Abstract
The dynamics of molecules in living cells hamper precise imaging of molecular patterns by functional and super resolution microscopy. Circumventing lethal chemical fixation, an on-stage cryo-arrest was developed for consecutive imaging of molecular patterns within the same living, but arrested cells. The reversibility of consecutive cryo-arrests was demonstrated by the high survival rate of different cell lines and intact growth factor signaling that was not perturbed by stress response. Reversible cryo-arrest was applied to study the evolution of ligand-induced receptor tyrosine kinase activation at different scales. The nanoscale clustering of epidermal growth factor receptor (EGFR) in the plasma membrane was assessed by single molecule localization microscopy and endosomal microscale activity patterns of ephrin receptor type-A (EphA2) by fluorescence lifetime imaging microscopy. We thereby demonstrate that reversible cryo-arrest allows the precise determination of molecular patterns while conserving the dynamic capabilities of living cells.
Resolution in fluorescence-microscopy is limited by the time it takes to collect enough photons to obtain sufficient signal-to-noise for image reconstruction. However, molecular patterns in living cells are highly dynamic, which impedes imaging with long acquisition times. Irreversible chemical fixation can solve this problem, yet it is inefficient in immobilizing membranes, can lead to protein extraction, artificial clustering 1–3, disrupts the out-of-equilibrium physiological state and prohibits the evolution of molecular patterns within cells.
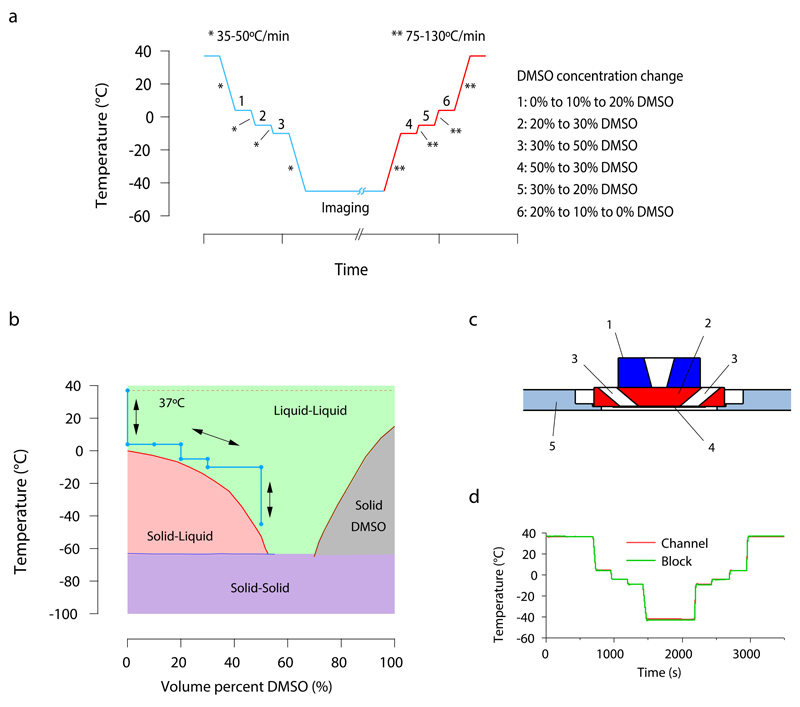
We therefore sought to reversibly arrest cells on a microscope by cooling to -45°C. For this, dimethyl sulfoxide (DMSO) was used to prevent formation of lethal ice in cells 4 that scatters light and displaces organic material 5–7. DMSO is more efficient in lowering the freezing point compared to other cell permeable cryoprotective agents 8,9. At 37°C it is cytotoxic 10,11, which is markedly reduced at lower temperatures 4,12. For reversible in situ cryo-arrest, cytotoxic effects of DMSO were avoided by adapting its concentration during cooling and warming, thereby maintaining the aqueous solution in a stable ice-free phase 8,12. This cryo-cycle reversibly arrested cells in a state compatible with life and allowed the observation of dynamic molecular patterns in the same cell at different time-points without restrictions in photon acquisition times.
