moreover, the supplement or depletion of nutrients (through fasting or calorie restriction [CR]), which can also influence epigenetics via manipulating the concentration of metabolites, is also detailed in this part.
Metabolic products and epigenetics
Once glucose enters into cells, the major portion can be converted to acetyl-CoA through glycolytic pathways, along with the decreased NAD+/NADH ratio.61 NAD+ is a key cofactor in a variety of enzymatic reactions, such as sirtuin-mediated histone deacetylations. Unlike the other HDACs, sirtuins consume NAD+ and release nicotinamide, O-acetyl-ADP-ribose (AADPR), and the deacetylated substrate.24,25 Additionally, a minor portion of glucose involves into hexosamine biosynthetic pathways to produce GlcNAc, which can act as a substrate for histone GlcNAclation.61 It is well known that acetyl-CoA generated from glycolysis and fatty acid β-oxidation enters into TCA cycle to produce cellular energy. Meanwhile, acetyl-CoA is a substrate of HATs and behaves as an essential acetyl group donor in histone lysine acetylation reactions (Fig. 2). Since histone acetylation is tightly associated with chromatin structure and regulates gene expression, acetyl-CoA links energy metabolism with chromatin dynamics and transcription. The study also indicated that the glucose-induced histone acylation is in a dose-dependent manner.62 α-Ketoglutarate (α-KG), an intermediate generated from TCA cycle, can function as a cofactor for DNA demethylation reactions by TET. The TET proteins (including TET 1, 2, and 3) utilize oxygen and α-KG to catalyze multiple oxidation reactions, converting 5mC to 5hmC and finally to 5-carboxylcytosine (5caC).8,63 Inactivation of other intermediates in the TCA cycle, such as succinate dehydrogenase and fumarate hydratase, leads to accumulation of their substrates and then inhibits TET catalyzing reactions.64 Regarding histone demethylation, LSD1 is proposed to catalyze demethylation through an amine oxidation reaction that uses flavin adenine dinucleotide (FAD) as a cofactor. JHDM requires Fe(II) and α-KG as cofactors to demethylate lysine of histones through an oxidative mechanism.27 Thus, the glycolysis and TCA cycle are able to connect energy pathways with epigenetic modifications to regulate gene transcription.
Figure 2.
Crosstalk between cell metabolism and epigenetics. As glucose comes into glycolytic reactions, a minor portion involves in hexosamine biosynthetic pathway to produce GlcNAc, which can act as the substrate for histone GlcNAcylation by O-GlcNAc transferase. The majority of glucose is converted to acetyl-CoA and passed into TCA cycle with the alteration of NAD+/NADH pair. NAD+ is a key cofactor for reactions catalyzed by sirtuins. Acetyl-CoA is used as an acetyl group donor for histone acetylation catalyzed by HATs. Other intermediates from TCA cycle, such as α-KG, can function as cofactors for DNA and histone demethylation reactions by TET proteins and JHDM, respectively. Lysine-specific demethylase 1 (LSD1) is proposed to catalyze demethylation using FAD.
Nutrients and epigenetics
Nutrients are essential components in foods that are used by organisms to survive and grow. Among them, most of the carbohydrates, proteins, and fats are the energy supplier for supporting daily metabolism and body activities. While for other nutrient components like vitamin, docosahexoenoic acid (DHA) and dietary fibers are also indispensible for human health. Here, we discuss how these nutrients affect health and disease progression via the epigenetic regulation.65–67
Folate (also called folic acid), a water-soluble vitamin B, is extensively studied for its important function in DNA synthesis, DNA repair, and DNA methylation.68 Moreover, folate is widely used as food supplementation during pregnancy and infancy to decrease the risk of neural tube defects. Because humans cannot synthesize folates in a de novo manner, folate has to be provided and absorbed from diet to meet demands. Structurally, folate carries a methyl group and can feed into the one-carbon metabolism, in which folate goes through several reactions to generate S-adenosylmethionine (SAM), the universal methyl group donor for DNA and histone methylations (Fig. 3).68,69 Before and during pregnancy, insufficient folate supplementation for mothers can increase the risk of neural tube defects of infants. One underlying mechanism is that low dietary folate can lead to abnormal DNA methylation via one-carbon metabolism.70,71 Steegers-Theunissen et al72 investigated whether periconceptional folate supplementation of the mother was related to methylation at the differentially methylated region (DMR) of the insulin-like growth factor 2 gene (IGF2) of the child. Children whose mother used folate 400 μg per day showed about 4.5% higher methylation of the IGF2 DMR than those who were not exposed to folate (absolute methylation was 0.495 [SE 0.004] and 0.474 [SE 0.007], respectively; P = 0.014). Interestingly, an inverse association between IGF2 DMR methylation and birth weight was observed. Their study indicates that periconceptional folate supplementation is associated with epigenetic changes in IGF2 in the child affected by intrauterine growth and development as reflected in birth weight.
