Abstract
Phytohormones control the development and growth of plants, as well as their response to biotic and abiotic stress. The seven most well-studied phytohormone classes defined today are as follows: auxins, ethylene, cytokinin, abscisic acid, jasmonic acid, gibberellins, and brassinosteroids. The basic principle of hormone regulation is conserved in all plants, but recent results suggest adaptations of synthesis, transport, or signaling pathways to the architecture and growth environment of different plant species. Thus, we aimed to define the extent to which information from the model plant Arabidopsis thaliana is transferable to other plants such as Solanum lycopersicum. We extracted the co-orthologues of genes coding for major pathway enzymes in A. thaliana from the translated genomes of 12 species from the clade Viridiplantae. Based on predicted domain architecture and localization of the identified proteins from all 13 species, we inspected the conservation of phytohormone pathways. The comparison was complemented by expression analysis of (co-) orthologous genes in S. lycopersicum. Altogether, this information allowed the assignment of putative functional equivalents between A. thaliana and S. lycopersicum but also pointed to some variations between the pathways in eudicots, monocots, mosses, and green algae. These results provide first insights into the conservation of the various phytohormone pathways between the model system A. thaliana and crop plants such as tomato. We conclude that orthologue prediction in combination with analysis of functional domain architecture and intracellular localization and expression studies are sufficient tools to transfer information from model plants to other plant species. Our results support the notion that hormone synthesis, transport, and response for most part of the pathways are conserved, and species-specific variations can be found.
Keywords: phytohormone biosynthesis, transport, signaling, orthologue search, domain analysis, expression profiling, pathway conservation
Introduction
A handful of small chemical compounds and their derivatives, the so-called phytohormones, coordinate the genetically determined growth and development of plants as well as the incessant integration of environmental signals.1–3 Typically, phytohormones act at low concentrations. Further, at organismic level, the action of these compounds is separated frequently from their biosynthesis in a spatiotemporal manner.4–6 Thereby, transport over short and long distances in the plant contributes to the distribution of hormones in different tissues, thus forming distinct morphogenetic gradients for each hormone.7,8 Short- and long-term responses are observed at target cells after perceiving the signal by binding to specific hormone receptors and integrating it into the cellular metabolism. This signal recognition finally leads to adaptation or differentiation of specific functions during growth and development and ensures maintenance of tissue and organ integrity.9,10 It is widely accepted that the ratio among the levels of different hormones determine the outcome of their activity and function.2,11 In part, this is attributed to the mutual effects on hormone-specific synthesis, transportation, signaling, and response pathways.
The role of phytohormones in plant physiology, growth, and development of numerous species has been explored since the late 19th century.12 However, our current knowledge on the molecular mechanisms underlying the biosynthesis and distribution of these compounds as well as signaling and cellular responses is still limited to only few model plants.1,13,14 Most of the information exists from studies on the model system A. thaliana.15–21 Although basic features of pathways involved in hormone biosynthesis and action are considered to be conserved, initial results suggest variations in different plant species. These variations are likely the consequences of optimizing these pathways with respect to differences in plant architecture and interaction with the environment, including both biotic and abiotic factors as well as breeding of highly productive cultivars for agricultural purposes.22,23
During the last decade, biochemical and cell biological analyses as well as the availability of information of the whole genome of A. thaliana24 yielded vast information on the molecular pathways for hormone synthesis, transport, and signaling.25 Generalization of data for the entire plant kingdom is still challenging, as information for other plant species is by far not as advanced as for A. thaliana. Thus, it is required to predict putative proteins involved in phytohormone pathways in different plant species by bioinformatics analysis, which can be validated subsequently in further experiments. In addition, it is of great importance to assess to which extent the information from the model plant A. thaliana can be transferred to other plants. This will be the base to establish species-specific variations. The identification of all genes contributing to the plant-specific regulatory phytohormone networks is a challenge of the current research. Such knowledge can be a valuable tool for improvement of plant productivity by more targeted species-specific breeding programs. Here, we focus on the pathways of seven phytohormone classes: auxin, ethylene, cytokinin, abscisic acid (ABA), jasmonic acid (JA), gibberellin (GA), and brassinosteroid (BR).
Auxin is a key regulator of many growth processes during plant life cycle and was the first phytohormone detected as a growth-promoting compound involved in the regulation of cell division and elongation, cell differentiation, photo- and gravitropism, apical dominance, flowering, and senescence.26–30 Indole-3-acetic acid (IAA) was identified as the major naturally occurring auxin in plants.31 IAA is mainly synthesized in shoot meristems and young tissues. Maintenance of auxin homeostasis requires the continuous transport of IAA conjugates through the entire plant.32 This is achieved by long-distance transport in the phloem toward the root tip and by local cell-to-cell transport mechanisms over shorter distances forced by chemiosmotic gradients.
Ethylene, which is the simplest alkene (C2H4), was the first gaseous biological signaling molecule discovered. In 1901, Neljubow33 reported that ethylene was the active compound in illuminating gas that caused altered growth of pea seedlings.34 In addition, seed germination, seedling growth, organ development and senescence, leaf and petal abscission, fruit ripening, and stress and pathogen responses are among the many processes governed at least in part by ethylene.35 The easy-to-score “triple response” phenotype of dark-grown A. thaliana seedlings exposed to ethylene enabled the identification of ethylene-insensitive and constitutive-response mutants.36 The analysis of these mutants led to the description of a primarily linear model for ethylene signal transduction, which starts with hormone perception and ends in transcriptional regulation.37,38 Current models, however, suggest the existence of a more complex pathway with both positive and negative regulatory feedback loops by several phosphorylation cascades, feedback-regulated transcriptional networks, and protein and mRNA turnover regulatory modules.39,40
Searching for substances promoting cell division in plant tissue cultures led to the discovery of adenine derivatives. Kinetin (6-furfurylaminopurine) was the active compound contained in autoclaved herring sperm DNA,41 and zeatin was identified as the naturally occurring cytokinin in maize endosperm.42,43 Besides its proposed activity in cell division, cytokinins are involved in the control of most aspects of plant growth and development, eg, shoot initiation and growth, apical dominance, sink/source relationships, photomorphogenesis, gametophyte development, and leaf senescence.18,44 Pathways deriving from purine and isopentenyl metabolism in meristems and differentiating young tissues are the major sources of cytokinin biosynthesis in plants.18,45,46 Transport over short and long distances contribute to the spatial distribution of the hormone within the plant. The signal transduction pathway in cytokinin perception and signaling is reminiscent to bacterial two-component phosphorelays.47
ABA was discovered as a growth inhibitor accumulated in abscising cotton fruit, therefore originally named as “abscisin”, and during development of dormancy in sycamore trees, leading to the name “dormin”.48 ABA has an inhibitory effect on growth processes including cell division and is a counterpart of growth-promoting hormones such as GA, auxin, or cytokinin. It is further involved in the regulation of seasonal dormancy in resting tissues and of drought, salt, and cold stress tolerance mechanisms, providing interesting agricultural aspects.49–52 This led to the assignment of ABA as a global player in the regulation of growth and developmental processes including storage of proteins as well as biotic stress response.53,54
JA and its biosynthetic precursor compounds are lipid derivatives synthesized from α-linolenic acid, first identified from the oil of Jasminum grandiflorum.55 First described physiological responses to JAMe (JA methyl ester) were growth-inhibiting and senescence-promoting effects.56,57 Later, altered gene expression and accumulation of jasmonate-induced proteins after external application of JA or its methyl ester have been reported.58,59 This and the finding of the multiple actions of this compound as an intrinsic and volatile signal molecule in plants in biotic and abiotic stress response and in developmental programs stimulated its intensive exploration during the last decades.60,61
More than 130 different GAs have been identified so far.62 The majority of these compounds are biologically inactive and represent intermediates of GA biosynthesis or catabolism. Only few, eg, GA1, GA3, GA4, and GA7, are bioactive in plant growth and development.20,63 Besides stem elongation, GAs are essential for fundamental developmental processes, such as seed germination, trichome development, pollen maturation, and induction of flowering.64 GA deficiency causes dwarfism and late-flowering phenotypes. Unraveling the molecular basis of GA biosynthesis, action, and signaling pathways over the past decades64–68 has a major impact on agriculture, with huge improvements in productivity, especially since the introduction of the semidwarf varieties during the “green revolution” in the 1960s.69
BRs are the least explored group of phytohormones and originally isolated from Brassica napus pollen.70 Brassinolide (BL) and its immediate precursor castasterone (CS) are the biological active compounds among numerous related phytosteroids.71 The essential function of BRs in the control of growth and developmental processes became evident after the identification and characterization of mutants in BR biosynthesis and signaling, which exhibited severe dwarf-like growth phenotypes, altered leaf morphology, reduced male fertility, and de-etiolated phenotypes in dark-grown seedlings.72 The biosynthetic pathway of BR was described mainly on the base of A. thaliana and cell cultures of Catharanthus roseus, while major key steps have been reported for other plants, such as rice, pea, and tomato.73–76 More recent findings provided insights into the complexity of the regulation of BR biosynthesis and homeostasis in a spatiotemporal manner, crosstalk with other hormone signaling pathways and integration of environmental signals.77–80
We analyzed the proteins involved in the pathways of these seven phytohormone classes by gathering genome information of 13 different species (Fig. 1), representing green algae, moss, monocots, and eudicots in the clade of Viridiplantae. Chlamydomonas reinhardtii represents a unicellular system, and comparison of findings for the green algae and for the other organisms studied allowed the identification of the components required only for multicellular systems. At first, we extracted information from literature for the biosynthetic pathways, transport mechanisms, and sensing systems in A. thaliana for the individual hormone classes. Next, we employed bioinformatic tools to identify orthologous genes in the selected plant species based on the information available for A. thaliana. From this groundwork, co-orthologues of the proteins involved in the various processes leading to hormone response were assigned, and the conservation of the pathways with respect to the knowledge governed from A. thaliana was evaluated by domain comparison and prediction of the intracellular localization of the putative identified proteins. Finally, we described these pathways in tomato in more detail by evaluating the expression profiles of the (co-)orthologous genes in different tissues based on publically available datasets. The results are discussed concerning evolution and functional equivalence of phytohormone pathways in plants. Extending our approach by analysis of expression patterns in different tissues of tomato gives insights in preferentially used pathways for the biosynthesis of phytohormones in this important crop plant.
Figure 1.
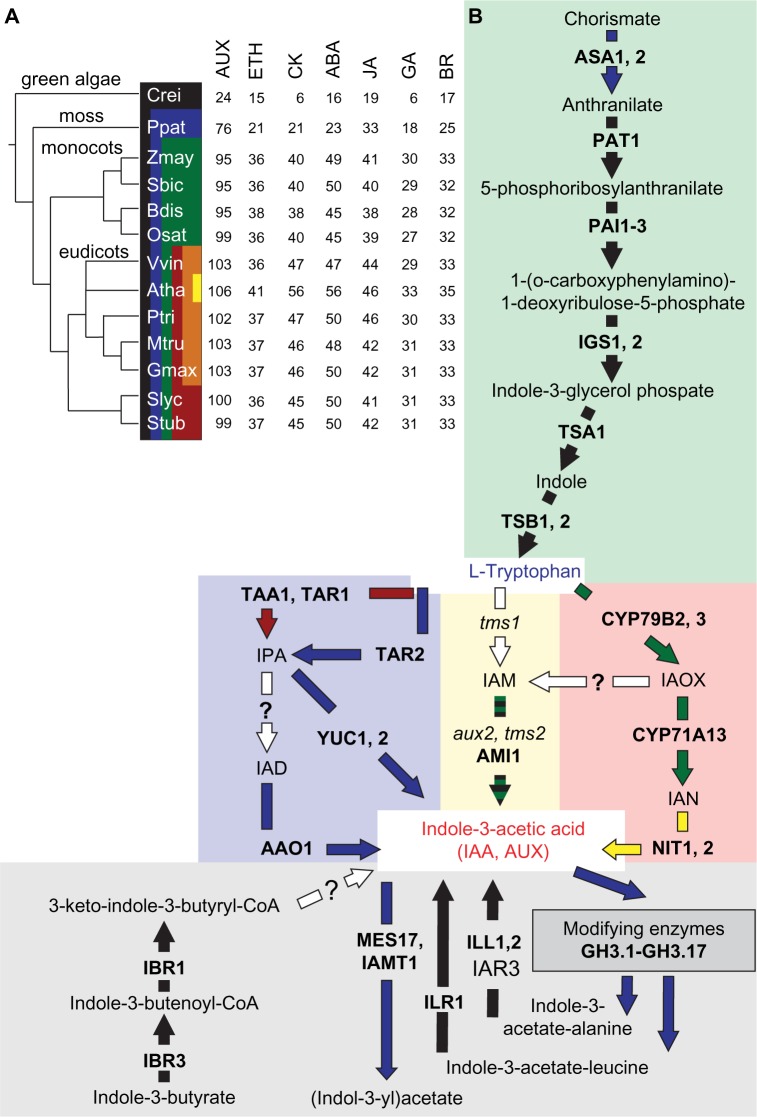
Biosynthesis pathway of auxin in selected species. (A) The phylogenetic relation of the selected 13 members of Viridiplantae. Species: Atha (A. thaliana), Gmax (G. max), Mtru (M. truncatula), Ptri (P. trichocarpa), Stub (S. tuberosum), Slyc (S. lycopersicum), Vvin (V. vinifera), Bdis (B. distachyon), Osat (O. sativa), Sbic (S. bicolor), Zmay (Z. mays), Ppat (P. patens), and Crei (C. reinhardtii). The color code indicates the distribution of the different clades demonstrating the distance from A. thaliana. The number of genes related to all analyzed phytohormone biosynthesis pathways in A. thaliana and of the identified co-orthologues in the other species are indicated. (B) Enzymes and intermediate products of the plastidial tryptophan biosynthesis pathway (green region) and distinct routes of Trp-dependent IAA biosynthesis in plants with IPA (blue region), IAM (yellow region), and TAM, IAOX and IAN (red region) as major biosynthesis intermediates. The route using IBA as precursor or storage intermediate and pathways of IAA conjugation are shown in the gray region. The arrows are colored according to the species in which the enzymes were found (taken from A) and multicolor dashed arrows indicate putative loss of co-orthologues genes in one clade during evolution.
Abbreviations: Proteins: AAO, Arabidopsis aldehyde oxidase; AMI, amidase; ASA, anthranilate synthase alpha subunit; CYP79B, cytochrome P450, family 79, subfamily B; GH, Gretchen Hagen; IBR, indole-3-butyric acid response; ILR1/ILL, IAA-leucine resistant; IAMT, IAA carboxylmethyltransferase; NIT, nitrilase; PAT, phosphoribosylanthranilate transferase; PAI, phosphoribosylanthranilate isomerase; IGS, indol-3-glycerol phosphate synthase 2; TSA, tryptophan synthase alpha chain; TSB, tryptophan synthase beta chain; TAA1, tryptophan aminotransferase of Arabidopsis; TAR, tryptophan aminotransferase related; YUC, YUCCA. Metabolites: IPA, indole-3-pyruvic acid; IAD, indole-3-acetaldehyde; IAM, indole-3-acetamide; IAOX, indole-3-acetaldoxime; IAN, indole-3-acetonitrile.
Methods
Curation of phytohormone pathway information
Genes involved in biosynthesis, transport, and signaling pathways of seven phytohormone classes for the reference model organism A. thaliana were extracted from the Arabidopsis Hormone Database version 2.0 (AHD2.0; http://ahd.cbi.pku.edu.cn/25). Cross-evaluation from the AHD2.0 was performed to enzymes belonging to biosynthetic pathways extracted from the AraCyc (part of Plant Metabolic Pathways; http://pathway.gramene.org/gramene/aracyc.shtml81). Next, putative candidates and complex enzyme schematic diagrams of the AHD2.0 were verified by an extensive literature screening for the seven different phytohormone classes in book chapters and reviews from the last 10 years. For enzymes assigned to be putatively involved in chemical reactions of the phytohormone pathways in Aracyc and/or AHD2.0, we searched for experimental evidence as well. A. thaliana gene identifier (AGI) and associated description was curated based on Gene Ontology (GO) information deposited in The Arabidopsis Information Resource database (TAIR10; http://www.arabidopsis.org/82). The identified genes were assigned to the categories biosynthesis, transport, or signaling.