We used reversible cryo-arrest to study the micro- and nano-scale organization of receptor tyrosine kinases (RTKs). RTK-mediated signaling is actuated by ligand-binding, which results in self-association and activation in the plasma membrane 13. RTKs are organized in nanoscale clusters 2,14–18, which are relevant for the reactivity of receptors and ligand-induced signal actuation 19,20. A full localization reconstruction by TIRF-PALM with nanometer resolution necessitates minutes, during which receptors will displace by tens of micrometers at 37°C due to their diffusion (2-5 × 10-2 μm2 s-1; ref. 21,22). Therefore, TIRF-PALM was performed on EGFR-mEOS2 expressing HeLa cells under reversible cryo-arrest, which enabled the observation of EGFR nanoscale cluster reorganization after growth factor stimulus in the same cell.
At the microscale, RTK-signal duration is controlled by vesicular dynamics 23,24, resulting in activity patterns difficult to resolve at physiological temperatures due to rapid vesicular motion. Eph receptors constitute the largest subfamily of RTKs that function as principal cell guidance cues during development 25. To study how vesicular dynamics controls the propagation of EphA2-signals inside the cell, we imaged its stimulus-induced activity patterns by the genetically encoded activity sensor LIFEA2 24 during consecutive cryo-arrests.
Results
Development and validation of reversible cryo-arrest
The time course of a reversible cryo-arrest cycle is depicted in figure 1a. Starting at 37°C, cells were cooled to 4°C, at which the culture medium was replaced with HEPES-buffered medium containing 20% DMSO in two incremental steps of 10%. Subsequently, the temperature was decreased stepwise to -45°C while raising the DMSO concentration to stay above the liquidus temperature. For warming back to 37°C the protocol was reversed (Fig. 1b).
Figure 1. Protocol for reversible cryo-arrest on a microscope stage.
(a) Time course of the cryo-arrest protocol. Temperature is changed stepwise between the physiological temperature of 37°C and the fixation temperature of -45°C and DMSO concentration is adapted as indicated on the right side of the figure (1-6). Media was exchanged with 3 μL s-1 to avoid cell detachment. * and ** represent the cooling and warming speeds indicated in the figure. (b) Binary phase diagram of DMSO-water mixtures (data adapted from 8) including the course of the cryo-arrest protocol (blue line, arrows). The green area indicates the temperature range in which the mixture is a stable liquid (and solidification is thermodynamically forbidden) (c) Side view sketch of the central part of the cryo-stage. (1) silver block with temperature control; (2) aluminum block; (3) medium inlet and outlet; (4) 100 μm thick channel with sample on a cover slide; (5) insulating polyvinyl chloride plate adapted for commercial microscopy stages. (d) Temperature recording during the course of an experiment. The temperature was measured directly in the channel with the help of a 50 μm thick thermocouple (red line). This temperature was compared to the temperature read out at the aluminum block of the stage (green line), as routinely performed during every experiment.
A cryo-stage dedicated to live cell microscopy was developed that enabled easy medium exchange. The central part of the stage was constructed from silver and aluminum with high thermal conductivity and the height of the flow-through chamber was limited to 100 μm to minimize the insulating layer of medium. A polyvinylchloride mounting was used to thermally isolate the stage from the microscope (Fig. 1c, Supplementary Fig. 1). This design allowed for maximum cooling rates of 35-50°C min-1, warming rates of 75-130°C min-1 and control of the chamber temperature with an accuracy of 1°C, reliably traced by a thermocouple (Fig. 1d).