Figure 3.
Involvement of dietary nutrients in epigenetics. Dietary intake of folate, vitamin B (2, 6, and 12), choline, and methionine (green color) regulate epigenetic modifications through involving one-carbon metabolism where the intermediate SAM is produced (red color) and subsequently be provided as the universal methyl group donor for DNA and histone methylations. One-carbon metabolism is described briefly as follows: folate is first converted to dihydrofolate, then to tetrahydrofolate (THF), which enters the cycle. Vitamin B6 is a cofactor in the conversion of THF to 5,10-methylene THF. The process of 5,10-methylene THF to 5-methyl THF is vitamin B2 dependent. 5-methyl THF serves as a methyl donor in a reaction converting homocysteine to methionine, in which vitamin B12 serves as a precursor to methionine synthase. In turn, methionine generated SAM. S-adenosylhomocysteine (SAH) that is converted from SAM participates in the generation of homocysteine. Choline is also involved in the production of methionine by converting it to betaine. The alteration of dietary methionine can also modulate the histone methylations through regulating SAM and SAH levels; the methionine restriction decreases histone modification, and this phenomenon can be sustained by diet.108
Components of vitamin B family (eg, 2, 6, and 12) and choline can also function as essential cofactors in one-carbon metabolism, which provides the methyl donor for chromatin methylation (Fig. 3).66,67 Severe deficiency of vitamin B12 induces promoter hypomethylation of the cystathionine betasynthase gene and represses its transcription in rats, which cannot be recovered by dietary supplementation of methionine to the B12 deficient rats. The human studies also show a link between vitamin B12 deficiency and the increased risk of various cancers (eg, colon and breast cancers).73 Choline is essential for fetal neurogenesis such as hippocampal development and memory function throughout life. During mouse embryonic development, choline deprivation caused hypermethylation of CpG3 within the promoter region of calbindin 1 (Calb1) and CpG7 within the repressor element 1 (RE1) binding site, and the binding of transcriptional repressor neuron restrictive silencing factor to RE1 decreased by 45%. These changes led to increasing of both gene and protein expressions of calbindin1, the early marker of neuronal differentiation.74 These findings suggest that during the early fetal period, a deficiency in methyl donors can alter DNA methylation and thereby affect fetal development.
DHA, an omega-3 factty acid, is a primary structural component for human brain, cerebral cortex, sperm, and retina, which has a wide range of effects to human health, including against Alzheimer’s disease and cancer.75,76 Sadli et al77 reported that the addition of DHA to M17 neuroblastoma culture medium showed elevated levels of H3K9ac and reduced levels of HDAC1, 2, and 3, suggesting that DHA promotes gene expression.
Dietary fibers are the indigestible component of food derived from plants, which are helpful for the digestion and absorption of nutrients.78,79 It has been demonstrated that dietary fibers facilitate the generation of short-chain fatty acids (butyrate), and there is a positive linear correlation between butyrate level and histone acetylation in colon epithelial cell.80 In the last decade, several effects of butyrate are clarified, including anti-inflammatory effects, effects on OB, IR, and inherited disorders.81–84 One of the major functions of butyrate that is involved in the epigenetic regulation of gene expression is inhibiting HDACs.85,86 Specifically, butyrate treatment of cells results in histone hyperacetylation, and butyrate itself inhibits class I HDAC activity in mammalian cells. These findings suggest that dietary fibers are an effective food component that is able to regulate epigenetic modifications.
CR and epigenetics
CR simulates human dieting, which is considered as the most effective environmental manipulation for extending life span so far. This phenomenon has also been experimentally proved in multiple species that range from worm to rodent.87 CR can induce wide metabolic changes (gluconeogenesis, fat mobilization, etc.) and undergoes weight loss in human subjects. Huang et al found that weight loss is associated with DNA methylation.88 The DNA methylation pattern in human individuals with normal weight, OB, and successful weight loss maintainers is different, and the pattern in successful weight loss maintainer is more closely resembled than those in normal weight. These findings suggest a relationship between OB, weight loss, and DNA methylation. Moreover, extensive studies reported the crucial role of CR on regulating SIRT1 activities, which is an NAD+-dependent deacetylase. Mechanistically, CR increases NAD+/NADH ratio and induces SIRT1 expression. The upregulated SIRT1 further suppresses the downstream PPARγ, then downregulates its target gene aP2, which encodes an assisting fat storage protein, thereby promoting fat mobilization into the blood.89,90 Therefore, CR regulation on metabolism through epigenetic mechanisms opens new avenues for related diseases therapy and precision nutrition supply.