Orthologue search and functional domain annotation
The plant genomes of (i) A. thaliana (TAIR10; https://www.arabidopsis.org/), (ii) Brachypodium distachyon (bradi1.2 with GAEVAL; http://www.plantgdb.org), (iii) C. reinhardtii (JGI v4 with GAEVAL; http://www.plantgdb.org), (iv) Glycine max (Glyma1; http://www.plantgdb.org), (v) Medicago truncatula (Mt3.5v5 http://jcvi.org), (vi) Oryza sativa (MSU Version 7.0 with GAEVAL; http://www.plantgdb.org), (vii) Physcomitrella patens (Phypa1.6; http://phytozome.net), (viii) Populus trichocarpa (Ptr v2.0 with GAEVAL; http://www.plantgdb.org), (ix) Solanum lycopersicum (ITAG2.4; https://solgenomics.net/organism/Solanum_lycopersicum/genome), (x) Solanum tuberosum (PGSC v3.4; http://potatogenome.net), (xi) Sorghum bicolor (JGI Sbi1; http://www.plantgdb.org), (xii) Vitis vinifera (Genescope 12X; http://genoscope.cns.fr), and (xiii) Zea mays (B73 RefGen v2; http://www.plantgdb.org), which have been downloaded from PlantGDB,83 have verified gene annotation in relation to alternative splicing and gene fusions/fissions by the gene annotation evaluation algorithm (GAEVAL 82).84 For the orthologue search via OrthoMCL85 and InParanoid,86 we selected these plant species to sample the tree of the super kingdom Viridiplantae by at least one species of the phylum Chlorophyta (C. reinhardtii), one species of the class Bryopsida (P. patens), and four species of the branch of monocots (S. bicolor, B. distachyon, O. sativa, and Z. mays) of which O. sativa and Z. mays are of large agricultural relevance. Besides economic importance, the seven eudicot species (A. thaliana, G. max, M. truncatula, P. trichocarpa, S. tuberosum, S. lycopersicum, and V. vinifera) were chosen as representative model organisms of distinct plant families or with broad interest in specific plant research fields (Fig. 1A).87,88 G. max was chosen due to agricultural relevance and S. tuberosum and V. vinifera were chosen due to their phylogenetic relation to S. lycopersicum and comparison with previous studies.89
The strategy to merge results of different orthologue algorithms has been justified in Paul et al.87 OrthoMCL was used to determine cliques of likely orthologous genes (CLOGs) for more than two species, and results were verified using the pairwise approach of InParanoid version 8.86 Genes were further analyzed when occurring in at least one of the two approaches. OrthoMCL filtered out poor-quality sequences based on the protein sequence length (<10 amino acids) and percentage of stop codons (marked by asterisks in the amino acid sequence; >20%). We searched in all CLOGs for all extracted genes of A. thaliana coding for enzymes involved in biosynthesis, transport, and signaling pathways of the seven phytohormone classes (373 genes based on our curation of phytohormone pathway information; Supplementary Tables 1–14).
Prediction of domain architecture
Domain prediction of proteins of all 13 plant species was performed using Pfam database (version 26.0),90 WebMGA,91 and HMMsearch.92 Customized Python scripts (www.python.org) were used to analyze domain architecture of the protein and conservation of domains of the co-orthologues in a particular orthologous group. Within one orthologous group, the domain architecture of the A. thaliana reference protein sequence was compared to the other co-orthologues concerning the order of the domains, the absence of domains, or the occurrence of new ones. The name of the PFAM domain is indicated when discussed, and the detailed description of the individual domains is available in the PFAM database (http://pfam.sanger.ac.uk/).
Prediction of protein localization
The prediction of the localization of identified proteins was performed by MultiLoc2, TargetP, and Predotar, which enable automated prediction by submitting ≥2 sequences at once or using local versions of the software. The predictor MultiLoc293 distinguishes between 11 different cellular compartments (extracellular, nucleus, Golgi, endoplasmic reticulum (ER), mitochondrion, plastid, plasma membrane, peroxisome, vacuole, cytosol, and cytoskeleton). The localization results of MultiLoc2 for vacuole, ER, Golgi, and plasma membrane are merged and represented as endomembranes. TargetP94 and Predotar95 are widely used programs and can distinguish between chloroplast, mitochondria, and secretory pathway localization and are used for a more accurate prediction of the localization of proteins in these plant compartments.
Transcriptome profiling
Existing S. lycopersicum transcriptome expression data (RNA-seq libraries; GSE33507)89 were obtained from the gene expression omnibus (GEO).96 The RNA-seq data contain S. lycopersicum (cv. Moneymaker) libraries from different organs: root, stem, leaf, flower, and fruits at three developmental stages (mature green, breaker stage, and 10 days after turning red).
Results and Discussion
General procedure for information extraction
We extracted the information for the phytohormone pathways established for A. thaliana (“Methods” section) and assigned co-orthologues to the proteins involved in these pathways in 13 photosynthetic eukaryotes including Arabidopsis (Fig. 1A, Supplementary Tables 1–7). In contrast to other orthologue studies in plants such as the study by Wu et al.97, our approach was not based on a single plant clade like Euasterid but attempt coverage of the whole Viridiplantae super kingdom by using model organisms from algae, mosses, monocots, and eudicots. The orthologue search was based on proteomes, and by this, the referenced tools OrthoMCL and InParanoid allowed detection of co-orthologues and putative paralogs without being restricted to single-copy orthologous genes.97 The approach in our study allowed the detection of CLOGs for phytohormone biosynthesis, transport, and signaling. Identified co-orthologues and co-orthologues (multiple co-orthologues of the same species)98 in one CLOG were analyzed for the presence of conserved functional domains found in the A. thaliana bait as it was performed in earlier studies.88,99 Co-orthologues containing all functional domains were assumed to be functionally related; however, we also considered proteins with additional or missing domains as relevant co-orthologues due to domain “stealing, swapping, or swiveling”.100 Additionally, putative targeting signals for intracellular localization were predicted to verify that the proteins within a CLOG share enzyme functionality and similar intracellular localization of the A. thaliana bait. This limited the number of co-orthologues with putative function in phytohormone pathways, because most pathways or parts of them are defined to specific organelles (Supplementary Tables 8–14). Focusing on tomato, the expression patterns of co-orthologues in the phytohormone pathways in tissues and fruit developmental stages gave hints for rate-limiting steps and tissue-specific expression. Publically available expression values in different tissues and fruit developmental stages based on “reads per kilobase per million mapped reads” (RPKM) normalized datasets were extracted from the GEO96 and Sol Genomics Network (SGN; Supplementary Tables 15–21).101 The expression data of co-orthologues in CLOGs were categorized as low expression (RPKM < 5), moderate expression (RPKM < 100), and high expression (RPKM ≥ 100). All information are deposited in a database,102 focusing on the assignment of the recently published tomato genome.89
The indole-3-pyruvic acid pathway is a major, but not the only, route of auxin biosynthesis in tomato
Two major pathways of IAA biosynthesis have been proposed so far.26 The tryptophan (Trp)-independent pathway derives from the plastidial Trp-biosynthesis route, which is initiated with the conversion of chorismate to anthranilate (Fig. 1B, green region). This pathway consumes immediate precursors of Trp, ie, indole-3-glycerol phosphate or indole for the production of IAA. However, the molecular identity of the required enzyme remains unknown up to now. The Trp-dependent IAA synthesis unifies several routes of IAA biosynthesis deriving from Trp. These routes vary in the formation of distinct intermediates leading to the names of the particular pathways, ie, the indole-3-pyruvic acid (IPA) pathway (Fig. 1B, blue area), the indoleacetamine (IAM) pathway (Fig. 1B, yellow area), and the indole-3-acetaldoxime (IAOX) pathway (Fig. 1B, red area). The high number of intermediates and identified genes proposed to be involved in their enzymatic processing (Supplementary Tables 1 and 8) provides a high complexity to auxin biosynthesis concerning different possible biosynthetic pathways, which is still under debate.26
The IPA pathway includes two enzymatic reactions to convert Trp to IAA.21,103,104 Either TAA1/TAR1 or TAR2 catalyzes the first reaction. While we could observe TAR2 co-orthologues in all plants except for green algae, TAA1/TAR1 was specific for eudicots (Fig. 1B). Remarkably, tomato co-orthologues to TAA1/TAR1 were not expressed in any of the analyzed tissues (Fig. 2). The second is catalyzed by YUC1 and YUC2, two proteins encoded by the YUCCA gene family of flavin monooxygenases. Co-orthologues of all these genes were found in all species with the exception of C. reinhardtii (Fig. 1B, Supplementary Tables 1, 8, and 15). Again, the expression of tomato YUC1, 2 co-orthologues, was low in most of the tissues (RPKM < 5) compared to other genes of the synthesis pathway (Fig. 2). This might point to conversion of IPA to indole-3-acetaldehyde (IAD) by an indole-3-caboxylase, an enzymatic activity described for IAA synthesis in plant growth-promoting rhizobacteria species, which has not been identified in plants yet. Co-orthologues of AAO1, the proposed aldehyde oxidase activity required for the subsequent conversion of IAD to IAA, were detected by our analysis in all plants, and their moderate expression in tomato exceeded that of YUC co-orthologues (RPKM > 5; Fig. 2). Nevertheless, it needs to be mentioned that broad substrate specificity was observed for the AAO1 multigene family that might link its activity to ABA synthesis as well, which is still discussed.105,106
Figure 2.
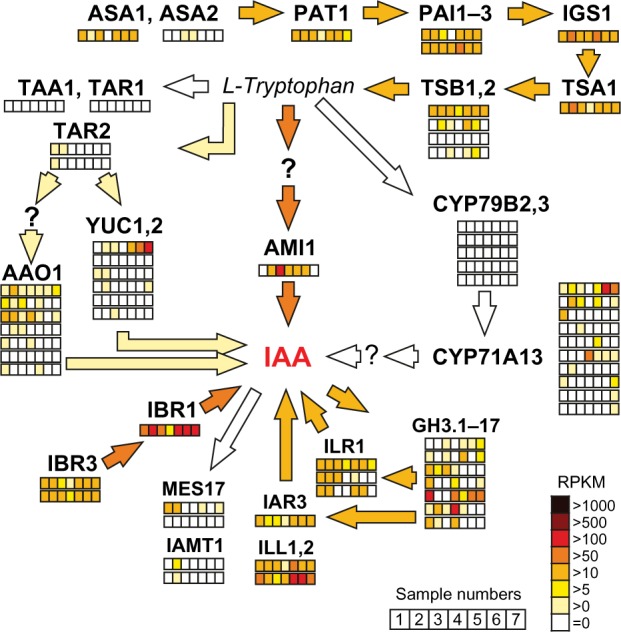
Expression of tomato genes encoding Trp and IAA biosynthesis enzymes. Major routes and enzyme activities involved in auxin biosynthesis are represented as in Figure 1B with focus on the expression of corresponding co-orthologues, genes identified in the tomato genome. Relative transcript levels in different tissues and developmental stages are shown in the color code as indicated in the right lower corner.
Notes: Analyzed samples in each row from left to right are: 1, root; 2, stem; 3, leaf; 4, flower; 5, fruit – mature green; 6, fruit – breaker stage; 7, fruit – ripening 10 days after turning red. Number and order of the rows at indicated co-orthologues corresponds to the presentation of expression data in Supplementary Table 15. Genes coding for enzyme activities not yet identified in tomato are indicated by question mark. Color intensities of the arrows from light to dark orange indicate the general pathway expression at low, moderate, and high levels, respectively.
The IAM pathway also predicts two steps for the conversion of Trp to IAA with IAM as an intermediate product (Fig. 1B). The pathway resembles the conversion of Trp to IAA found in Agrobacterium strains.107 In our study, only co-orthologues of AMI1, the enzyme that catalyzes the second step,108,109 were identified in all plants except for P. patens. AMI co-orthologues were highly expressed in tomato leaves compared to other organs (Fig. 2). In contrast, proteins similar to the bacterial proteins encoded by aux1/iaaM/tms1 genes were not identified. Recently, the conversion of IAOX to IAM was suggested as an alternative route to generate IAM.110
The activity of YUCCA enzymes is assigned to the IAOX pathway for converting tryptamine (TAM) into IAOX (Fig. 1B). However, we detected neither tomato co-orthologues to A. thaliana NIT1, 2 enzymes converting tryptophan to TAM nor to enzymes converting indole-3-acetonitril (IAN) to IAA (Fig. 1B). This observation stands in line with discussion that the IAOX pathway is present in Brassicaceae only.111 Furthermore, the identified co-orthologues of the cytochrome P450 oxidases CYP79B2/B3 involved in IAOX production in A. thaliana110 were also not expressed in the examined tissues in tomato (Fig. 2, Supplementary Table 15). This supports the current model that the IPA pathway is the major route of auxin biosynthesis in tomato. Nevertheless, we cannot exclude that several Trp-dependent auxin biosynthesis pathways may coexist and operate in different tissues.103
IAA conjugation, storage, and degradation is conserved among species
The mechanism of stimulation of adventitious root formation by indol-3-butyric acid (IBA) is well established. Further, IBA is a naturally occurring IAA precursor in many plant species, which requires a peroxisomal β-oxidation activity and the IBA synthase112 for its conversion to the biologically active IAA (Fig. 1B, gray region). The compound IBA appears as reversible auxin storage form, which is transported independent of IAA.113 In tomato, the orthologue to IBR1 and the two IBR3 co-orthologues involved in conversion of IBA to IAA were expressed in nearly all analyzed tissues at moderate levels (Fig. 2).
Both IAA and IBA are found in conjugated forms either with amino acids like alanine (Ala) or leucine (Leu) or in ester-linked conjugates with glucose.27 Co-orthologues for IAA-specific GH3 proteins were present in all plant species, except for C. reinhardtii (Fig. 1B). The conjugates either serve as hydrolyzable storage forms or play a role in IAA degradation.26 Several IAA-leucine resistant (ILR) and ILR-like (ILL) proteins, which contribute to the release of active IAA from amino acid conjugates, exist in A. thaliana and are grouped in CLOGs containing co-orthologues of all selected plant species. Tomato co-orthologues were expressed in all analyzed tissues (Fig. 2). Another ILL protein (ILL3) has been shown to be involved in auxin biosynthesis in P. euphratica,114 but was not included in the CLOG of the other ILL proteins. We observed that ILL3 co-orthologues exist in monocots and eudicots only, and the corresponding tomato gene was expressed in all tissues.
Irreversible oxidation of IAA is the major target for IAA inactivation and occurs on conjugated forms as well. The accumulation of oxIAA observed after IAA application indicates that this catabolic pathway is involved in the regulation of bioactive auxin levels in plants. Finally, conversion of IAA in its methyl ester form by IAMT1 and MES17 results in a nonpolar modified form of IAA, which can probably be transported independent of auxin transporters. We observed that the required enzymes, however, were only moderately expressed in tomato (Fig. 2), and IAMT1 co-orthologues could be found only in P. patens, eudicots and rice, but not in any other of the selected monocots or C. reinhardtii (Fig. 1B, Supplementary Table 1). This might suggest that this enzyme was lost specifically in some monocots.
The directional cellular auxin transport system is specific to multicellular organisms
Besides long-distance phloem transport, the directed cell-to-cell transport of IAA is essential for the regulation of auxin homeostasis.115 Key regulators are PIN-type auxin transport proteins (Fig. 3A), which are distributed asymmetrically along the plasma membrane. As expected, these proteins could be detected in multicellular organisms only (Fig. 3B), and most of them were not expressed in the tomato fruit (Supplementary Table 15). The polar orientated localization of the transporter changes dynamically in response to light or physical stimuli such as gravity and defines the direction and velocity of cellular auxin transport. Release of IAA into the low pH environment of the apoplast has been shown to lead to its protonation into IAAH. AUX1/LAX1 influx carriers localized at the opposite side of the next cell facilitate uptake of the apolar IAAH by the adjacent cell. In line with its function in long-distance transport, AUX1 orthologue in tomato was only moderately expressed in roots, stem, and leaves (Supplementary Table 15), while at least one LAX1 co-orthologue was moderately expressed in all tomato tissues. According to the chemiosmosis model, IAA is deprotonated and trapped in the neutral cytosolic compartment until exported by PIN proteins or other auxin transport mechanisms consisting of P-glycoproteins of the ABCB transporter family (ABCB/PGP). While most PIN proteins are plasma membrane proteins, PIN5, PIN8, and PIN-LIKE proteins are localized at the ER membrane and regulate the intracellular distribution of IAA.116 Consequently, in our analysis, PIN5 and PIN8 were grouped into two distinct CLOGs containing none of the other PIN genes (PIN1–4, PIN6–7). Further, co-orthologues of PIN5 and PIN8 were found only in monocots and eudicots and tended to occur as single-copy genes (Fig. 3A, Supplementary Tables 1 and 8). With respect to their function in intracellular transport, co-orthologues to all other PINs and NRT1.1 existed in all plants, but not in C. reinhardtii, and the number of co-orthologues varied between 3 and 14 (Fig. 3B).
Figure 3.