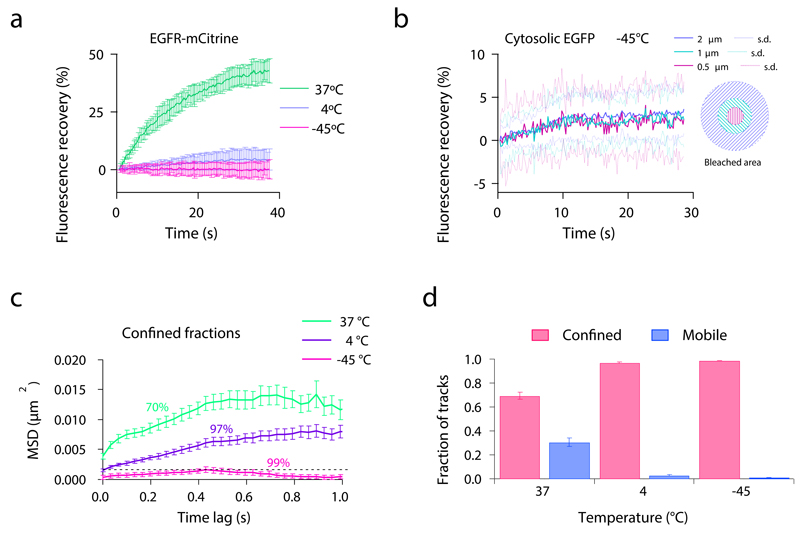
To evaluate the efficiency of protein immobilization in cells during cryo-arrest, we analyzed the diffusion of cytoplasmic eGFP and transmembrane EGFR tagged with mCitrine (EGFR-mCitrine). Fluorescence recovery after photobleaching revealed that the diffusion of EGFR-mCitrine was dramatically reduced after cooling to 4°C and not measurable at -45°C (Fig. 2a). Single particle tracking of EGFR either by Cy3-labeled SNAP-EGFR (SNAP-tag fused to EGFR 20) or streptavidin-Quantum Dots conjugated to biotinylated EGF 26 confirmed the immobility of this transmembrane receptor at -45°C within the experimental timeframe (Fig. 2c, d, Supplementary Fig. 2, Supplementary Video 1 & 2). The fluorescence recovery of cytoplasmic eGFP at -45°C was strongly reduced to ~3%. This recovery was equally fast in three concentric areas around the center of the bleached spot (Fig. 2b), showing that it was due to photo-physical effects such as dark-state recovery 27 and not diffusion.
Figure 2. Protein diffusion before and during cryo-arrest.
(a) Fluorescence recovery after photobleaching (FRAP) of EGFR-mCitrine expressed in HeLa cells at different temperatures during the cryo-arrest protocol. Shown are fluorescence recovery curves obtained at 37°C (n=11 cells), 4°C (n=19) and -45°C (n=19) after bleaching a 2 μm spot on the plasma membrane. (b) FRAP of cytoplasmic eGFP expressed in HeLa cells at -45°C. Shown are the fluorescence recovery curves measured in concentric areas within the bleached spot (an innermost circle with a diameter of 0.5 μm, an intermediate circle with 1 μm and the full bleached area with a diameter of 2 μm; n=20 cells. (c, d) Single molecule tracking of Cy3-SNAP-EGFR in HeLa cells measured at 37°C (n=10 cells), 4°C (n=14) and -45°C (n=14). (c) Mean squared displacement (MSD) of the confined fractions of Cy3-SNAP-EGFR. Numbers represent the percentage of molecules that are included in this confined fraction. (d) Fractions of detected molecule tracks that are classified as confined and mobile. (a, b) mean ± s.d., (c, d) mean ± s.e.m. All data was obtained from 3 independent experiments.
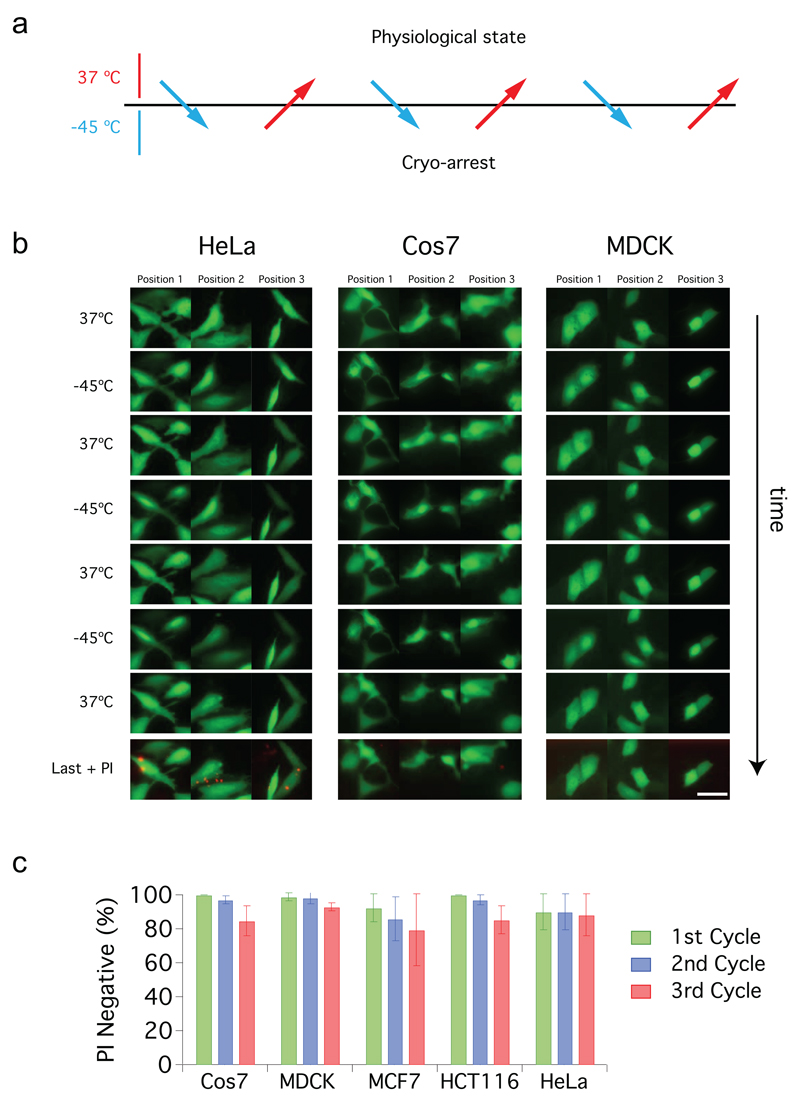