Conclusions
Epigenetics influence gene expression with no alterations of DNA sequence and might be inherited by next generation. The major molecular machinery of epigenetics includes DNA methylation, histone posttranslational modifications, and miRNAs. Modifications of chromatin structure ultimately regulate transcription. Increasing evidence demonstrates that metabolic diseases are highly associated with epigenetic alterations. It is well established that metabolic products from cellular metabolic processes like TCA cycle and glycolysis are involved in the regulation of chromatin methylation and acetylation. Moreover, dietary nutrients (such as folate, vitamin B family, DHA, and dietary fibers) play important roles in DNA methylation and histone modifications by directly inhibiting related enzymes or by changing the availability of substrates for those enzymatic reactions.
In the past two decades, scientists have generated a variety of valuable data and knowledge about the basis of epigenetics and cellular metabolism. Here, we discussed the reciprocal regulation between these two phenomena, which eventually leads us to a better understanding of metabolic-related disease progression and pathogenesis. Nevertheless, due to our incomplete knowledge from current studies, we need to make investigations on two key issues. (1) How nutrients and environmental factors are sensed and signaled to the epigenetic machinery? Here, we discussed sparse cases and steps of how nutrients are able to direct epigenetic modifications. However, the detailed molecular mechanisms are still missing for several important signaling pathways. (2) How does the cell response to the metabolic changes at a systematic level? Since the whole cellular network is highly orchestrated and complicated, it is basically impossible to understand the connection between metabolite and epigenetics by focusing on a single pathway. In order to achieve a deeper understanding of these puzzles, there is no doubt that the conventional genetic and cellular analysis should be upgraded and combined with recently developed high-throughput sequencing strategies. In future, we expect more exciting discoveries and methods to thrive and help us understand the molecular regulation of epigenetics and cellular metabolism, which may provide a better way to maintain our health through nutritional modulation besides pharmacotherapies.
Footnotes
ACADEMIC EDITOR: Christian Bronner, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2705 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Provenance: the authors were invited to submit this paper.
Author Contributions
Wrote the first draft of the manuscript: WX. Contributed to the writing of the manuscript: ZY and FX. Jointly developed the structure and arguments for the paper: FW. Made critical revisions and approved final version: ZY and FX. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Kietrys AM, Kool ET. Epigenetics: a new methyl mark on messengers. Nature. 2016;530(7591):423–424. doi: 10.1038/530423a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73(5):770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15(17):2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 6.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 7.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 8.Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7(1):9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Even-Faitelson L, Hassan-Zadeh V, Baghestani Z, Bazett-Jones DP. Coming to terms with chromatin structure. Chromosoma. 2016;125(1):95–110. doi: 10.1007/s00412-015-0534-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Zhu P, Li G. New insights into the helical structure of 30-nm chromatin fibers. Protein Cell. 2014;5(7):489–491. doi: 10.1007/s13238-014-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Dong X, Greven MC, Kundaje A, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13(9):R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng C, Alexander R, Min R, et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22(9):1658–1667. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Jin G, Zhou X. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 2015;43(8):3873–3885. doi: 10.1093/nar/gkv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Zhao W, Zhou X. Modeling co-occupancy of transcription factors using chromatin features. Nucleic Acids Res. 2016;44(5):e49. doi: 10.1093/nar/gkv1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 19.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol Chem. 2008;389(4):353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 20.Lau OD, Kundu TK, Soccio RE, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5(3):589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11(2):155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 22.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 23.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20(8):615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 26.Sabari BR, Tang Z, Huang H, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12(4):321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Pan G, Tian S, Nie J, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Swanson SK, Gogol M, et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol Cell. 2015;60(3):408–421. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5(8):837–840. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 33.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 34.Bandiera S, Hatem E, Lyonnet S, Henrion-Caude A. microRNAs in diseases: from candidate to modifier genes. Clin Genet. 2010;77(4):306–313. doi: 10.1111/j.1399-0004.2010.01370.x. [DOI] [PubMed] [Google Scholar]