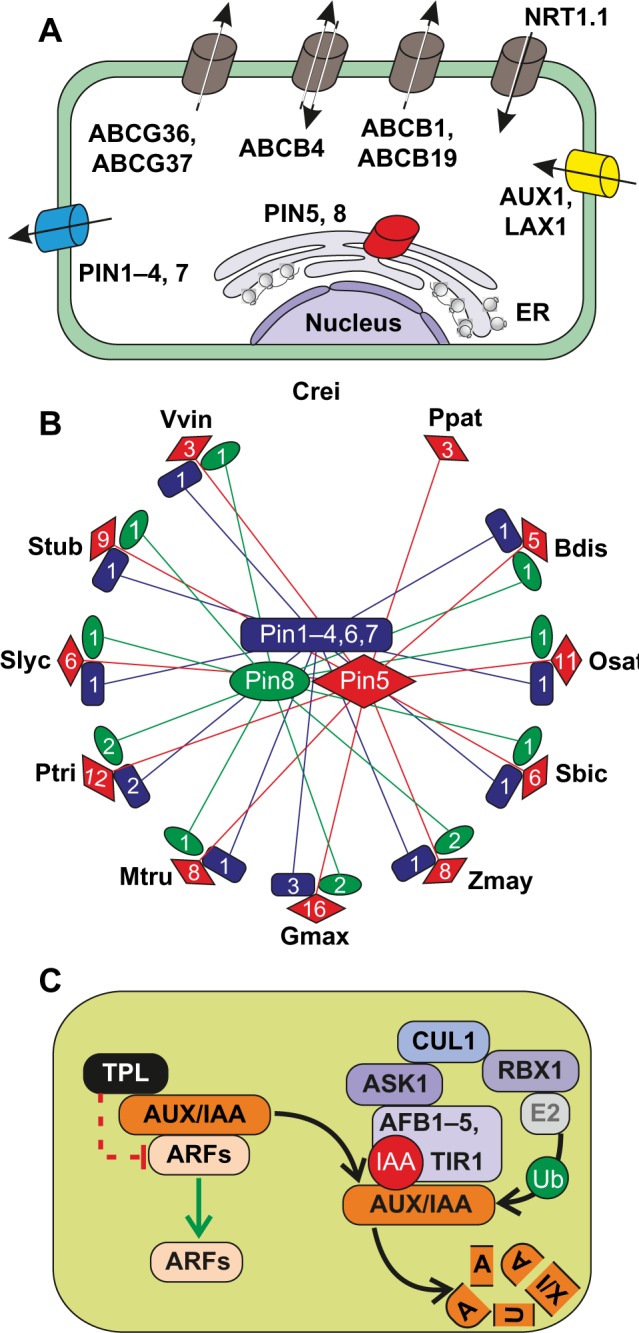
Auxin transport and signaling pathways. (A) Survey and localization of main transporters involved in inter- and intracellular auxin transport processes. (B) Number and relationship of PIN gene family members (Supplementary Table 8) are illustrated for the 13 selected plant species (abbreviations s–e Fig. 1) by red diamonds (Pin5), green ellipses (Pin8), and blue rectangles (Pin1–4, 6, 7). (C) Principle mechanisms and factors involved in auxin signaling in plants. For further details and explanations, see text. The bar-headed dashed line in red indicates indirect suppression mechanisms.
Abbreviations: Proteins: AUX, auxin resistant; LAX, like auxin resistant; ABCB, ATP-binding cassette B; ABCG, ATP-binding cassette G; NRT, nitrate transporter; PIN, PIN-formed; IAA, indole-3-acetic acid inducible; AUX, auxin inducible; TPL, TOPLESS; TPR, TOPLESS related; ARF, auxin-responsive factor; TIR, transport inhibitor response; AFB, auxin signaling F-box protein; ASK, Arabidopsis SKP1 homolog; CUL, Cullin; RBX, RING-box.
Auxin perception is tightly linked to the regulation of auxin-responsive gene
Two classes of interacting transcription factors are involved in the control of auxin-regulated gene expression (Fig. 3C115–117). AUX/IAA transcriptional repressors were found to be present in all monocots and eudicots and were represented by a single CLOG (Supplementary Tables 1 and 8) with varying numbers of co-orthologues ranging from 5 in tomato to 15 in A. thaliana. Remarkably, one tomato orthologue was found to be highly expressed only in fruits (Solyc09g065850), while all others were not expressed in this tissue (Supplementary Table 15). AUX/IAAs generally consist of four functional domains. The “N-terminal domain I” harbors an ethylene response factor (ERF)-associated amphiphilic repressor (EAR) motif required for recruitment of TOPLESS (TPL), which is acting as a transcriptional corepressor in the absence of auxin. Interestingly, co-orthologues to TPL were identified in all analyzed plant genomes except in C. reinhardtii. For P. patens, we could identify two TPL co-orthologues but no co-orthologues to AUX/IAA (Supplementary Table 1). Domain II of AUX/IAA proteins is required for the control of their auxin-stimulated degradation, and domains III and IV participate in the formation of homo- and hetero- oligomeric complexes including interactions with members of the second class of transcription regulators, the auxin- responsive factors (ARFs). ARFs recognize and bind through their B3 DNA-binding domain-specific auxin-response elements and act as either transcriptional repressors or activators.118 In A. thaliana, the ARF subfamily consists of 15 members while we identified only six ARF co-orthologues in the genome of tomato (Supplementary Table 15).
In the presence of auxin, the activation of ARFs is controlled by degradation of AUX/IAAs and release of TPL-mediated repression of DNA-binding and transcriptional activation (Fig. 3C). Degradation of AUX/IAAs is triggered by binding of auxin to one of the F-box proteins TIR1/AFB1–5, the substrate recognition subunit in the SCFTIR/AFB E3 ubiquitin ligase complex, which per se is acting as an intracellular auxin receptor in plants. The stability of the E3 complex is regulated by modification of the scaffold subunit Cullin1 (CUL1) through covalent binding of the ubiquitin-related small protein RUB1. Our results suggest that the regulatory regime of auxin signaling can be transferred to all phototrophic eukaryotes containing co-orthologues with the exception for C. reinhardtii (Supplementary Tables 1 and 8) in which, if present, auxin signaling must be differently perceived. Although all components of the basic SCF complex were also present in the green algae co-orthologues of the TIR/AFB, F-Box proteins were not present.
ACC synthases form a diverse gene family within the ethylene pathway
The biosynthesis pathway of ethylene consists of only three enzymes (Fig. 4). S-adenosylmethionine is synthesized from methionine by S-adenosyl-l-methionine synthetase (SAM1) and is converted to 1-aminocyclopropane-1-carboxylate (ACC) by ACC synthase (ACS). Our orthologue search shows that each selected plant species contains at least one orthologue for SAM1. More interestingly, the A. thaliana genome encodes 12 ACS-like genes, which could be divided into three CLOGs. ACS3 is an A. thaliana-specific pseudogene with a short protein sequence, whereas ACS10 and ACS12 belonged to the same CLOG (Supplementary Table 2), and it has been shown that these aminotransferases can complement the Escherichia coli aminotransferase mutant DL39.119 Thus, there are nine authentic ACS genes in the A. thaliana genome,120 which were grouped in one CLOG containing co-orthologues from green algae, monocots, and eudicots but without co-orthologues from the moss P. patens. In tomato, there are at least eight characterized ACS genes,121 but we could assign 13 co-orthologues to the nine A. thaliana ACS genes involved in ethylene synthesis (Supplementary Table 16). In the last step of ethylene biosynthesis, ACC oxidase (ACO, four genes in A. thaliana) converts ACC to ethylene. Remarkably, ACO2 and ACO3 build their own CLOG without other plant co-orthologues and ACO1 and ACO4 further fell in different CLOGs. Overall, our analysis revealed six genes in the tomato genome in the three CLOGs of ACOs, which has been shown in earlier results.121 Coherent with the importance of ethylene for fruit development, the high expression of two genes (RPKM > 1000; Solyc01g095080, Solyc05g050010) was observed during ripening (Fig. 4121). The current model considers the formation of ACC as the rate-limiting step. SAM1 and EFE/ACO co-orthologues were highly expressed in all tissues in tomato, while the ACS co-orthologues showed only moderate expression (Fig. 4; Supplementary Table 16). However, the activity of ACS is regulated by phosphorylation at protein level as it was documented for SlACS2 (Solyc01g095080).122
Figure 4.
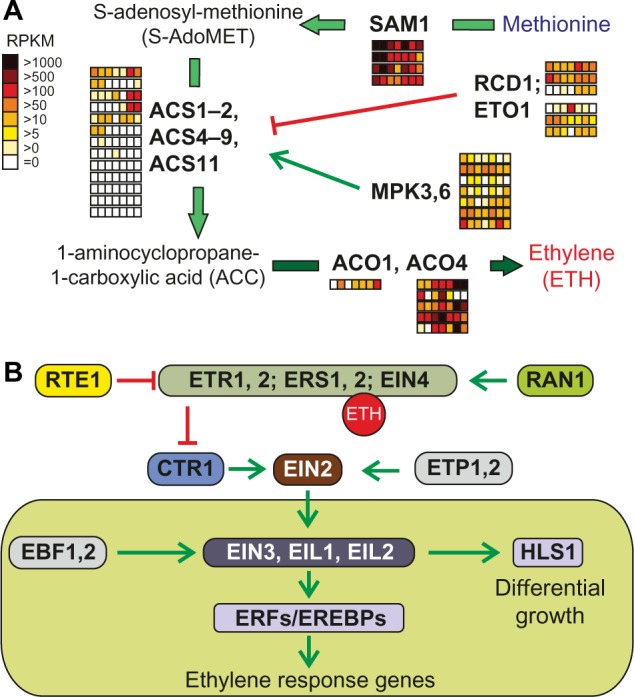
Pathways of ethylene synthesis and signaling. (A) Enzymes and intermediate products of the ethylene production unify to methionine (blue) as precursor for ethylene (red) synthesis. The enzymatic step catalyzed by ACS is inhibited by the action of RCD1 and ETO1 or activated by MPK3 and MPK6. Further details are presented in the text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). Expression of the identified genes in tomato is shown as explained in Figure 2. (B) The components involved in ethylene signaling are represented as interaction scheme, at which activation of downstream components is indicated by green arrows and inhibition by red bar-headed lines.
Abbreviations: Proteins: SAM, S-adenosylmethionine synthetase; ACS, ACC synthase; RCD, radical-induced cell death; ETO, ethylene overproducer; MPK, mitogen-activated protein kinase; ACO, ACC oxidase; ETR/ERS, ethylene response; EIN, ethylene-insensitive; RAN, RAS-related nuclear protein; RTE, reversion-to-ethylene sensitivity; CTR, constitutive triple response; EDR, enhanced disease resistance; EIN, ethylene-insensitive; ETP, EIN2 targeting protein; EIL, ethylene-insensitive 3-like; EBF, EIN3-binding F-box protein; HLS, HOOKLESS.
The ethylene signaling cascade starts with binding of ethylene to ER-localized receptors with protein kinase activities.50,123,124 In all monocots, eudicots, and mosses, ethylene receptors represent a gene family that is composed of ETR1, ERS1, ETR2, ERS2, and EIN4 in A. thaliana (Supplementary Tables 2 and 9). Our analysis revealed the presence of seven co-orthologues in S. lycopersicum, from which five have been previously described (SlETR1, -2, -4, -5, and Never ripe), while two have not been yet tested in tomato (SlETR6 and -7125; Supplementary Table 16).
In contrast to the orthologue search, previous sequence analysis divided ethylene receptors into subfamily I and II. Subfamily I members tend to have higher similarity to histidine kinases, whereas subfamily II members have acquired serine kinase activities.126 The ethylene signaling in A. thaliana is more dependent on subfamily I members, which cannot be functionally replaced by subfamily II members.127 In turn, reduced expression of the subfamily II receptor genes, SlETR4 or SlETR6, in tomato results in substantially increased ethylene sensitivity.124 This phenotype cannot be restored to that of wild-type by overexpression of the subfamily I receptor. Thus, the subfamily II receptors might have a more important function in tomato than in A. thaliana.
In the absence of ethylene, the receptors activate the negative regulator CTR1 (constitutive triple response1), thus repressing the activity of downstream ethylene signaling components.39 In A. thaliana, the Ser/Thr protein kinase CTR1 is localized at the ER membrane due to its physical interaction with the receptors128 and directly phosphorylates the C-terminal domain of EIN2 in the absence of the ethylene signal. Our orthologue search detected co-orthologues of the two A. thaliana CTR1 genes only in monocots and eudicots, while in line with former studies, four co-orthologues were found in tomato (Supplementary Table 16124).
The physical movement of EIN2 C-terminal region from the ER membrane to the nucleus allows the ethylene signal to activate the downstream transcription factors EIN3 and EIN3-like (EILs) in the nucleus. Phosphorylation at specific sites in EIN2 region regulates translocation.39 In tomato, we identified six co-orthologues to these transcription factors (EIN4/EIL1, four co-orthologues; EIL3, two co-orthologues; Supplementary Tables 2 and 9). The ethylene signal stabilizes EIN3 and EIL transcription factors, which are short-lived proteins in the absence of ethylene, and consequently activates ERFs, inducing various physiological responses.129
Recent advances propose a more complex route that includes feedback-regulated transcriptional networks as well as protein and mRNA turnover regulatory modules. According to this regime, ETP1 and ETP2 control EIN2 levels, whereas EBF1 and EBF2 (EIN3-binding factor) regulate the levels of EIN3 in response to the ethylene signal.39 Interestingly, the orthologous group including ETP1 and ETP2 contains 134 additional co-orthologues of A. thaliana, most of them containing both or at least one F-Box and F-Box associated (FBA_1) domains (Supplementary Table 9). Remarkably, only one representative of B. distachyon and S. tuberosum was detected in the same CLOG, which could be due to the fact that these F-Box proteins evolve very fast or they are species-specific in their primary sequence. Four orthologues were identified for EBF1 and EBF2 in tomato and at least three of them showed moderate or high expression in all tissues (Supplementary Tables 2 and 16). However, A. thaliana ETP1 and ETP2 were grouped in one CLOG containing 134 additional putative co-orthologues but only one orthologue of B. distachyon and S. tuberosum each. With respect to the high number of co-orthologues only in Arabidopsis and the low conservation within the super kingdom of Viridiplantae, the functional relation of all the 136 co-orthologues in the ethylene signaling pathway is unlikely.
Cytokinin biosynthesis seems to be specific for mono-cots and eudicots
Enzymes encoded by isopentenyltransferase (IPT) genes catalyze the initial step of cytokinin biosynthesis. Earlier work based on sequence alignments by ClustalW130 proposed that IPTs likely evolved in plants, although similar sequences exist in some cyanobacteria.45 In contrast, our orthologue search approach (see “Methods” section) confirmed the occurrence of IPT genes in all monocots and eudicots, but not in mosses or algae. The only exception was tRNA-IPT (IPT9), which was detected in all selected plant species. Nevertheless, functional equivalent enzymes for IPT in species like green algae are expected, because cytokinin has been detected in the green alga Chara globularis and putative cytokinin receptors have been found in different green algae.131,132 We identified six co-orthologues in tomato for the nine A. thaliana IPTs, from which only one showed high expression in all tissues (Supplementary Table 17). IPT utilizes adenosine tri- or diphosphate (ATP/ADP) and dimethylallyl-diphosphate (DAMPP), the reactive isomer of isopentenyl diphosphate, to synthesize N6-(Δ2-isopentenyl) adenosine-5′-triphosphate and -diphosphate ribonucleotides iPRTP and iPRDP, respectively (Fig. 5A133). Both compounds are converted subsequently into the corresponding trans-zeatin ribonucleotides (tZRTP/tZRDP) by cytokinin trans-hydroxylases, such as the cytochrome P450-dependent monooxygenases CYP735A1 and CYP735A2. Both enzymes were identified in A. thaliana and found to be conserved in monocots and eudicots. In tomato, two co-orthologues were detected in one CLOG, which showed different expression patterns in tissues and developmental stages (Supplementary Table 17). Only CYP735A1 showed moderate expression in root, flower, and mature fruit, whereas CYP735A2 was not expressed in any of these tissues. The formation of cytokinin ribonucleosides and the free bases iP and tZ has been originally assumed to be catalyzed by 5′ ribonucleotide phosphohydrolases and adenosine nucleosidases.134,135 However, the identification of the LONELY GUY (LOG) family of enzymes in rice and A. thaliana suggests the direct conversion into the bioactive forms of the two cytokinins. Interestingly, one CLOG containing LOG1, LOG3, and LOG4 was found to be conserved in all plant species but the remaining six A. thaliana LOG genes were clustered in different CLOGs and could be divided into Arabidopsis specific (LOG2, LOG6, and LOG9), eudicot specific (LOG7), or monocot/eudicot specific (LOG5 and LOG8). In summary, we identified four co-orthologues of the LOG family in tomato (Supplementary Table 10). Remarkably, almost all co-orthologues of IPT, CYP735A, and LOG showed low expression in fruit ripening stages (Supplementary Table 17), which on the one hand suggests that cytokinins are not produced in fruits at high levels, but on the other hand, studies on the metabolite cytokinin showed that it has an unclear influence on fruit development.136
Figure 5.