Survival of five different cell lines exceeded 80% after three consecutive cryo-arrest cycles as assessed by propidium iodide (PI) staining (Fig. 3). In parallel, we used cytoplasmic eGFP to monitor cellular morphology. Surviving adherent cells maintained their typical shape, whereas the minor fraction of PI-positive cells exhibited morphological changes such as blebbing, detached or lost their fluorescence during or after cryo-arrests (Fig. 3b).
Figure 3. Survival of mammalian cells during repeated cryo-arrest cycles.
(a) General scheme of an imaging experiment using three consecutive cycles of reversible cryo-arrest. (b) Effect of three consecutive cycles of cryo-arrest on cell survival and morphology. Representative images of HeLa, Cos7 and MDCK cells transfected with eGFP. After each cycle cells were stained with propidium iodide (PI) to mark dead cells. In the last row an overlay of eGFP (green) and PI staining (red) after the last cycle is shown. Scale bar: 20 µm. (c) Quantification of the fraction of PI-negative cells during the course of the three consecutive cycles of cryo-arrest for the indicated cell lines (Cos7 n=64, HeLa n=70, HCT116 n=100, MCF7 n=77 and MDCK n=73 cells). All data is shown as mean ± s.d. from 3 independent experiments.
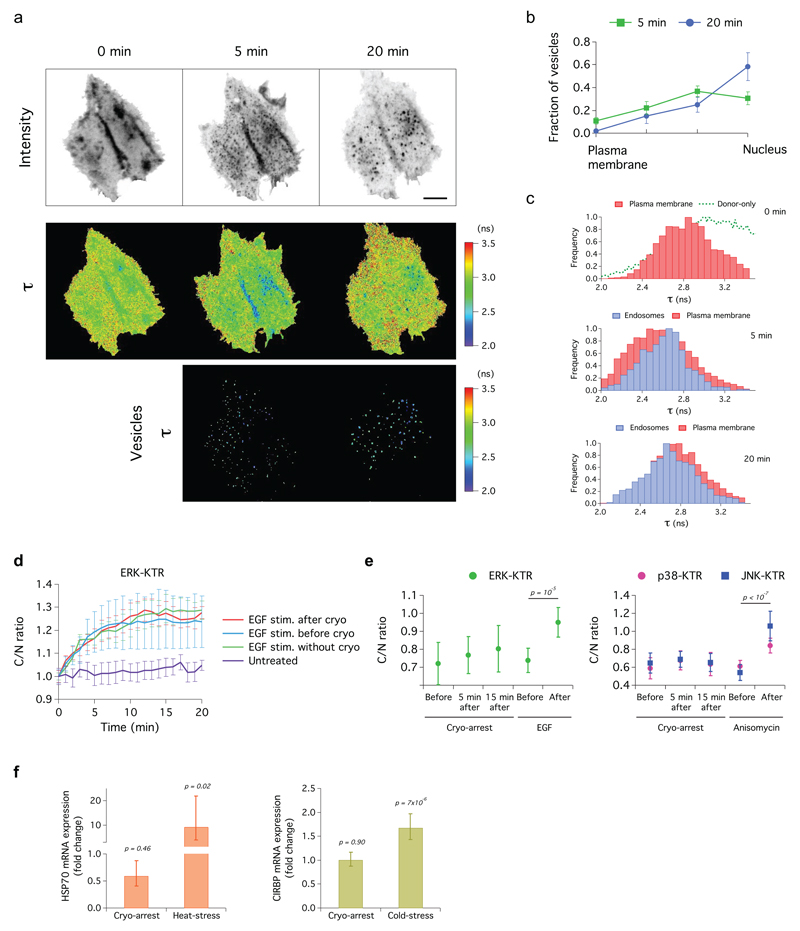
To study if EGF-signaling is affected by cryo-arrest, we analyzed the activity of EGFR and its downstream effector, the extracellular-signal related kinase (ERK). We assessed EGFR-activity in HeLa cells co-expressing EGFR-mCitrine and a phosphotyrosine binding domain fused to mCherry (PTB-mCherry). Stimulation with a 5-min pulse of EGF after one cryo-arrest cycle resulted in endocytosis of EGFR-mCitrine that continued over two additional cryo-arrests at 5 min and 20 min after EGF-stimulus (Fig. 4a, b). This shows that ligand-induced receptor endocytosis was fully functional after three cryo-arrests. The fluorescence lifetime of EGFR-mCitrine decreased in the plasma membrane and in small endosomes that became clearly detectable during cryo-arrest after 5 min of EGF-stimulation (Fig. 4c). This FRET-induced fluorescence lifetime decrease indicates the molecular association of EGFR-mCitrine and PTB-mCherry due to EGFR-phosphorylation 28. 