- 35.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. FEBS J. 2011;278(7):1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 36.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389(3):305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 37.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes. 2012;61(2):542–546. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Mello VD, Pulkkinen L, Lalli M, Kolehmainen M, Pihlajamaki J, Uusitupa M. DNA methylation in obesity and type 2 diabetes. Ann Med. 2014;46(3):103–113. doi: 10.3109/07853890.2013.857259. [DOI] [PubMed] [Google Scholar]
- 40.Kwak SH, Park KS. Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med. 2016;48:e220. doi: 10.1038/emm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhawan S, Tschen SI, Zeng C, et al. DNA methylation directs functional maturation of pancreatic beta cells. J Clin Invest. 2015;125(7):2851–2860. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuroda A, Rauch TA, Todorov I, et al. Insulin gene expression is regulated by DNA methylation. PLoS One. 2009;4(9):e6953. doi: 10.1371/journal.pone.0006953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang BT, Dayeh TA, Kirkpatrick CL, et al. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54(2):360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barres R, Zierath JR. DNA methylation in metabolic disorders. Am J Clin Nutr. 2011;93(4):897S–900S. doi: 10.3945/ajcn.110.001933. [DOI] [PubMed] [Google Scholar]
- 45.Ahmadian M, Suh JM, Hah N, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokomori N, Tawata M, Onaya T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. Diabetes. 1999;48(4):685–690. doi: 10.2337/diabetes.48.4.685. [DOI] [PubMed] [Google Scholar]
- 47.Sugii S, Evans RM. Epigenetic codes of PPARgamma in metabolic disease. FEBS Lett. 2011;585(13):2121–2128. [Google Scholar]
- 48.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunjan A, Singh RK. Epigenetic therapy: targeting histones and their modifications in human disease. Future Med Chem. 2010;2(4):543–548. doi: 10.4155/fmc.10.18. [DOI] [PubMed] [Google Scholar]
- 50.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 51.LeRoith D, Fonseca V, Vinik A. Metabolic memory in diabetes—focus on insulin. Diabetes Metab Res Rev. 2005;21(2):85–90. doi: 10.1002/dmrr.530. [DOI] [PubMed] [Google Scholar]
- 52.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105(26):9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vecellio M, Spallotta F, Nanni S, et al. The histone acetylase activator pentadecylidenemalonate 1b rescues proliferation and differentiation in the human cardiac mesenchymal cells of type 2 diabetic patients. Diabetes. 2014;63(6):2132–2147. doi: 10.2337/db13-0731. [DOI] [PubMed] [Google Scholar]
- 54.Khan S, Jena G, Tikoo K, Kumar V. Valproate attenuates the proteinuria, podocyte and renal injury by facilitating autophagy and inactivation of NF-kappaB/iNOS signaling in diabetic rat. Biochimie. 2015;110:1–16. doi: 10.1016/j.biochi.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 55.Khan S, Jena G, Tikoo K. Sodium valproate ameliorates diabetes-induced fibrosis and renal damage by the inhibition of histone deacetylases in diabetic rat. Exp Mol Pathol. 2015;98(2):230–239. doi: 10.1016/j.yexmp.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Lu M, Zhang Q, Deng M, et al. An analysis of human microRNA and disease associations. PLoS One. 2008;3(10):e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qabaja A, Alshalalfa M, Bismar TA, Alhajj R. Protein network-based Lasso regression model for the construction of disease-miRNA functional interactions. EURASIP J Bioinform Syst Biol. 2013;2013(1):3. doi: 10.1186/1687-4153-2013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86(3):410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 60.Choi SE, Fu T, Seok S, et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12(6):1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015;3:10. doi: 10.1186/s40170-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoekstra AS, de Graaff MA, Briaire-de Bruijn IH, et al. Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget. 2015;6(36):38777–38788. doi: 10.18632/oncotarget.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallou-Kabani C, Junien C. Nutritional epigenomics of metabolic syndrome: new perspective against the epidemic. Diabetes. 2005;54(7):1899–1906. doi: 10.2337/diabetes.54.7.1899. [DOI] [PubMed] [Google Scholar]
- 66.Junien C, Gallou-Kabani C, Vige A, Gross MS. Nutritional epigenomics of metabolic syndrome. Med Sci (Paris) 2005;21:44–52. SecNo. [PubMed] [Google Scholar]
- 67.Burdge GC, Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Annu Rev Nutr. 2010;30:315–339. doi: 10.1146/annurev.nutr.012809.104751. [DOI] [PubMed] [Google Scholar]
- 68.Choi SW, Friso S. Epigenetics: a new bridge between nutrition and health. Adv Nutr. 2010;1(1):8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23(8):853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Put NM, van Straaten HW, Trijbels FJ, Blom HJ. Folate, homocysteine and neural tube defects: an overview. Exp Biol Med (Maywood) 2001;226(4):243–270. doi: 10.1177/153537020122600402. [DOI] [PubMed] [Google Scholar]