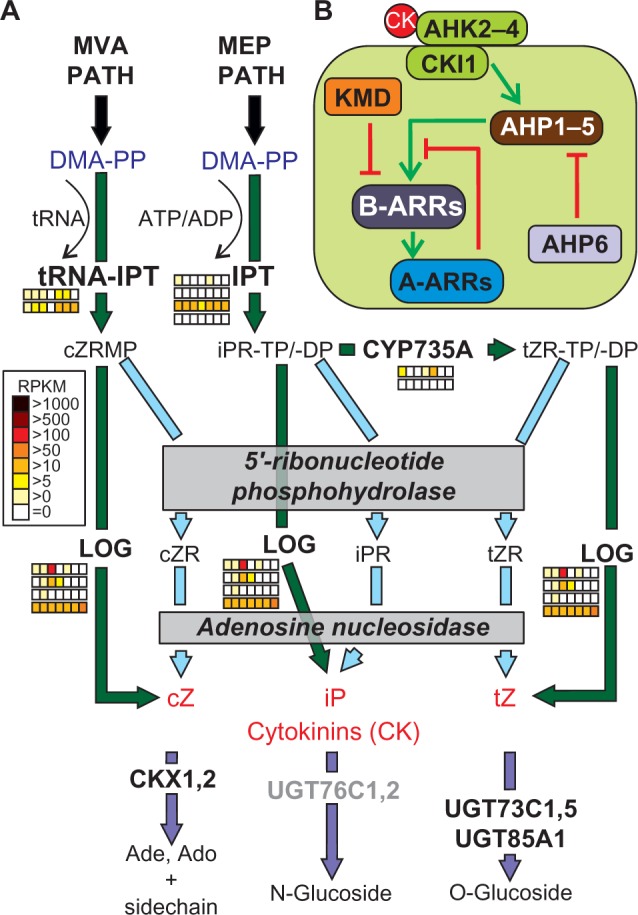
Pathways of cytokinin synthesis and signaling. (A) The precursor of cytokinin synthesis is DMA-PP, which is derived from the MVA or MEP pathways. The intermediate products cZRMP, iPRTP/-DP, or tZRTP/-DP are converted to the final products such as cZ, iP, or tZ (red) by the action of LOG or by yet not clearly defined 5′-ribonucleotide phosphohydrolases and adenosine nucleosidases. For further details, see text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). Expression of the identified co-orthologues in tomato is shown as explained in Figure 2. Genes coding for enzyme activities not expressed by any orthologue are colored in gray. (B) The components involved in cytokinin signaling are presented as interaction scheme.
Abbreviations: Proteins: IPT, isopentenyltransferase; CYP735A, cytochrome P450, family 735, subfamily A; LOG, LONELY GUY; UGT73C, UDP-glucosyl transferase 73C; UGT85A, UDP-glucosyl transferase 85A; CKX, cytokinin oxidase/dehydrogenase; AHK, Arabidopsis histidine kinase; CKI, cytokinin-independent; AHP, histidine-containing phosphotransmitter; ARR2, response regulator; KMD, KISS ME DEADLY. Metabolites: ATP, adenosine triphosphate; ADP, adenosine diphosphate; DMA-PP, dimethylallyl pyrophosphate; cZRMP, cZR 5′ monophosphate; iPRTP, iPR 5′ triphosphate; tZRTP, tZR 5′ triphosphate; cZR, cis-zeatin riboside; iPR, isopentenylribose; tZR, trans-zeatin riboside; cZ, cis-zeatin; iP, isopentenyladenine; tZ, trans-zeatin.
Although found in substantial amounts, cis-zeatin (cZ) has no biological hormone activity and cannot be converted sufficiently into trans-zeatin (tZ).137 Moreover, the synthesis of cZ seems to be completely independent of the tZ synthesis pathway because two of the nine IPT genes in A. thaliana (IPT2 and IPT9) are specifically involved in the formation of cZRMP by utilizing AMP and DMA-PP.45 We were able to assign one specific tomato orthologue for both enzymes (IPT2: Solyc11g066960; IPT9: Solyc12g014190), suggesting that this pathway is also present in tomato. cZRMP is mainly derived from the cytosolic mevalonate (MVA) pathway of IPP synthesis, while the tZ pathway preferentially consumes IPP derived from the plastidial methylerythritol phosphate (MEP) pathway.
Conjugation to glucose and irreversible cleavage are the two major reactions efficiently reducing the levels of bioactive cytokinins (Fig. 5A18). N-glycosyltransferases catalyze the glycosylation of nitrogen residues in the purine ring, while O-glycosylation occurs on the oxygen in the side chain of tZ or dihydrozeatin (dhZ). The latter reaction likely produces storage forms that can be rapidly reactivated by β-glucosidases.138 Removal of the unsaturated side chains of tZ and iP by the activity of cytokinin oxidases (CKX) leads to their irreversible inactivation and is important for the regulation of active cytokinin levels and limitation of cytokinin effects. The members of both IPT and CKX gene families show remarkably distinct expression patterns in A. thaliana and expression of several CKX genes can be stimulated upon cytokinin application.139 For tomato, only one orthologue of IPT5 and another of IPT9 were found to be expressed in all tissues, while transcripts of the other IPT co-orthologues were absent in most of the tissues. None of the co-orthologues of the tomato CKX gene family was expressed in all tissues, and a high variance in the expression patterns was observed as it was shown for A. thaliana in previous studies (Supplementary Table 17).139 Interestingly, with the exception of the CKX family, we could show that always one gene of each enzyme family of the cytokinin biosynthesis path in tomato was expressed in every tissue. In summary, for the green algae C. reinhardtii and the moss P. patens, no co-orthologues were identified for cytokinin trans-hydroxylases and glucosyltransferases, while C. reinhardtii also lacked the CKX family. Furthermore, only the tRNA-IPT (IPT9) and a small set of LOG family genes were found to be present in the unicellular algae.
Cytokinin transport, perception, and signaling
tZ-ribonucleoside (tZR) is preferentially transported toward the shoot through the xylem, while the rootward transport of cytokinins occurs through the phloem. Initially, it was discussed that cytokinin receptors (AHK2, 3, and 4 in A. thaliana), with typical domain structure of membrane-bound histidine sensor kinases, represent the extracellular recognition site for cytokinin binding. Recent studies identified most of the receptor proteins in the ER membrane, which opened the discussion whether perception of cytokinins occurs in the ER lumen rather than on the cell surface.140–142 AHK2, 3, and 4 are proposed to form heterodimeric receptor complexes upon cytokinin perception, while AHK1 is located in a distinct orthologous group,143 which was confirmed by our orthologue search. Cytokinin response1 (CRE1) and Woodenleg (WOL1) are allelic to AHK4 and were identified in mutant screens for cytokinin phenotypes.144 Cytokinin insensitive1 (CKI1) is one additional, more distantly related histidine kinase in A. thaliana145; however, its involvement in cytokinin signaling remains to be shown. In tomato, three co-orthologues were also present in the CLOG of AHK2–4 (Supplementary Table 17), which were expressed at high levels in all analyzed tissues. In contrast, for the tomato orthologue of CKI1, no expression was observed (Supplementary Table 17).
Binding of cytokinins triggers the activation of the histidine kinase domain of AHK on the cytosolic site, leading to its autophosphorylation. Components acting downstream in the phosphorelay signaling pathway include histidine-containing phosphotransfer proteins, named AHP in A. thaliana. The AHP family consists of five members (AHP1–5) and one pseudo AHP considered as a negative regulator (AHP6). We detected four co-orthologues of AHP in tomato falling in four different CLOGs. One of the four co-orthologues was expressed in all tissues (Supplementary Table 17). For the AHP4 and AHP6 co-orthologues, no expression was detected. AHP has been shown to function in the transfer of the signal to the nucleus to regulate the activity of response regulators (ARRs) acting as transcription factors. ARRs form a large family with 15 proteins in A. thaliana, which can be divided according to their domain structure into type-A AARs acting as repressors (ARR3–9, 15–17) and type-B ARRs acting as activators (ARR1–2, 10–12).140,146,147 In tomato, six type-A AAR and five type-B AAR co-orthologues exist (Fig. 5B; Supplementary Tables 3 and 17). For ARR16 and ARR17, only a weak response to cytokinin was reported.148 These two AARs were found only in eudicots in our analysis, which could indicate redundant or newly developed functions in cytokinin response. The stability of some of the type-B AARs is controlled by their recruitment to specific SCF E3 ubiquitin ligase complexes containing one of the four F-box proteins KISS ME DEADLY (KMD1–KMD4). Two co-orthologues of KMD were detected in tomato (Fig. 5B, Supplementary Table 3).
Type-A ARRs do not possess a defined output domain, and most of them are transcriptionally induced in response to cytokinins (Supplementary Table 10). Interestingly, for the six co-orthologues in tomato, we observed that each orthologue corresponded to a pair of Arabidopsis type-A ARRs except for the group containing ARR8 and ARR9. Furthermore, in tomato type-A ARRs were expressed in all analyzed tissues with two exceptions. The orthologue of ARR5/6 was not expressed in tomato fruit, while the orthologue of ARR16/17 was only expressed in flowers and fruits (Supplementary Table 17). Altogether, the function of type-A ARRs as negative regulators of cytokinin response by interfering with the function of positive regulators in the signal transduction pathway is widely assumed. One exception so far is ARR4, which has been shown in Arabidopsis to interact positively with phytochrome B,149 and it will be interesting to see whether this particular function is conserved in tomato and other plant species as well.
Putative rate limitations in biosynthesis of ABA in tomato
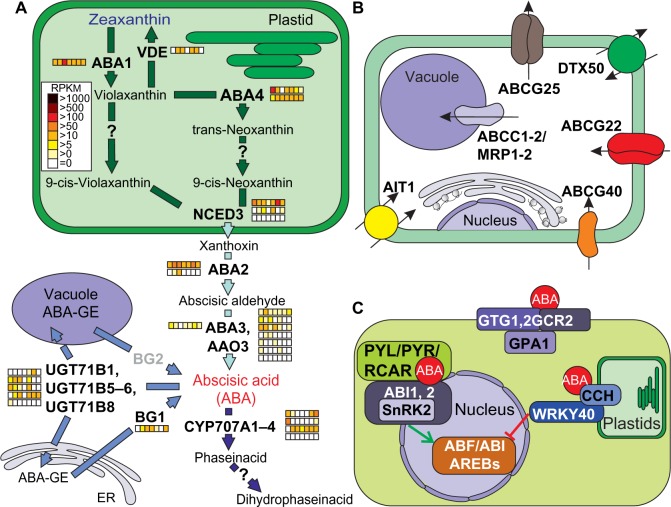
ABA synthesis is initiated in plastids and derives from the metabolism of carotenoids.17,150 Moreover, ABA-specific defects are connected to mutations in genes acting downstream of zeaxanthin synthesis. Zeatin epoxidase (ZEP), also called ABA1, catalyzes the formation of violaxanthin, and this step is reversed by the action of violaxanthin de-epoxidase (VDE; Fig. 6A). The process known as xanthophyll cycle is assumed to provide sufficient amounts of zeaxanthin for an increased photo-oxidative protection under high-light conditions. Each of the two enzymes is encoded by a single gene in Arabidopsis and has one orthologue in tomato, whereas VDE1 was not found in C. reinhardtii (Supplementary Tables 4 and 11). In contrast to ZEP1, which was at least moderately expressed in all tissues (Supplementary Table 18), VDE1 was not expressed in roots, flowers, and ripe fruits.
Figure 6.
Pathways of abscisic acid synthesis, transport, and signaling.
Notes: (A) Enzymes and intermediate products of the plastidial, endoplasmatic, and cytosolic pathways for abscisic acid production unify to the plastidal production of zeaxanthin (blue) as precursor for abscisic acid (red) synthesis in the cytosol. The storage pathway is catalyzed by BG1 and BG2 as well as by members of the UGT71B gene family. For further details, see text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). The expression of the identified genes in tomato is shown as explained in Figure 2. Genes coding for enzyme activities not expressed by any orthologue are indicated in gray. (B) Survey and localization of main transporters involved in uptake and intracellular distribution of abscisic acid. (C) The components involved in abscisic acid signaling are represented as interaction scheme.
Abbreviations: Proteins: ABA, ABA deficient; VDE1, violaxanthin de-epoxidase; NCED, nine-cis-epoxycarotenoid dioxygenase; CYP707A, cytochrome P450, family 707, subfamily A; UGT71B, UDP-glucosyl transferase 71B; BG, beta-1,3-glucanase; ABCG, ATP-binding casette G25; DTX, detoxification efflux carrier; ABCG, ATP-binding cassette G; AIT, ABA-importing transporter; ABCC1, ATP-binding cassette C; GTG, GPCR-type G protein; GCR, G protein coupled receptor; GPA, G PROTEIN alpha subunit; CCH, conditional chlorina; WRKY, WRKY DNA-binding protein; PYL1, PYR1-like 1, PYR, pyrabactin resistance; RCAR, regulatory component of ABA receptor; SNRK, sucrose nonfermenting 1(SNF1)-related protein kinase; ABI, ABA insensitive; ABF, abscisic acid responsive elements-binding factor.
Formation of 9-cis-violaxanthin and 9-cis-neoxanthin via its intermediate trans-neoxanthin are the last steps occurring in plastids before cleavage by 9-cis-epoxicarotenoid dioxygenase (NCED; five genes in A. thaliana151) and releasing of xanthoxin from the C40 precursor into the cytosol. While the enzyme-encoding genes are known for the ABA4-dependent formation (ABA4, AT1G67080) of trans-neoxanthin, the activities required for the isomerization to the 9-cis derivatives remain to be identified. Two co-orthologues of ABA4 were identified in tomato, which are globally expressed (Fig. 6A; Supplementary Table 18). The subsequent cleavage of xanthophyll by NCED is the rate-limiting step in ABA synthesis,17 and expression of NCED is tightly regulated in response to stress and developmental signals. In contrast to previous results,151 we observed a localization signal for only three (NCED 2, 3, 5) NCEDs in A. thaliana. The number of co-orthologues in the 13 plant species ranges from none in C. reinhardtii to nine in Z. mays, all containing the carotenoid oxygenase-specific domain (retinal pigment epithelial membrane (RPE)66, Supplementary Tables 4 and 11). In tomato, three NCED co-orthologues were present and one appeared to be globally expressed (Fig. 6A; Supplementary Table 18).
The final two steps of ABA synthesis occur in the cytoplasm and include the conversion of xanthoxin to ABA aldehyde catalyzed by SDR1/ABA2 and the formation of the carboxyl group mainly by abscisic acid oxidase 3 (AAO3), which is one of the three AAOs in A. thaliana.152 All three enzymes depend on a molybdenum cofactor.153 Consequently, a mutant of the sulfurase in A. thaliana that produces the functional cofactor was named aba3, because strong ABA deficiency phenotypes were caused due to abolishing all AAO activities.154 In tomato, we identified two co-orthologues of ABA2, six of AAO3, and one of ABA3 (Fig. 6A; Supplementary Tables 4 and 18). However, not all co-orthologues of AAO3 might be involved in ABA synthesis. On the one hand, three of them showed only moderate expression (Fig. 6A), while on the other hand, it has been shown that Arabidopsis AAO3 is the major AAO in ABA biosynthesis.152,153
ABA catabolism plays an important role in the regulation of ABA homeostasis, and currently, two pathways have been described.17 The first is initiated by 8′-hydroxylation of ABA, which leads to the isomerization of the intermediate 8′-OH-ABA to phaseic acid (PA) and the subsequent degradation via conversion to dihydrophaseic acid (Fig. 6A17). Four P450-type monooxygenases CYP707A were identified to catalyze the initial step in A. thaliana.155 All of them were present in the same CLOG together with four co-orthologues of tomato (Supplementary Tables 4 and 11; Solyc08g005610, Solyc08g075320, Solyc04g078900, and Solyc01g108210). The expression patterns showed spatial and temporal differences, indicating that individual members might have different importance during growth and development (Fig. 6A; Supplementary Table 18). The second pathway leads to the formation of glycose ester conjugates (ABA-GE) by the activity of several glycosyltransferases (UGT71Bs17). ABA-GE is considered as storage or transport form of ABA, which accumulates in vacuoles and the apoplast. Translocation to the ER and rapid reactivation of ABA by cleaving the glycose ester conjugates by β-glucosidases (BG1 and BG2) is assumed to occur in response to dehydration. The five tomato co-orthologues to UGT71B and the one to BG1 were found to be expressed in most tissues. BG2, which was assumed to catalyze the conversion of vacuolar stored ABA conjugates, was expressed at very low level (Fig. 6A; Supplementary Table 18).
Transport of ABA needs functional assignment within orthologous groups
Several transporters encoded by distinct transporter gene families are involved in the transport of ABA or ABA conjugates across the various membranes (Fig. 6B).156 The members of the subfamily G of the large ATP-binding cassette (ABC) protein family are one group of these transporters (ABCG).157 In A. thaliana, ABCG25 was the first ABA efflux carrier identified in the plasma membrane. It is expressed mainly in vascular tissues.158,159 ABCG22 and ABCG40 reside in guard cells and contribute to enhanced import of ABA into stomatal cells.160 The CLOGs of ABCG22 and ABCG40 contained five additional co-orthologues to Arabidopsis ABCG22/25 and 14 co-orthologues to ABCG40, for which the functional assignment remains to be elaborated. In total 31 ABCG co-orthologues have been detected in tomato (Supplementary Table 18).