20 min after EGF-stimulation, the fluorescence lifetime distributions shifted to higher values (Fig. 4c), showing the inactivation of the receptor by protein tyrosine phosphatase (PTP) activity 23,29. Three cryo-cycles without EGF-stimulation did not activate the receptor as apparent from the constant average fluorescence lifetime (Supplementary Fig. 3a). We assessed the activity of ERK by a fluorescent kinase translocation reporter (KTR) construct 30. The measurements showed no impairment of the response by cryo-arrest before or after EGF-stimulation and cryo-arrest alone did not induce an ERK-response (Fig. 4d, e; Supplementary Fig. 4). These experiments indicate that consecutive cryo-arrest cycles do not impair EGF-induced signaling activity.
Figure 4. EGF-signaling and stress response in HeLa cells during reversible cryo-arrest.
(a) FRET-FLIM time sequence measured at -45°C in cells co-expressing EGFR-mCitrine and PTB-mCherry during three consecutive cryo-arrests; before (0 min), 5 and 20 min after EGF stimulation. Upper row: fluorescence images of EGFR-mCitrine, middle row: mCitrine fluorescence lifetime (τ) maps, lower row: τ on endosomes (color-coding right). Scale bar: 10 µm. (b) Number of vesicles (mean ± s.e.m.) in 4 concentric radial bins of equivalent area (see Supplementary Fig. 9b). (c) τ distribution of EGFR-mCitrine in the plasma membrane (red) and in endosomes (blue), before, 5 and 20 min after EGF stimulation. Green dotted line: τ distribution of mCitrine in absence of PTB-mCherry. (d-e) Kinase activity of ERK, JNK and p38 reported by whole cell over nuclear fluorescence intensity (C/N) of their KTR constructs. (d) ERK activity upon EGF stimulation without cryo-arrest (EGF), upon 2 min EGF stimulation after cryo-arrest (cryo + EGF), upon cryo-arrest 2 min after EGF stimulation (EGF + cryo) and unstimulated cells (untreated). (e) ERK, JNK and p38 activity before and after cryo-arrest or treatment with EGF (ERK) or anisomycin (JNK, 50 ng mL-1 and p38, 5 µg mL-1). (f) HSP70 and CIRBP mRNA levels 15 min after cryo-arrest, after 30 min at 42°C (heat stress) or after 24 h at 32°C (cold stress). Expression is represented as fold change relative to control at 37°C. (c-f) data represents mean ± s.d.; p-values from two-sided student’s t-test. EGF: 200 ng mL-1. n: see online methods.