- 71.Blom HJ, Smulders Y. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J Inherit Metab Dis. 2011;34(1):75–81. doi: 10.1007/s10545-010-9177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, et al. Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS One. 2009;4(11):e7845. doi: 10.1371/journal.pone.0007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poirier LA, Wise CK, Delongchamp RR, Sinha R. Blood determinations of S-adenosylmethionine, S-adenosylhomocysteine, and homocysteine: correlations with diet. Cancer Epidemiol Biomarkers Prev. 2001;10(6):649–655. [PubMed] [Google Scholar]
- 74.Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene. FASEB J. 2010;24(1):184–195. doi: 10.1096/fj.09-140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Connor WE. Importance of n-3 fatty acids in health and disease. Am J Clin Nutr. 2000;71(1 suppl):171S–175S. doi: 10.1093/ajcn/71.1.171S. [DOI] [PubMed] [Google Scholar]
- 76.Lauritzen L, Hansen HS, Jorgensen MH, Michaelsen KF. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog Lipid Res. 2001;40(1–2):1–94. doi: 10.1016/s0163-7827(00)00017-5. [DOI] [PubMed] [Google Scholar]
- 77.Sadli N, Ackland ML, De Mel D, Sinclair AJ, Suphioglu C. Effects of zinc and DHA on the epigenetic regulation of human neuronal cells. Cell Physiol Biochem. 2012;29(1–2):87–98. doi: 10.1159/000337590. [DOI] [PubMed] [Google Scholar]
- 78.Marlett JA, McBurney MI, Slavin JL, American Dietetic A Position of the American Dietetic Association: health implications of dietary fiber. J Am Diet Assoc. 2002;102(7):993–1000. doi: 10.1016/s0002-8223(02)90228-2. [DOI] [PubMed] [Google Scholar]
- 79.Anderson JW, Chen WJ. Plant fiber. Carbohydrate and lipid metabolism. Am J Clin Nutr. 1979;32(2):346–363. doi: 10.1093/ajcn/32.2.346. [DOI] [PubMed] [Google Scholar]
- 80.Boffa LC, Lupton JR, Mariani MR, et al. Modulation of colonic epithelial cell proliferation, histone acetylation, and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer Res. 1992;52(21):5906–5912. [PubMed] [Google Scholar]
- 81.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 82.Saemann MD, Bohmig GA, Osterreicher CH, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14(15):2380–2382. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 83.Cavaglieri CR, Nishiyama A, Fernandes LC, Curi R, Miles EA, Calder PC. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003;73(13):1683–1690. doi: 10.1016/s0024-3205(03)00490-9. [DOI] [PubMed] [Google Scholar]
- 84.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4(1):4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cang S, Ma Y, Liu D. New clinical developments in histone deacetylase inhibitors for epigenetic therapy of cancer. J Hematol Oncol. 2009;2:22. doi: 10.1186/1756-8722-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4(1):13–18. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 87.Sohal RS, Forster MJ. Caloric restriction and the aging process: a critique. Free Radic Biol Med. 2014;73:366–382. doi: 10.1016/j.freeradbiomed.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang YT, Maccani JZ, Hawley NL, Wing RR, Kelsey KT, McCaffery JM. Epigenetic patterns in successful weight loss maintainers: a pilot study. Int J Obes (Lond) 2015;39(5):865–868. doi: 10.1038/ijo.2014.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6(4):298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 90.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hazen SL, Smith JD. An antiatherosclerotic signaling cascade involving intestinal microbiota, microRNA-10b, and ABCA1/ABCG1-mediated reverse cholesterol transport. Circ Res. 2012;111(8):948–950. doi: 10.1161/CIRCRESAHA.112.277277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 93.Melkman-Zehavi T, Oren R, Kredo-Russo S, et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J. 2011;30(5):835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karbiener M, Pisani DF, Frontini A, et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells. 2014;32(6):1578–1590. doi: 10.1002/stem.1603. [DOI] [PubMed] [Google Scholar]
- 95.Vickers KC, Shoucri BM, Levin MG, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57(2):533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107(27):12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 99.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 100.Lee EK, Lee MJ, Abdelmohsen K, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31(4):626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 102.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 103.Grueter CE, van Rooij E, Johnson BA, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149(3):671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakanishi N, Nakagawa Y, Tokushige N, et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385(4):492–496. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 105.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51(6):1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299(2):E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mentch SJ, Mehrmohamadi M, Huang L, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22(5):861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]