The ABA-importing transporter 1 (AIT1), originally characterized as nitrate transporter (NRT1.2), is involved in cellular import of ABA in the vascular tissues of inflorescence stems, leaves, and roots.7 In the genome of tomato, four co-orthologues to AIT1 were identified (Supplementary Table 18). DTX50 is an ABC transporter of the detoxification efflux carriers/multidrug and toxic compound extrusion (DTX/MATE) family shown to function in efficient ABA export after drought stress.161 DTX50 plays an important role in the removal of excess ABA levels to prevent hyperaccumulation of the hormone.157 Again, four co-orthologues were detected in the genome of tomato (Supplementary Table 18). Finally, two members of the ABCC subfamily in A. thaliana, AtABCC1/MRT1 and AtABCC2/MRT2 (multidrug resistance–associated protein), have been shown to transport ABA-GE across the tonoplast into the vacuole. Both belong to the same CLOG in which 14 tomato genes were also clustered.
Three ABA-specific receptor types associated with distinct cellular localizations have been described (Fig. 6C).17,48,162 One type is localized to the ER, Golgi, and peripheral plasma membranes and consists of the G protein coupled receptor proteins GTG1, GTG2, and GCR2. Interaction with the G-alpha subunit GPA1 regulates the binding of ABA. Signal transduction from these receptors mainly targets the regulation of membrane channels involved in Ca2+ transport and control of water permeability, eg, in guard cells. We identified three tomato co-orthologues of GCR2, and one for GTG1 and GTG2. Interestingly, the latter was only expressed in flower and fruit tissues (Supplementary Table 18).
The subunit H of the Mg2+ chelatase complex (CCH) localized in plastids was proposed as an additional ABA receptor.163 The tomato orthologue of CCH was found to be highly expressed in all tissues (Fig. 6C; Supplementary Table 18). Binding of ABA enhances the interaction of subunit H on the cytosolic side with a group of transcription factors from the WRKY family, thereby preventing their translocation to the nucleus. In the absence of ABA, the transcription factors act as negative regulators of ABA response genes.
The third class of ABA receptors consists of soluble proteins seen as entrance components of the “core ABA signaling pathway” (Fig. 6C48). These receptors belong to a gene family (14 genes in A. thaliana) known as pyrobactin resistant (PYR), PYR-like (PYL), or regulatory component of ABA receptor (RCAR) which was represented in our study by eight co-orthologues in S. lycopersicum (Supplementary Table 4). The proteins control the activity of downstream components of the signaling pathway by the interaction with protein phosphatase 2C (PP2C), which acts as a negative regulator of ABA response. This inhibition is maintained by the interaction with the active ABI1, 2 enzymes containing the PP2C domain (conserved in all 13 species, Supplementary Table 11). Further, SNF1-related kinases (SnRKs; seven co-orthologues in tomato) regulate the ABA signaling in the absence of ABA or at low concentrations of ABA. This interaction prevents the phosphorylation and activation of numerous transcription factors, ion channels, and other mediators of ABA responses. Alternating binding of PYR/PYL/RCAR to ABI1, 2 controls release and activity of the SnRKs and thereby the activation of a transcription factor cascade cooperating in the regulation of numerous ABA response genes.164
Differences in the localization of biosynthesis enzymes from JA in tomato
Jasmonates are lipid-derived compounds synthesized from α-linolenic acid released from plastid membranes by one of the seven different branches of the lipoxygenase (LOX) pathway.165 Subsequently, β-oxidation occurs in peroxisomes and the final step of JA synthesis takes place in the cytoplasm.166 Defective in anther dehiscense 1 (DAD1), phospholipase1 (PLA1), and phospholipase 2 (PLA2) are involved in α-linolenic acid production.61 In A. thaliana, dad1 mutant results in a male sterile phenotype,167 indicating that DAD1 is responsible for JA biosynthesis in flowers, while in leaves JA is synthesized by the DGL (Dongle) gene.168,169 Both enzyme CLOGs contained no co-orthologues from C. reinhardtii and P. patens; thus, it is likely that they evolved in the ancestor of monocots and eudicots (Supplementary Table 5). We identified two co-orthologues for DGL and one orthologue for DAD1 in tomato, but none of them were expressed in the tissues and developmental stages of fruits analyzed (Fig. 7A; Supplementary Table 19). This suggests that another pathway for linolenic acid in tomato may exist or that the pathway is only activated under stress conditions.
Figure 7.
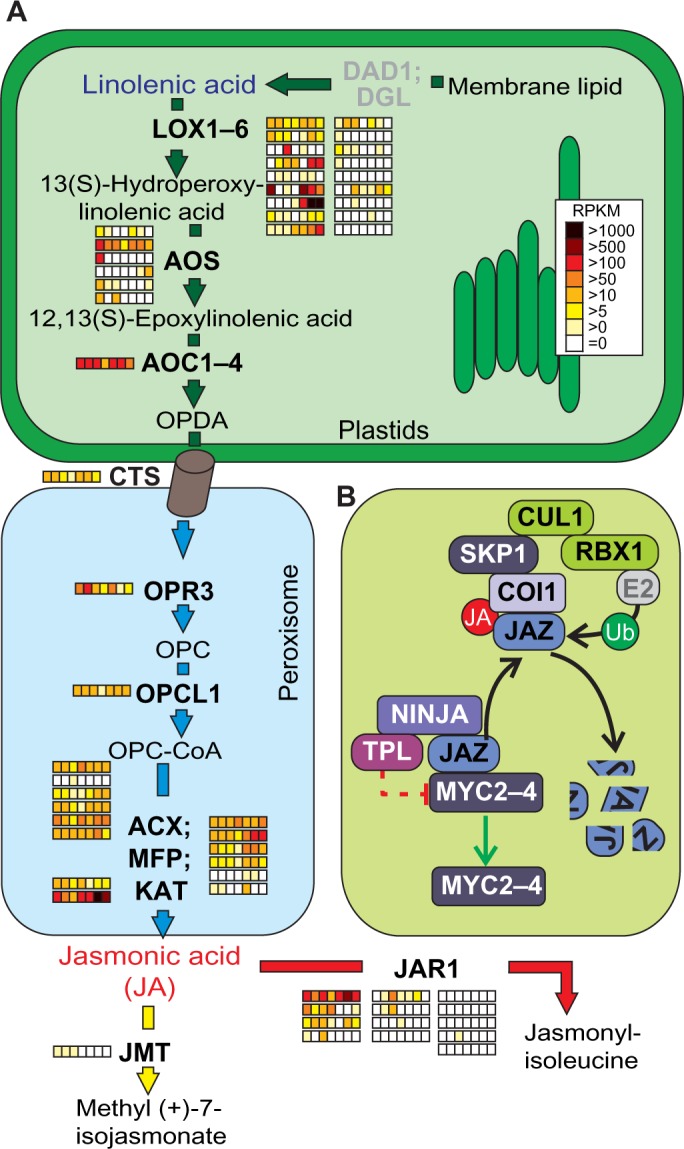
Pathways of jasmonic acid synthesis and signaling.
Notes: (A) Enzymes and intermediate products of the plastidial and peroxisomal pathways for jasmonic acid production starting from linolenic acid as precursor and ending with delivering of jasmonic acid from the peroxisome into the cytosol. The last steps catalyzed by JMT and JAR1 mark the conversion of jasmonic acid to its methyl ester as volatile transport form and jasmonyl-isoleucine as biological active form, respectively. For further details, see text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). Expression of the identified co-orthologues in tomato is shown as explained in Figure 2. Genes coding for enzyme activities not expressed by any orthologue are indicated in gray. (B) The components involved in jasmonic acid signaling are represented as interaction scheme. The bar-headed dashed line in red indicates indirect suppression mechanisms.
Abbreviations: Proteins: DAD, defective anther dehiscence; DGL, DONGLE; LOX, lipoxygenase 2; AOS, allene oxide synthase; AOC, allene oxide cyclase; CTS, COMATOSE; OPR, oxophytodienoate-reductase; OPCL, OPC-8:0 CoA ligase; ACX, acyl-CoA oxidase; MFP2, multifunctional protein; AIM, abnormal inflorescence meristem; KAT, 3-keto-acyl-CoA thiolase; JMT, jasmonic acid carboxyl methyltransferase; JAR, jasmonate resistant; ST2a, sulfotransferase 2A; JAZ, jasmonate-ZIM-domain protein; COI, coronatine-insensitive; NINJA, novel interactor of JAZ; TPL, TOPLESS; SKP1, S-phase kinase-associated protein 1; CUL, Cullin; RBX, RING-box. Metabolites: OPC, 3-oxo-2-(20(Z)-pentenyl)-cyclopentane; OPC-CoA, 3-oxo-2-(20(Z)-pentenyl)-cyclopentane-coenzyme A.
α-linolenic acid (18:3) acts as a fatty acid substrate for synthesis of 13(S)-hydroperoxylinolenic acid by LOXs.170 Except in C. reinhardtii, all plant species contained more than six co-orthologues, with a maximum of 43 co-orthologues in G. max. In total, 18 co-orthologues were identified in tomato; however, a chloroplast targeting signal was only predicted for three of them (Solyc05g014790, Solyc03g122340, and Solyc01g006560), which most likely indicated their involvement in jasmonate synthesis (Fig. 7A; Supplementary Table 12). At least one of them was expressed in each tissue and Solyc01 g006560 was highly expressed in leaves. Allene oxide synthase (AOS; six co-orthologues in tomato)171 catalyzes the dehydration of 13(S)-hydroperoxylinolenic acid to 12,13(S)-epoxylinolenic acid, which is converted to cis(+)-12-oxophytodienoic acid (OPDA) likely by allene oxide cyclase (AOC). Interestingly, we detected only two co-orthologues of AOS containing a chloroplast targeting signal (Solyc04g079730 and Solyc11g069800) in tomato. At least one of them was expressed in all tissues, similar to the unique tomato AOC orthologue (Fig. 7A). In A. thaliana, transcription of AOC is induced within two hours after wounding and occurs in anthers and pollen grains,172 while we observed moderate expression of the tomato orthologue in all tissues (Supplementary Table 19173,174).
OPDA is transported from plastids to peroxisomes by the action of CTS,175 and the expression of the tomato orthologue was at least low or moderate in all tissues (Fig. 7A; Supplementary Table 19). The OPDA reductase (OPR3) catalyzes the reduction of OPDA to 3-oxo-2-(2′(Z)-pentenyl)-cyclopentane-1-octanoic acid (OPC176) and the orthologue in tomato showed high variance in expression from low to high levels in the tissues analyzed (Fig. 7A). OPDA-CoA intermediate is formed in peroxisomes by 4-coumarate:CoA ligase like (4-CL-like) enzymes such as OPCL1, whose orthologue in tomato is globally expressed (Fig. 7A).177 The subsequent β-oxidation to synthesize JA occurs in three cycles178 by the action of an acyl-CoA oxidase (ACX1), a multifunctional protein (MFP), and an L-3-ketoacyl CoA thiolase (KAT).60 While two peroxisome targeted co-orthologues of ACX1 and six of MFP were identified in tomato, we could not predict a peroxisomal targeting signal in one of the two co-orthologues of KAT (Fig. 7A; Supplementary Table 12). Interestingly, MFP2 and AIM1 were located in one orthologous group in A. thaliana while six co-orthologues were found in tomato of which the peroxisomal-located enzymes Solyc12g007170, Solyc12g099440, Solyc07g019670, and Solyc01g066620 were at least low expressed in all tissues except Solyc01g066620 (Supplementary Table 19).
JA is subsequently secreted into the cytoplasm, where it can be further utilized either by S-adenosyl-l-methionine:jasmonic acid carboxyl methyltransferase (JMT) to form the volatile JA methyl ester (methyl (+)-7-isojasmonate) or conjugated to isoleucine by the activity of JAR1, which converts JA into the biologically active jasmonyl-isoleucine.4,179,180 JMT was only present in eudicots and represented by a single orthologue in tomato, which exhibited a low expression in root, stem, and leaf. In tomato, (+)-7-iso-JA-Ile is exclusively synthesized upon wounding, and in SlJAR1-RNAi lines, JA-Ile is downregulated by 50%–75%, confirming the importance of JAR1 for the conversion of JA to JA-Ile.181 Remarkably, we observed that the CLOG of JAR1 contained 13 tomato co-orthologues, and one of them (Solyc01g095580) was highly expressed in all tissues except of flowers and stems. Interestingly, the co-orthologue Solyc10g011660showed a high expression in flowers and stems, and by this, we could compensate Solyc01g095580 in both tissues (Fig. 7A; Supplementary Tables 5, 12, and 19). Thus, it needs to be experimentally proven whether Solyc01g095580 is the essential JAR1 orthologue in jasmonate signaling and whether rapid induction of the gene is caused by stress induction during the harvesting procedure.
Jasmonate response co-orthologues show differences between tomato and Arabidopsis
The main components of JA response are transcription factors of the JA-ZIM-domain (JAZ) repressor family, calcium-related signaling molecules, and JA-related transcription activators. Ca2+-dependent phosphorylation and MAPK cascades are also involved in the regulation of JA biosynthesis.61 Among the targets of JA signaling are JA biosynthesis genes that form a positive feedback system.182
We observed that the regulation of transcriptional changes triggered by JA synthesis was conserved in eudicots and monocots (Fig. 7B; Supplementary Tables 5 and 12).183–185 The primary regulators are MYC transcription factors and JAZ repressors are associated in corepressor complexes with NINJA and TPL (Topless; Fig. 7B). None of the eight JAZ repressors in A. thaliana were represented in green algae and moss, and JAZ5, 6, and 9 occurred only in eudicots or were found specifically in A. thaliana (Supplementary Table 5). Degradation of JAZ repressors is mediated by binding of JA-Ile to the F-box protein coronatine-insensitive 1 (COI1) bound to the SCFCol1 E3 ubiquitin ligase complex (Fig. 7B), which in turn allows the expression of early JA response genes.186–188 JAZ proteins also bind to MYB21 and MYB24, two transcription factors encoded by genes strongly induced by JA in flower tissues that control stamen development and pollen maturation in A. thaliana.189,190 For tomato, only three JAZ co-orthologues were identified in two distinct CLOGs, showing the same domain architecture containing a TIFY (transcription factor domain) and a CCT_2 (short plant protein motif) domain (Supplementary Table 12). For two co-orthologues (Solyc07g042170 and Solyc03g118540), high expression levels were found in roots and Solyc07g042170 was at least moderately expressed in all other tissues (Supplementary Table 19).
Impairment of JA signaling leads to severe phenotypes like male sterility in A. thaliana, while in the tomato mutant, jai1-1 female sterility was observed.191 In line with the essential role of JAI1, the tomato orthologue was expressed in all tissues. Interestingly, expression of the COI1 orthologue was not observed in tomato flowers (Supplementary Table 19),165 although it is interacting with JAI1.
Low-expressed enzymes in tomato influence GA bio-synthetic pathway
GAs derive from the diterpene geranylgeranyl diphosphate (GGDP) synthesized in plastids from the isoprenoid precursor isopentenyl pyrophosphate, which is provided mainly by the plastid-resident MEP pathway, but also by the cytosolic MVA pathway (Fig. 8A).192 Our analysis showed that both pathways were conserved in mosses, monocots, and eudicots, but the MVA pathway was missing in the green algae C. reinhardtii.193,194 Remarkably, the co-orthologues from the MEP and MVA pathway showed moderate or high expression in all tomato tissues, whereas the expression of co-orthologues catalyzing the subsequent steps of GA biosynthesis was low in tomato or even not present in some tissues (Fig. 8A, Supplementary Table 20). The chloroplast-localized ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS), which are involved in formation of ent-kaurene, were detected in all plants except in the green algae (Fig. 8A, Supplementary Table 6). Interestingly, expression of tomato CPS and KS co-orthologues was detected if at all at low levels only (Fig. 8A, Supplementary Table 20). GA1 (CPS) and GA2 (KS) in A. thaliana were discriminated on the basis of their domain architecture, which could be confirmed by our orthologue search due to the fact that they fall into two different CLOGs. Inspection of the domain architecture of co-orthologues within the two CLOGs indicated that orthologue search and functional domain architecture might differ, but it could also exemplify an interesting case of domain stealing.100 For instance, some of the monocot co-orthologues in the CPS group contain a Prenyltrans_1 domain. This domain is absent in the CPS of A. thaliana, but present in the KS orthologue (Supplementary Table 13).
Figure 8.
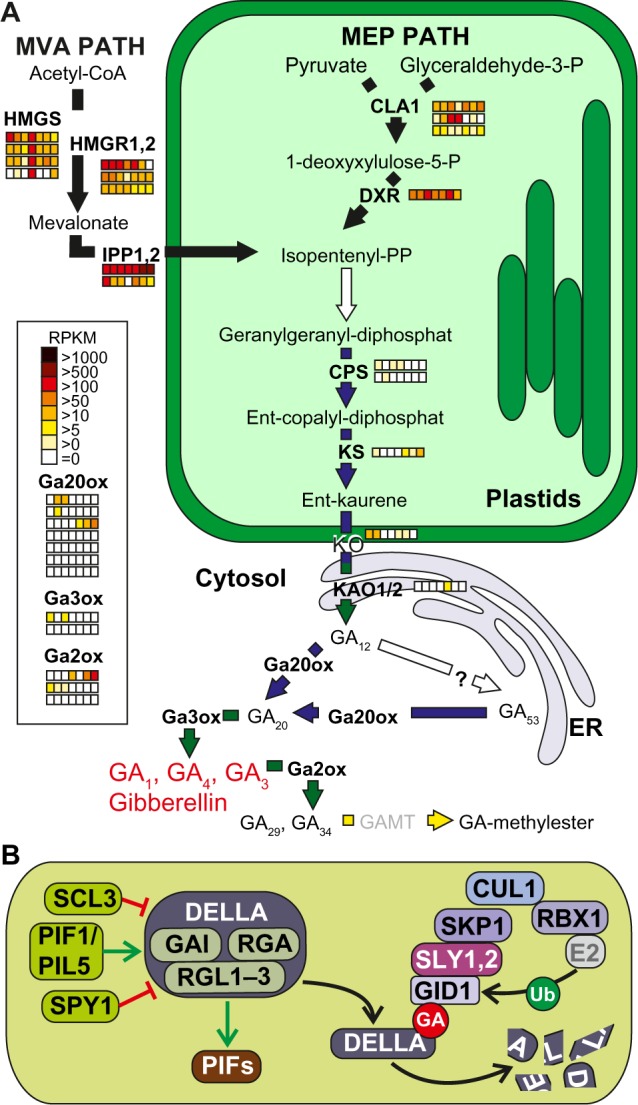
Pathways of gibberellin synthesis and signaling. (A) Enzymes and intermediate products of the plastidial and cytosolic pathways for isopentenyl pyrophosphate production unify to the plastidal production of ent-kauren as precursor for gibberellin synthesis (GA1, GA4, and GA3) in the cytosol. GA2ox and GAMT mark the biological inactivation of gibberellin. For further details, see text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). Expression of the identified genes in tomato is shown as explained in Figure 2. (B) The components involved in gibberellin signaling are presented as interaction scheme. Enzyme activities not expressed by any orthologue gene in tomato are indicated in gray.
Abbreviations: Proteins: CLA, cloroplastos alterados; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; IPP, isopentenyl diphosphate isomerase; CPS, ent-copalyl diphosphate synthetase, KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO1, ent-kaurenoic acid oxydase; GA20ox, gibberellin 20-oxidase; GA3ox1, gibberellin 3-oxidase; GA2ox, gibberellin 2-oxidase; GAMT, gibberellin methyltransferase; GID, GA insensitive dwarf; GAI, gibberellic acid insensitive; RGA, repressor of GA1–3; RGL, RGA-like; SLY, SLEEPY; PIF, PHY-interacting factor; SPY, SPINDLY; SCL, scarecrow-like.
Conversion of ent-kaurene to GA12 requires the transport of ent-kaurene into the cytoplasm and several oxidation steps. The first step is catalyzed by the ent-kaurene oxidase (KO), localized in the outer envelope of plastids, which is involved in the simultaneous transfer of ent-kaurenoic acid to the ER.195 Subsequent oxidization requires the ER-associated ent-kaurenoic acid oxidase (KAO) for conversion to GA12. However, the nature of the enzyme activity involved in the formation of GA53 by hydroxylation on C-13 is still not clear. The subsequent hydroxylation on C-20 of GA12 or GA53 is catalyzed by cytoplasmic GA20 oxoglutarate-dependent dioxygenases (GA20ox). The activity of other oxoglutarate-dependent dioxygenases like the GA3ox further leads to the production of bioactive GA compounds such as GA1, GA4, or GA3. Deactivation of GAs by C-2 hydroxylation is catalyzed by GA2 oxidases (GA2ox). As in the case of KS, KAO1 and KAO2, co-orthologues of GA3ox and GA2ox were not found in P. patens and C. reinhardtii (Supplementary Table 6), while their expression was low in most of the analyzed tissues (Supplementary Table 20).
Oxidization and conversion of GAs into their epoxides by cytochrome P450 monooxygenases as well as formation of GA methyl esters by the activity of GA methyl transferases (GAMTs) are discussed as other mechanisms of GA deactivation.66 However, these mechanisms appear to be species-specific as co-orthologues to the A. thaliana GAMT were not identified by our approach in any of the selected species except for V. vinifera (Supplementary Table 6).
GA perception and signaling is absent in green algae and mosses
Current models for GA perception and signaling consider three key components.196 The first is the soluble, nuclear-localized GA receptor GID1,197 which we identified in monocots and eudicots (Supplementary Table 6). The second are the growth repressing DELLA proteins GAI, RGA, and RGL1, 2, and 3 identified in A. thaliana198 as well as SLR1 found in rice,199 which form a subfamily of the plant-specific GRAS transcription regulators. All of these proteins were grouped in one CLOG also containing co-orthologues from the moss in addition to monocots and eudicots. However, in contrast to A. thaliana that has five GRAS enzymes, many eudicots and monocots contained only one orthologue. The third are the F-box proteins SLY1 and SLY2,200 which are called GID2 in rice (Fig. 8B201). No co-orthologues of these factors were detected in the moss and the green algae. By this, the GA signaling pathway seems to have emerged after mosses. However, co-orthologues of SLY were not found in V. vinifera (Supplementary Table 6). Remarkably, SLY2 co-orthologues were not expressed in the analyzed tomato tissues under normal conditions (Supplementary Table 20).
In the absence of GA, DELLA proteins act as central repressors of GA response by suppressing the activity of PIF transcription factors (PIF3, 4), which are involved in the regulation of growth-promoting GA-responsive genes.202,203 Binding of GA to the receptor GID1 triggers its interaction with DELLA proteins and causes their release from repression complexes.204 In turn, simultaneous binding of SLY1/GID2 recruits DELLA proteins for ubiquitination by the SLY1/GID2-dependent E3-ligase complex and subsequent degradation by the proteasome.204 Furthermore, the level of DELLA proteins is also controlled by the activity of the phytochrome-interacting factor PIF1/PIL5, and the regulation of DELLA activity is done by proteins SCL3 and SPY1.67,205 In general, PIFs have been evolved specifically in different species, as PIF6 was present only in A. thaliana and we observed PIF1 co-orthologues only in some eudicots (S. tuberosum, S. lycopersicum, M. truncatula, and G. max; Supplementary Tables 6 and 13). Further, our analysis for expression in tomato revealed that most of the enzymes were expressed at low levels under normal conditions (Supplementary Table 20).
BR synthesis is conserved in Viridiplantae until synthesis of campesterol
As for all other phytosteriods, the biosynthesis of BR derives from isopentenyl pyrophosphate (isopentenyl PP; Fig. 9A), which is conjugated to the triterpene squalene (Fig. 9A).16,206,207 By the action of CAS1, SMT1, CYP51, FACKEL, HYD1, DWF7, DWF5, and DWF1, campesterol is synthesized,73 which is the direct C28 precursor of the BR specific pathway. In line with the fact that campesterol is not exclusively a precursor for BR, the pathway was conserved in all analyzed plants with the exception of FACKEL and DWF5 (Fig. 9A, Supplementary Tables 7 and 14). The enzyme FACKEL was only present in Z. mays, S. tuberosum, and A. thaliana, whereas DWF5 co-orthologues were only missing in C. reinhardtii. In tomato, CAS1 co-orthologues showed a moderate expression at maximum and could potentially limit the rate of campesterol synthesis. In contrast, SMT1, CYP51, and DWF5 showed the tendency to be highly expressed in most of the tissues (Supplementary Table 21). Furthermore, HYD1 orthologue was not expressed in flower tissue.
Figure 9.
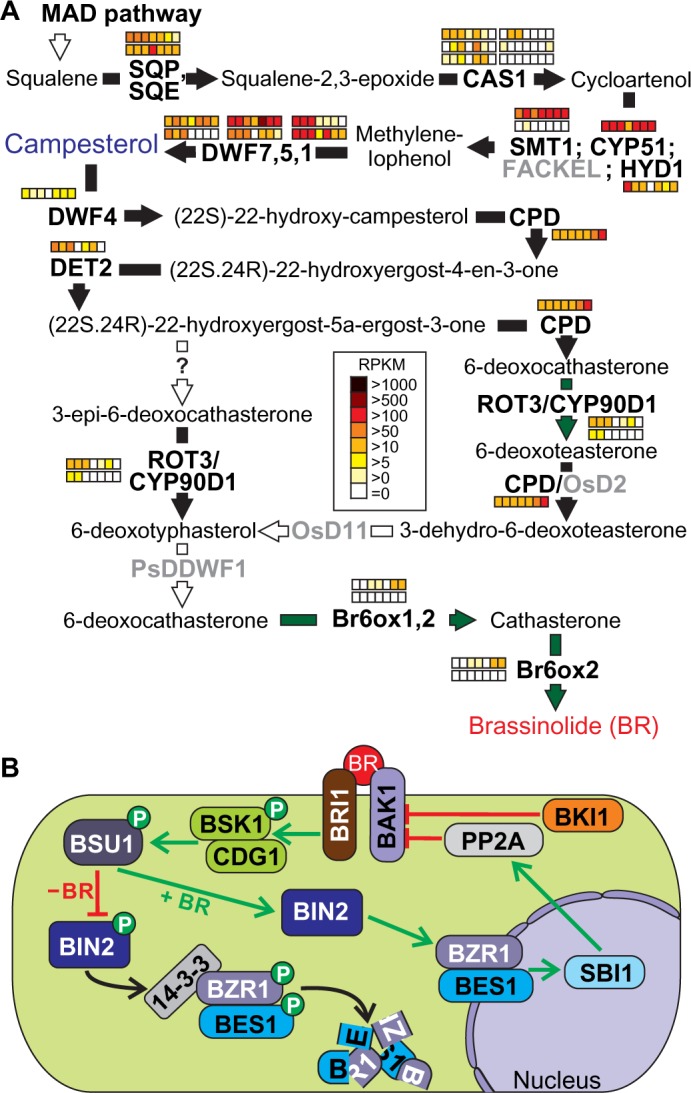
Pathways of brassinosteroide synthesis and signaling.
Notes: (A) Brassinosteroide synthesis uses the precursor compound campesterol, which is derived from the MAD pathway. Two major routes are shown leading to the formation of the bioactive brassinosteroids castasterone and brassinolide. For further details, see text. The arrows are colored according to the species in which the enzymes were found (Fig. 1A). Expression of the identified genes in tomato is shown as explained in Figure 2. (B) The components involved in brassinosteroid signaling are presented as interaction scheme. Enzyme activities not expressed by any orthologue gene in Arabidopsis or tomato are indicated in gray. D2 and D11 were only detected in O. sativa and DDWF1 was identified in Pisum sativum.
Abbreviations: Proteins: SQP, squalene monooxygenase; SQE, squalene epoxidase; CAS, cycloartenol synthase; SMT, sterol methyltransferase; CYP, cytochrome P450; HYD, Hydra; DWF, dwarf; CPD, constitutive photomorphogenic dwarf; DET2, de-etiolated; ROT, rotundifolia; Br6ox, brassinosteroid-6-oxidase; BRI1, brassinosteroid-insensitive; BAK1, BRI1-associated receptor kinase; BKI, BRI1 kinase inhibitor; BSK, brassinosteroid-signaling kinase; CDG, constitutive differential growth; CDL, CDG1-LIKE; BSU, BRI1 suppressor; BIN, brassinosteroid-insensitive; BZR, brassinazole-resistant; BES, BRI1-EMS-suppressor 1; SBI, suppressor of BRI; PP2A, serine/threonine protein phosphatase 2A.
The subsequent processing of BR synthesis starting from campesterol as a basic precursor is not yet fully described and the exact order of enzymatic reactions in this part of the BR pathway is still under debate. For our analysis, we depicted the two pathways described by Zhao and Li,76 which are branching during the action of DET2, DWF4 (annotated as CYP724B2 and CYP90B3 in tomato, respectively), and CPD (annotated as CYP90A1 in A. thaliana). Remarkably, only DET2 co-orthologues were found in the green algae and the moss included in our analysis (Supplementary Table 14). For eudicots, the reaction catalyzed by the P450 monooxygenase DWF4 has been proposed as a rate-limiting step in BR synthesis,208 and transcriptome profiling of tomato co-orthologues revealed only low expression in all tissues for DWF4. In contrast, CPD showed moderate or high expression in all tissues (Supplementary Table 21). For co-orthologues in both routes, mostly no or only low expression was observed in flower tissue. The two routes lead to the production of 6-deoxotyphasterol and the only two additional enzymes identified in A. thaliana are ROT3 (CYP90C1) and CYP90D1, but it is unknown yet whether both enzymes indeed participate in both pathways.
The conversion of 6-deoxotyphasterol to 6-deoxocathasterone is catalyzed by DDWF1, which is only described in pea,209 while the corresponding gene in A. thaliana has not been identified yet. Finally, oxidation on the C-6 by BR6ox leads to CS, the immediate precursor of BL. Br6ox (CYP85A1 and CYP85A2 in A. thaliana) has been originally identified as an extreme dwarf (dx) allele of the tomato DWARF locus (D). CYP85A2 (Br6ox2), but not CYP85A1 (Br6ox1), drives the conversion of CS to BL. Expression analysis of tomato co-orthologues in different tissues showed that only one co-orthologue was expressed in both cases. For the co-orthologues of Br6ox, only moderate expression was detected in fruit ripening stages, whereas for ROT3/CYP90D1, moderate expression was observed in root, stem, and leaf (Supplementary Table 21).
Alternative modes of BR signaling in monocots and moss
Because long-distance transport is not shown for BR or derivatives so far, it is widely assumed that they act in close vicinity to the places of synthesis.210 Binding of BRs to the extracellular leucine-rich repeat domain of the serine/threonine receptor kinase BRI1 initiates the release of the inhibitor protein BKI1 and formation of heterodimers with BAK1 (Fig. 9B211,212). While we detected co-orthologues of BAK1 and BRI1 in all plants except the green algae C. reinhardtii, the inhibitor BKI1 was identified only in eudicots, which suggests an alternative mode of regulation in monocots (Supplementary Table 7). Interestingly, the CLOG of BAK1 contained seven to 30 co-orthologues per species in eudicots and monocots. However, the identified co-orthologues showed a large heterogeneity in their domain architecture (Supplementary Table 14).
Autophosphorylation and sequential transphosphorylation of the cytoplasmic kinase domains fully activate the receptor complex and triggers the downstream signal transduction by activating BSK1, which in turn phosphorylates BSU1. Interestingly, BSK1 was low expressed during fruit ripening and in the mature fruit (Supplementary Table 21). BSU1 acts as a protein phosphatase and inactivates the serine/threonine kinase BIN2 by dephosphorylation. In the absence of BR, BIN2 is constitutively active and controls the phosphorylation of BZR1 and BES1/BZR2, two transcription factors involved in the regulation of BR-responsive genes. We observed that this regulatory network was mainly conserved in all plants except for green algae. Both transcription factors are kept inactive by either rapid proteasomal degradation or cytoplasmic retention through interaction with 14-3-3.74 Only BZR1 and BES1/BZR2 were not identified in P. patens, which suggests that species-specific transcription factors might exist in moss (Supplementary Table 7).
Besides controlling the expression of growth-promoting genes, BZR1 and BZR2 also feedback the expression of upstream signaling components and genes involved in BR biosynthesis. Among these, SBI1 mediates the methylation of PP2A, which in turn is translocated to the plasma membrane and promotes the inactivation of the internalized BRI1 receptor by dephosphorylation (Fig. 9B). In contrast to PP2A, SBI1 was not identified in the eudicot S. tuberosum, which could be related to the incomplete genome draft or an analogous redundant function.
Conclusion
We applied an integrated bioinformatics approach combining orthologue search, domain analysis, prediction of protein localization based on signal sequence detection, and expression analysis to transfer the accumulated knowledge on hormone pathways from the model plant A. thaliana to other plant systems. In general, the orthologue search revealed that most of the analyzed pathways, especially the signaling pathways, were only partially covered by co-orthologues from the green algae C. reinhardtii. Further, the biosynthetic pathways in the moss P. patens were more complete than in the green algae compared to the model plant A. thaliana. However, for example, the biosynthetic pathway of GA seemed to be conserved in P. patens only within the first part (until production of GGDP), while downstream, some acting enzymes such as KS, KAO1, GAMT1, 2, GA3ox1, and GA2ox1 could not be detected. Interestingly, concerning the signaling of GA, we observed enzymes of the DELLA complex as well as proteins activated by DELLA (PIF3) and proteins regulating DELLA (SCL3 and SPY). Thus, alterations of the pathways in moss could be expected as well. Remarkably, the analysis of the conservation of phytohormone pathways in 13 different species (Fig. 1) leads to the proposal of evolutionary routes and distinctions of pathways between, eg, eudicots and monocots or between multicellular and unicellular photosynthetic eukaryotes. For example, GH3 enzymes and PIN transporter involved in auxin pathways were not present in C. reinhardtii (Figs. 1 and 3), as these enzymes mark features of multicellular systems. In turn, TAA1/TAR1 proteins involved in auxin synthesis were only present in eudicots and thus have evolved very late in plant diversification. A similar situation was found for BKI1, involved in BR signaling (Fig. 9). This suggests that eudicots have an increased complexity of signal perception. Moreover, the complexity of central components in jasmonic acid (JAZ5, JAZ5, and JAZ9; Fig. 7) or GA signaling (PIF1; Fig. 8) appears to be larger in eudicots than in monocots. In contrast, we could show that the two auxin transporters PIN5 and PIN8 (Fig. 3), IPTs involved in cytokinin biosynthesis (Fig. 5), and DAD1 and DGL1 involved in JA biosynthesis (Fig. 7) are proteins that can likely be traced back to the common ancestor of monocots and eudicots. Further, the existence of Brassicaceae-specific routes, like the IAOX pathway for auxin, could be confirmed based on the results presented here.
Remarkably, the application of the combination of orthologue search and functional domain prediction of the CLOGs led to the identification of putative domain-stealing events during the evolution of the GA synthesis pathway (Fig. 8). We observed a domain exchange in some species between the two subsequently acting enzymes CPS and KS involved in GA biosynthesis, which suggests that these enzymes operate in a larger complex. Both CLOGs (CPS and KS) contained proteins with at least one terpene synthase domain, but the CPS orthologue in A. thaliana is missing the functional domain to transfer allylic prenyl groups, which is present in some CPS co-orthologues of monocots (S. bicolor, Z. mays), whereas the KS orthologue of A. thaliana contains the prenyltransferase domain.
The localization prediction in combination with the orthologue search had the advantage to allow the dissection of large CLOGs and categorized these enzymes according to their localization to be putatively involved in hormone synthesis. For example, we detected 18 and 6 co-orthologues of the LOX and AOS gene families, respectively, involved in JA synthesis. Prediction of the protein localization limited the number of enzymes with likely equivalent function in JA synthesis to three LOX and two AOS co-orthologues (Fig. 7). Therefore, this approach provides an advantage for the assignment of protein members in equivalent pathways. Additionally, inspection of the expression profile provides information on putative active pathways in the case that multiple pathways exist. For example, three alternative routes for conversion of tryptophan to auxin exist, but only genes coding for enzymes of two routes were expressed in the analyzed tissues of tomato (Fig. 2).
Expression analysis of the orthologous enzymes involved in phytohormone biosynthesis, transport, and signaling in different tissues of tomato gave insights in the expression patterns of orthologue groups containing more than one enzyme of the same species and allowed conclusions on functions depending on the developmental stage or tissue. Interestingly, the co-orthologues of DGL and DAD1 in the JA biosynthesis pathway (Fig. 7) as well as the co-orthologues of BG2 in the ABA biosynthetic pathway (Fig. 6) were not expressed in tomato. Moreover, the expression at specific developmental stages could help to assign the function to specific co-orthologues. For example, Br6ox involved in BR synthesis had a high expression during fruit ripening, while ROT3 showed the highest expression in root, stem, and leaf tissues (Fig. 9). Further, the ABA receptor GTG2 was only expressed in flower and fruit tissues of tomato (Fig. 6), and a similar observation was presented for ACS and ACO involved in ethylene synthesis (Fig. 4), which was in line with the importance of the two phytohormones ABA and ethylene during reproduction and fruit ripening.
In general, we can conclude that the information derived from the model plant A. thaliana can largely be transferred to eudicots, while some distinctions, particularly in the signaling mechanisms, became obvious in comparison between monocots and A. thaliana. In turn, the pathways in the moss P. patens and green algae C. reinhardtii are in parts distinct or even missing and the transfer of information from A. thaliana to these species is mostly difficult. Our results provide the platform for future work on experimental confirmation of the phytohormone pathways in a broader variety of plant species and for the identification of species-specific components.
Supplementary Material
Supplementary Tables 1–7. Survey of genes involved in phytohormone biosynthesis, transport, and signaling in A. thaliana and their co-orthologues in selected Viridiplantae. Analyzed phytohormones are auxin (S1), ethylene (S2), cytokinin (S3), abscisic acid (S4), jasmonic acid (S5), gibberellin (S6), and brassinosteroid (S7). The corresponding AGI number, gene, and mutant names, description in database, number of species containing co-orthologues, number of co-orthologues in the indicated plant species and the overall distribution in the groups of eudicots, monocots, mosses, and algae are indicated. The selected plant species are: Atha, A. thaliana; Bdis, B. distachyon; Crei, C. reinhardtii; Gmax, G. max; Mtru, M. truncatula; Osat, O. sativa; Ppat, P. patens; Ptri, P. trichocarpa; Sbic, S. bicolor; Slyc, S. lycopersicum; Stub, S. tuberosum; Vvin, V. vinifera; Zmay, Z. mays.
Supplementary Tables 8–14. Survey of co-orthologues involved in phytohormone biosynthesis, transport, and signaling in the analyzed plant genomes with prediction of intracellular localization and domain structure. Analyzed phytohormones are auxin (S8), ethylene (S9), cytokinin (S10), abscisic acid (S11), jasmonic acid (S12), gibberellin (S13), and brassinosteroid (S14). The selected plant species are: Atha, A. thaliana; Bdis, B. distachyon; Crei, C. reinhardtii; Gmax, G. max; Mtru, M. truncatula; Osat, O. sativa; Ppat, P. patens; Ptri, P. trichocarpa; Sbic, S. bicolor; Slyc, S. lycopersicum; Stub, S. tuberosum; Vvin, V. vinifera; Zmay, Z. mays.
Supplementary Tables 15–21. Expression of tomato co-orthologues involved in phytohormone biosynthesis, transport, and signaling in different tissues: root, stem, leaf, flower, and fruits at the three developmental stages, namely, mature green (fruit_mature), breaker stage (berry_breaker), and 10 days after turning red (berry_10 days). Analyzed phytohormones are auxin (S15), ethylene (S16), cytokinin (S17), abscisic acid (S18), jasmonic acid (S19), gibberellin (S20), and brassinosteroid (S21).
Acknowledgments
We would like to thank our colleagues Dr. S. Fragkostefanakis, Dr. R. Pernil, and U. Bodensohn for critical reading and helpful discussions during preparation and revision of the manuscript.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Associate Editor
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2730 words, excluding any confidential comments to the academic editor.
FUNDING: We acknowledge funding to ES by the Deutsche Forschungsgemeinschaft (SFB902) and by the Cluster of Excellence Macromolecular Complexes (EXC) as well as to ES, K-DS, NF, and MLC by the ERC (SPOT-ITN, FP7, Marie-Curie-training program). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the bioinformatics approaches: ES, SS. Searched the databases and analyzed the data: SS. Contributed to writing of the manuscript: SS, KDS, NF, SJ, MLC, ES. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Gray WM. Hormonal regulation of plant growth and development. PLoS Biol. 2004;2(9):1270–3. doi: 10.1371/journal.pbio.0020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McAtee P, Karim S, Schaffer R, David K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci. 2013;4(79):1–7. doi: 10.3389/fpls.2013.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamm P, Kumar PP. The phytohormone signal network regulating elongation growth during shade avoidance. J Exp Bot. 2010;61(11):2889–903. doi: 10.1093/jxb/erq147. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca S, Rosado A, Vaughan-Hirsch J, Bishopp A, Chini A. Molecular locks and keys: the role of small molecules in phytohormone research. Front Plant Sci. 2014;5(709):1–16. doi: 10.3389/fpls.2014.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YH, Irving HR. Developing a model of plant hormone interactions. Plant Signal Behav. 2011;6(4):494–500. doi: 10.4161/psb.6.4.14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells DM, Laplaze L, Bennett MJ, Vernoux T. Biosensors for phytohormone quantification: challenges, solutions, and opportunities. Trends Plant Sci. 2013;18(5):244–9. doi: 10.1016/j.tplants.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Borghi L, Kang J, Ko D, Lee Y, Martinoia E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem Soc Trans. 2015;43:924–30. doi: 10.1042/BST20150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010;2(6):1–16. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol. 2010;17(6):642–5. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparks E, Wachsman G, Benfey PN. Spatiotemporal signalling in plant development. Nat Rev Genet. 2013;14(9):631–44. doi: 10.1038/nrg3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohli A, Sreenivasulu N, Lakshmanan P, Kumar PP. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013;32(7):945–57. doi: 10.1007/s00299-013-1461-y. [DOI] [PubMed] [Google Scholar]
- 12.Darwin CD, Darwin F. In: The Power of Movement in Plants. Murray John., editor. William Clowes and Sons; London: 1880. [Google Scholar]
- 13.Ahammed GJ, Xia XJ, Li X, Shi K, Yu JQ, Zhou YH. Role of brassinosteroid in plant adaptation to abiotic stresses and its interplay with other hormones. Curr Protein Pept Sci. 2015;16(5):462–73. doi: 10.2174/1389203716666150330141427. [DOI] [PubMed] [Google Scholar]
- 14.Wani SH, Kumar V, Shriram V, Sah SK. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016;4(3):162–76. [Google Scholar]
- 15.Acosta IF, Farmer EE. Jasmonates. Arabidopsis Book. 2010;8:e0129. doi: 10.1199/tab.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clouse SD. Brassinosteroids. Arabidopsis Book. 2011;9:e0151. doi: 10.1199/tab.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book. 2014;12:e0168. doi: 10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaller GE, Kieber JJ. Ethylene. Arabidopsis Book. 2002;1:e0071. doi: 10.1199/tab.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun TP. Gibberellin metabolism, perception and signaling pathways. Arabidopsis Arabidopsis Book. 2008;6:e0103. doi: 10.1199/tab.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y. Auxin biosynthesis. Arabidopsis Book. 2014;12:e0173. doi: 10.1199/tab.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7(8):1267–87. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SJ, Jiang K, Tal L, et al. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet. 2014;46(12):1337–42. doi: 10.1038/ng.3131. [DOI] [PubMed] [Google Scholar]
- 24.Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Z, Liu X, Peng Z, et al. AHD2.0: an update version of Arabidopsis hormone database for plant systematic studies. Nucleic Acids Res. 2011;39(Database issue):D1123–9. doi: 10.1093/nar/gkq1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64(9):2541–55. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig-Muller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot. 2011;62(6):1757–73. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- 28.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63(8):2853–72. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- 29.Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7(11):847–59. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 30.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95(5):707–35. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Went FW, Thimann KV. Phytohormones. The Macmillan Company; New York, NY: 1937. [Google Scholar]
- 32.Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol Plant. 2012;5(4):772–86. doi: 10.1093/mp/ssr109. [DOI] [PubMed] [Google Scholar]
- 33.Neljubow D. Über die horizontale Nutation der Stengel von Pisum sativum und einiger anderer Pflanzen. Beih bot Zbl. 1901;10:128–39. [Google Scholar]
- 34.Gane R. Production of ethylene by some ripening fruits. Nature. 1934;134:1008–8. [Google Scholar]
- 35.Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Binder BM. The ethylene receptors: complex perception for a simple gas. Plant Sci. 2008;175(1–2):8–17. [Google Scholar]
- 37.Iqbal N, Trivellini A, Masood A, Ferrante A, Khan NA. Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol Biochem. 2013;73:128–38. doi: 10.1016/j.plaphy.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Schaller GE. Ethylene and the regulation of plant development. BMC Biol. 2012;10(9):1–3. doi: 10.1186/1741-7007-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16(5):554–60. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Stepanova AN, Alonso JM. Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol. 2009;12(5):548–55. doi: 10.1016/j.pbi.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Miller CO, Skoog F, Vonsaltza MH, Strong FM. Kinetin, a cell division factor from deoxyribonucleic acid. J Am Chem Soc. 1955;77(5):1392. [Google Scholar]
- 42.Letham DS. Regulators of cell-division in plant-tissues.15. Cytokinins from Zea-Mays. Phytochemistry. 1973;12(10):2445–55. [Google Scholar]
- 43.Parker CW, Letham DS. Regulators of cell-division in plant-tissues.16. Metabolism of zeatin by radish cotyledons and hypocotyls. Planta. 1973;114(3):199–218. doi: 10.1007/BF00389036. [DOI] [PubMed] [Google Scholar]
- 44.Hwang I, Sheen J, Muller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–80. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 45.Frebort I, Kowalska M, Hluska T, Frebortova J, Galuszka P. Evolution of cytokinin biosynthesis and degradation. J Exp Bot. 2011;62(8):2431–52. doi: 10.1093/jxb/err004. [DOI] [PubMed] [Google Scholar]
- 46.Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 47.Schaller GE, Kieber JJ, Shiu SH. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book. 2008;6:e0112. doi: 10.1199/tab.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–79. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 49.Cline MG, Oh C. A reappraisal of the role of abscisic acid and its interaction with auxin in apical dominance. Ann Bot. 2006;98(4):891–7. doi: 10.1093/aob/mcl173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma B, Yin CC, He SJ, et al. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 2014;10(10):1–16. doi: 10.1371/journal.pgen.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sah SK, Reddy KR, Li J. Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci. 2016;7(571):1–26. doi: 10.3389/fpls.2016.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp RE, LeNoble ME. ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot. 2002;53(366):33–7. [PubMed] [Google Scholar]
- 53.Cheng ZJ, Zhao XY, Shao XX, et al. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell. 2014;26(3):1053–68. doi: 10.1105/tpc.113.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chinnusamy V, Gong Z, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol. 2008;50(10):1187–95. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demole E, Lederer E, Mercier D. Isolement et determination de la structure du jasmonate de methyle, constituant odorant caracteristique de lessence de jasmin. Helv Chim Acta. 1962;45(2):675–85. [Google Scholar]
- 56.Dathe W, Ronsch H, Preiss A, Schade W, Sembdner G, Schreiber K. Endogenous plant hormones of the broad bean, Vicia faba L (−)-jasmonic acid, a plant-growth inhibitor in pericarp. Planta. 1981;153(6):530–5. doi: 10.1007/BF00385537. [DOI] [PubMed] [Google Scholar]
- 57.Ueda J, Kato J. Isolation and Identification of a senescence-promoting substance from wormwood (Artemisia absinthium L) Plant Physiol. 1980;66(2):246–9. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci U S A. 1990;87(19):7713–6. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weidhase RA, Kramell HM, Lehmann J, Liebisch HW, Lerbs W, Parthier B. Methyljasmonate-induced changes in the polypeptide pattern of senescing barley leaf segments. Plant Sci. 1987;51(2–3):177–86. [Google Scholar]
- 60.Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot. 2007;100(4):681–97. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111(6):1021–58. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yabuta T, Sumiki Y. On the crystal of gibberellin, a substance to promote plant growth. J Agric Chem Soc Japan. 1938;1526:14. [Google Scholar]
- 63.Lo SF, Yang SY, Chen KT, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20(10):2603–18. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot. 2009;60(4):1085–92. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 65.Daviere JM, Achard P. Gibberellin signaling in plants. Development. 2013;140(6):1147–51. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 66.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 67.Schwechheimer C. Understanding gibberellic acid signaling – are we there yet? Curr Opin Plant Biol. 2008;11(1):9–15. doi: 10.1016/j.pbi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–51. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 69.Hedden P. The genes of the green revolution. Trends Genet. 2003;19(1):5–9. doi: 10.1016/s0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 70.Grove MD, Spencer GF, Rohwedder WK, et al. Brassinolide, a plant growth- promoting steroid isolated from Brassica-napus pollen. Nature. 1979;281(5728):216–7. [Google Scholar]
- 71.Kutschera U, Wang ZY. Brassinosteroid action in flowering plants: a Darwinian perspective. J Exp Bot. 2012;63(10):3511–22. doi: 10.1093/jxb/ers065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clouse SD, Sasse JM. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–51. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 73.Fujioka S, Yokota T. Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol. 2003;54:137–64. doi: 10.1146/annurev.arplant.54.031902.134921. [DOI] [PubMed] [Google Scholar]
- 74.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Ann Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 75.Ye HX, Li L, Yin YH. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol. 2011;53(6):455–68. doi: 10.1111/j.1744-7909.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhao BL, Li J. Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol. 2012;54(10):746–59. doi: 10.1111/j.1744-7909.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- 77.Fridman Y, Savaldi-Goldstein S. Brassinosteroids in growth control: how, when and where. Plant Sci. 2013;209:24–31. doi: 10.1016/j.plantsci.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 78.Gudesblat GE, Russinova E. Plants grow on brassinosteroids. Curr Opin Plant Biol. 2011;14(5):530–7. doi: 10.1016/j.pbi.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 79.Guo HQ, Li L, Aluru M, Aluru S, Yin YH. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol. 2013;16(5):545–53. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development. 2013;140(8):1615–20. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132(2):453–60. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamesch P, Berardini TZ, Li D, et al. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue):D1202–10. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duvick J, Fu A, Muppirala U, et al. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–65. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong QF, Lawrence CJ, Schlueter SD, et al. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139(2):610–8. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonnhammer ELL, Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43(D1):D234–9. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paul P, Simm S, Mirus O, Scharf KD, Fragkostefanakis S, Schleiff E. The complexity of vesicle transport factors in plants examined by orthology search. PLoS One. 2014;9(5):e97745. doi: 10.1371/journal.pone.0097745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simm S, Fragkostefanakis S, Paul P, et al. Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum 2. Bioinform Biol Insights. 2015;9:1–17. doi: 10.4137/BBI.S20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sato S, Tabata S, Hirakawa H, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–41. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(D1):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu ST, Zhu ZW, Fu LM, Niu BF, Li WZ. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finn RD, Clements J, Arndt W, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–8. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blum T, Briesemeister S, Kohlbacher O. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300(4):1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 95.Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4(6):1581–90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 96.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(D1):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics. 2006;174(3):1407–20. doi: 10.1534/genetics.106.062455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 99.Ebersberger I, Simm S, Leisegang MS, et al. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014;42(3):1509–23. doi: 10.1093/nar/gkt1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heringa J, Taylor WR. Three-dimensional domain duplication, swapping and stealing. Curr Opin Struct Biol. 1997;7(3):416–21. doi: 10.1016/s0959-440x(97)80060-7. [DOI] [PubMed] [Google Scholar]
- 101.Fernandez-Pozo N, Menda N, Edwards JD, et al. The Sol Genomics Network (SGN) – from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43(Database issue):D1036–41. doi: 10.1093/nar/gku1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bostan H, Chiusano ML. NexGenEx-Tom: a gene expression platform to investigate the functionalities of the tomato genome. BMC Plant Biol. 2015;13:15. doi: 10.1186/s12870-014-0412-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tivendale ND, Ross JJ, Cohen JD. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014;19(1):44–51. doi: 10.1016/j.tplants.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 104.Zhao YD. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant. 2012;5(2):334–8. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mashiguchi K, Tanaka K, Sakai T, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(45):18512–7. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mendel RR, Kruse T. Cell biology of molybdenum in plants and humans. Biochim Biophys Acta. 2012;1823(9):1568–79. doi: 10.1016/j.bbamcr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 107.Yamada T, Palm CJ, Brooks B, Kosuge T. Nucleotide-sequences of the Pseudomonas savastanoi indoleacetic-acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci U S A. 1985;82(19):6522–6. doi: 10.1073/pnas.82.19.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 2009;583(2):487–92. doi: 10.1016/j.febslet.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 109.Pollmann S, Neu D, Weiler EW. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry. 2003;62(3):293–300. doi: 10.1016/s0031-9422(02)00563-0. [DOI] [PubMed] [Google Scholar]
- 110.Sugawara S, Hishiyama S, Jikumaru Y, et al. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106(13):5430–5. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275(43):33712–7. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- 112.Ludwigmuller J, Epstein E. Indole-3-butyric acid in Arabidopsis thaliana 3 In-vivo biosynthesis. Plant Growth Regul. 1994;14(1):7–14. [Google Scholar]
- 113.Strader LC, Bartel B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant. 2011;4(3):477–86. doi: 10.1093/mp/ssr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Junghans U, Polle A, Duchting P, et al. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006;29(8):1519–31. doi: 10.1111/j.1365-3040.2006.01529.x. [DOI] [PubMed] [Google Scholar]
- 115.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136(6):1005–16. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 116.Peer WA. From perception to attenuation: auxin signalling and responses. Curr Opin Plant Biol. 2013;16(5):561–8. doi: 10.1016/j.pbi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 117.Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot. 2013;64(9):2557–63. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- 118.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–60. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 119.Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem. 2003;278(49):49102–12. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- 120.Lin ZF, Zhong SL, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60(12):3311–36. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 121.Liu MC, Pirrello J, Chervin C, Roustan JP, Bouzayen M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 2015;169(4):2380–90. doi: 10.1104/pp.15.01361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H. Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J. 2010;64(1):140–50. doi: 10.1111/j.1365-313X.2010.04316.x. [DOI] [PubMed] [Google Scholar]
- 123.Broekgaarden C, Caarls L, Vos IA, Pieterse CMJ, Van Wees SCM. Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 2015;169(4):2371–9. doi: 10.1104/pp.15.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 125.Kamiyoshihara Y, Tieman DM, Huber DJ, Klee HJ. Ligand-induced alterations in the phosphorylation state of ethylene receptors in tomato fruit. Plant Physiol. 2012;160(1):488–97. doi: 10.1104/pp.112.202820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang W, Hall AE, O’Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A. 2003;100(1):352–7. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xie F, Liu Q, Wen CK. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 2006;142(2):492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gao ZY, Chen YF, Randlett MD, et al. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278(36):34725–32. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- 129.Koyama T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci. 2014;5:650. doi: 10.3389/fpls.2014.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thompson JD, Higgins DG, Gibson TJ. Clustal-W – improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gruhn N, Halawa M, Snel B, Seidl MF, Heyl A. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiol. 2014;165(1):227–37. doi: 10.1104/pp.113.228080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang W, Yamane H, Takahashi N, Chapman DJ, Phinney BO. Identification of a cytokinin in the green alga Chara globularis. Phytochemistry. 1989;28(2):337–8. [Google Scholar]
- 133.Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot. 2008;59(1):75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 134.Chen CM, Kristopeit SM. Metabolism of cytokinin – dephosphorylation of cytokinin ribonucleotide by 5′-nucleotidases from wheat-germ cytosol. Plant Physiol. 1981;67(3):494–8. doi: 10.1104/pp.67.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Polya GM. Regulation of a plant 5′(3′)-ribonucleotide phosphohydrolase by cyclic nucleotides and pyrimidine, purine, and cytokinin ribosides. Proc Natl Acad Sci U S A. 1974;71(4):1299–303. doi: 10.1073/pnas.71.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matsuo S, Kikuchi K, Fukuda M, Honda I, Imanishi S. Roles and regulation of cytokinins in tomato fruit development. J Exp Bot. 2012;63(15):5569–79. doi: 10.1093/jxb/ers207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012;160(1):319–31. doi: 10.1104/pp.112.196733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–49. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 139.Werner T, Kollmer I, Bartrina I, Holst K, Schmulling T. New insights into the biology of cytokinin degradation. Plant Biol. 2006;8(3):371–81. doi: 10.1055/s-2006-923928. [DOI] [PubMed] [Google Scholar]
- 140.Lomin SN, Krivosheev DM, Steklov MY, Osolodkin DI, Romanov GA. Receptor properties and features of cytokinin signaling. Acta Naturae. 2012;4(3):31–45. [PMC free article] [PubMed] [Google Scholar]
- 141.Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot. 2011;62(14):5149–59. doi: 10.1093/jxb/err220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmulling T. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156(4):1808–18. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tran LSP, Urao T, Qin F, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(51):20623–8. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134(16):2959–68. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Deng Y, Dong H, Mu J, et al. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell. 2010;22(4):1232–48. doi: 10.1105/tpc.108.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.El-Showk S, Ruonala R, Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140(7):1373–83. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- 147.To JPC, Haberer G, Ferreira FJ, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16(3):658–71. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.D’Agostino IB, Deruere J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124(4):1706–17. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mira-Rodado V, Sweere U, Grefen C, et al. Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J Exp Bot. 2007;58(10):2595–607. doi: 10.1093/jxb/erm087. [DOI] [PubMed] [Google Scholar]
- 150.Ruiz-Sola MA, Rodriguez-Concepcion M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tan BC, Joseph LM, Deng WT, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35(1):44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 152.Seo M, Peeters AJ, Koiwai H, et al. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci U S A. 2000;97(23):12908–13. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45(11):1694–703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- 154.Rodriguez-Gacio Mdel C, Matilla-Vazquez MA, Matilla AJ. Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal Behav. 2009;4(11):1035–49. doi: 10.4161/psb.4.11.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kushiro T, Okamoto M, Nakabayashi K, et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23(7):1647–56. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boursiac Y, Leran S, Corratge-Faillie C, Gojon A, Krouk G, Lacombe B. ABA transport and transporters. Trends Plant Sci. 2013;18(6):325–33. doi: 10.1016/j.tplants.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 157.Jarzyniak KM, Jasinski M. Membrane transporters and drought resistance – a complex issue. Front Plant Sci. 2014;5(687):1–15. doi: 10.3389/fpls.2014.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kuromori T, Miyaji T, Yabuuchi H, et al. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci U S A. 2010;107(5):2361–6. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kuromori T, Shinozaki K. ABA transport factors found in Arabidopsis ABC transporters. Plant Signal Behav. 2010;5(9):1124–6. doi: 10.4161/psb.5.9.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Merilo E, Jalakas P, Laanemets K, et al. Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol Plant. 2015;8(9):1321–33. doi: 10.1016/j.molp.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 161.Zhang HW, Zhu HF, Pan YJ, Yu YX, Luan S, Li LG. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant. 2014;7(10):1522–32. doi: 10.1093/mp/ssu063. [DOI] [PubMed] [Google Scholar]
- 162.Umezawa T, Nakashima K, Miyakawa T, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–39. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsuzuki T, Takahashi K, Inoue S, et al. Mg-chelatase H subunit affects ABA signaling in stomatal guard cells, but is not an ABA receptor in Arabidopsis thaliana. J Plant Res. 2011;124(4):527–38. doi: 10.1007/s10265-011-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Junker A, Hartmann A, Schreiber F, Baumlein H. An engineer’s view on regulation of seed development. Trends Plant Sci. 2010;15(6):303–7. doi: 10.1016/j.tplants.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 165.Dobritzsch S, Weyhe M, Schubert R, et al. Dissection of jasmonate functions in tomato stamen development by transcriptome and metabolome analyses. BMC Biol. 2015;13:28. doi: 10.1186/s12915-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. Plant Cell. 2002;14:S153–64. doi: 10.1105/tpc.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001;13(10):2191–209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hyun Y, Choi S, Hwang HJ, et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev Cell. 2008;14(2):183–92. doi: 10.1016/j.devcel.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 169.Yang WY, Devaiah SP, Pan XQ, Isaac G, Welti R, Wang XM. AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J Biol Chem. 2007;282(25):18116–28. doi: 10.1074/jbc.M700405200. [DOI] [PubMed] [Google Scholar]
- 170.Bannenberg G, Martinez M, Hamberg M, Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44(2):85–95. doi: 10.1007/s11745-008-3245-7. [DOI] [PubMed] [Google Scholar]
- 171.Schaller F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot. 2001;52(354):11–23. [PubMed] [Google Scholar]
- 172.Kubigsteltig I, Laudert D, Weiler EW. Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta. 1999;208(4):463–71. doi: 10.1007/s004250050583. [DOI] [PubMed] [Google Scholar]
- 173.Hause B, Stenzel I, Miersch O, et al. Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 2000;24(1):113–26. doi: 10.1046/j.1365-313x.2000.00861.x. [DOI] [PubMed] [Google Scholar]
- 174.Stenzel I, Otto M, Delker C, et al. ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: tissue- and organ-specific promoter activities and in vivo heteromerization. J Exp Bot. 2012;63(17):6125–38. doi: 10.1093/jxb/ers261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Theodoulou FL, Job K, Slocombe SP, et al. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005;137(3):835–40. doi: 10.1104/pp.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Strassner J, Schaller F, Frick UB, et al. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 2002;32(4):585–601. doi: 10.1046/j.1365-313x.2002.01449.x. [DOI] [PubMed] [Google Scholar]
- 177.Schneider K, Kienow L, Schmelzer E, et al. A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J Biol Chem. 2005;280(14):13962–72. doi: 10.1074/jbc.M413578200. [DOI] [PubMed] [Google Scholar]
- 178.Miersch O, Wasternack C. Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: endogenous jasmonates do not induce jasmonate biosynthesis. Biol Chem. 2000;381(8):715–22. doi: 10.1515/BC.2000.092. [DOI] [PubMed] [Google Scholar]
- 179.Fonseca S, Chini A, Hamberg M, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–50. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 180.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16(8):2117–27. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Suza WP, Rowe ML, Hamberg M, Staswick PE. A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta. 2010;231(3):717–28. doi: 10.1007/s00425-009-1080-6. [DOI] [PubMed] [Google Scholar]
- 182.Sasaki Y, Asamizu E, Shibata D, et al. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 2001;8(4):153–61. doi: 10.1093/dnares/8.4.153. [DOI] [PubMed] [Google Scholar]
- 183.Goossens A, Hakkinen ST, Laakso I, et al. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci U S A. 2003;100(14):8595–600. doi: 10.1073/pnas.1032967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pauwels L, Goossens A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pauwels L, Morreel K, De Witte E, et al. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci U S A. 2008;105(4):1380–5. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19(7):2225–45. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fernandez-Calvo P, Chini A, Fernandez-Barbero G, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16(7):1938–50. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mandaokar A, Thines B, Shin B, et al. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46(6):984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- 190.Song SS, Qi TC, Huang H, et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23(3):1000–13. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li L, Zhao YF, McCaig BC, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development (vol 16, pg 126, 2004) Plant Cell. 2004;16(3):783–3. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J Biol Chem. 2002;277(47):45188–94. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- 193.Lohr M, Schwender J, Polle JEW. Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae. Plant Sci. 2012;185:9–22. doi: 10.1016/j.plantsci.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 194.Vranova E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Ann Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 195.Helliwell CA, Sullivan JA, Mould RM, Gray JC, Peacock WJ, Dennis ES. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001;28(2):201–8. doi: 10.1046/j.1365-313x.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- 196.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21(5):1328–39. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sun TP. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154(2):567–70. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tyler L, Thomas SG, Hu J, et al. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135(2):1008–19. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell. 2008;20(9):2437–46. doi: 10.1105/tpc.108.061648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ariizumi T, Lawrence PK, Steber CM. The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011;155(2):765–75. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gomi K, Sasaki A, Itoh H, et al. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004;37(4):626–34. doi: 10.1111/j.1365-313x.2003.01990.x. [DOI] [PubMed] [Google Scholar]
- 202.Bai S, Yao T, Li M, et al. PIF3 is involved in the primary root growth inhibition of Arabidopsis induced by nitric oxide in the light. Mol Plant. 2014;7(4):616–25. doi: 10.1093/mp/sst142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Filo J, Wu A, Eliason E, Richardson T, Thines BC, Harmon FG. Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signal Behav. 2015;10(3):e992707. doi: 10.4161/15592324.2014.992707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21(9):R338–45. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 205.Schwechheimer C. Gibberellin signaling in plants – the extended version. Front Plant Sci. 2012;2:107. doi: 10.3389/fpls.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nes WD. Biosynthesis of cholesterol and other sterols. Chem Rev. 2011;111(10):6423–51. doi: 10.1021/cr200021m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Singh B, Sharma RA. Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications. 3. Biotech. 2015;5(2):129–51. doi: 10.1007/s13205-014-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fujioka S, Inoue T, Takatsuto S, Yanagisawa T, Yokota T, Sakurai A. Biological activities of biosynthetically-related congeners of brassinolide. Biosci Biotechnol Biochem. 1995;59:1973–5. [Google Scholar]
- 209.Nomura T, Ueno M, Yamada Y, Takatsuto S, Takeuchi Y, Yokota T. Roles of brassinosteroids and related mRNAs in pea seed growth and germination. Plant Physiol. 2007;143(4):1680–8. doi: 10.1104/pp.106.093096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Symons GM, Reid JB. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004;135(4):2196–206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.She J, Han ZF, Kim TW, et al. Structural insight into brassinosteroid perception by BRI1. Nature. 2011;474(7352):472–U496. doi: 10.1038/nature10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang J, Jiang J, Wang J, et al. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014;24(11):1328–41. doi: 10.1038/cr.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–7. Survey of genes involved in phytohormone biosynthesis, transport, and signaling in A. thaliana and their co-orthologues in selected Viridiplantae. Analyzed phytohormones are auxin (S1), ethylene (S2), cytokinin (S3), abscisic acid (S4), jasmonic acid (S5), gibberellin (S6), and brassinosteroid (S7). The corresponding AGI number, gene, and mutant names, description in database, number of species containing co-orthologues, number of co-orthologues in the indicated plant species and the overall distribution in the groups of eudicots, monocots, mosses, and algae are indicated. The selected plant species are: Atha, A. thaliana; Bdis, B. distachyon; Crei, C. reinhardtii; Gmax, G. max; Mtru, M. truncatula; Osat, O. sativa; Ppat, P. patens; Ptri, P. trichocarpa; Sbic, S. bicolor; Slyc, S. lycopersicum; Stub, S. tuberosum; Vvin, V. vinifera; Zmay, Z. mays.
Supplementary Tables 8–14. Survey of co-orthologues involved in phytohormone biosynthesis, transport, and signaling in the analyzed plant genomes with prediction of intracellular localization and domain structure. Analyzed phytohormones are auxin (S8), ethylene (S9), cytokinin (S10), abscisic acid (S11), jasmonic acid (S12), gibberellin (S13), and brassinosteroid (S14). The selected plant species are: Atha, A. thaliana; Bdis, B. distachyon; Crei, C. reinhardtii; Gmax, G. max; Mtru, M. truncatula; Osat, O. sativa; Ppat, P. patens; Ptri, P. trichocarpa; Sbic, S. bicolor; Slyc, S. lycopersicum; Stub, S. tuberosum; Vvin, V. vinifera; Zmay, Z. mays.
Supplementary Tables 15–21. Expression of tomato co-orthologues involved in phytohormone biosynthesis, transport, and signaling in different tissues: root, stem, leaf, flower, and fruits at the three developmental stages, namely, mature green (fruit_mature), breaker stage (berry_breaker), and 10 days after turning red (berry_10 days). Analyzed phytohormones are auxin (S15), ethylene (S16), cytokinin (S17), abscisic acid (S18), jasmonic acid (S19), gibberellin (S20), and brassinosteroid (S21).