To test if cryo-arrest elicits a stress-response, we measured the activity of the stress-activated protein kinase JNK and the cold-activated p38 MAPK 31 using their respective KTRs 30. These measurements showed only a slight and transient increase in p38-activity and no change in JNK-activity following cryo-arrest (Fig. 4e). Additionally, no changes in mRNA expression of the heat shock protein HSP70 or the cold-stress protein CIRBP 32 were found (Fig. 4f).
Imaging nanoscale organization of EGFR during cryo-arrest
We first evaluated if segregation of lipid-bilayer membranes into liquid-disordered and liquid-ordered domains 33 are introduced by cryo-arrest. Even though this effect was not observed in living cells 34, it could occur at temperatures below 0°C. This might influence the distribution of EGFR since it tends to partition into liquid-disordered domains 35. The distribution of the ordered domain marker DiOC18 and EGFR was maintained in HeLa cells between 37°C and -45°C (Supplementary Fig. 5). This shows that cryo-arrest did not lead to membrane phase separation and EGFR segregation on length scales observable by diffraction-limited light microscopy.
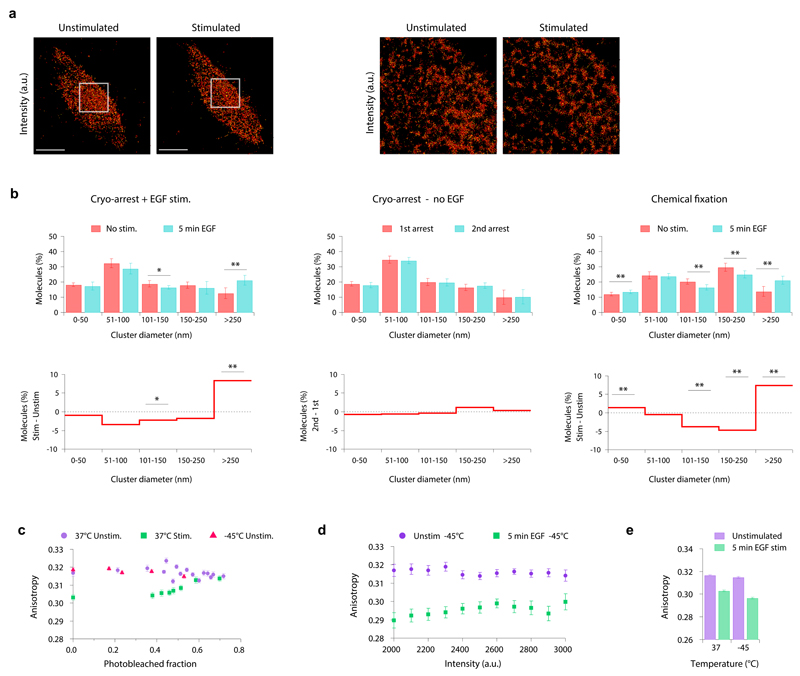
To investigate the nanoscale distribution of EGFR in the plasma membrane, we reversibly cryo-arrested HeLa cells expressing EGFR-mEos2 before and 5 min after EGF-stimulation. The distribution of EGFR-mEos2 at the basal membrane was determined by TIRF-PALM with a localization precision of 24 ± 8 nm (mean ± s.d.) (Supplementary Fig. 6). We grouped molecules in clusters of at least 5 molecules if the distance between neighboring molecules was less than 40 nm by using a density-based clustering algorithm 36. This analysis captures self-association and also local enrichment in plasma membrane domains, because the minimum distance exceeds the size of an EGFR-dimer (~12 nm). 61 ± 3 % of receptors appeared in clusters, with diameters ranging from tens to hundreds of nanometers (Fig. 5a, b and Supplementary Fig. 7a). Consecutive cryo-arrests without stimulation did not show any differences in clustering parameters, indicating that no clusters were induced or disrupted (Fig. 5b and Supplementary Fig. 7a). We performed the same analysis in formaldehyde-fixed cells where the distribution was clearly shifted to larger clusters (>100 nm). This was likely caused by formaldehyde-induced crosslinking (Fig. 5b and Supplementary Fig. 7a).
Figure 5. Nanoscale organization of EGFR in the plasma membrane.
(a, left) Representative TIRF-PALM image (localization intensity) of EGFR-mEos2 in the same cryo-arrested HeLa cell, before and 5 min after EGF stimulation. (a, right) detailed view of the selected areas marked in the left images. Scale bars: 10 µm (b) Density-based spatial cluster analysis using a neighborhood radius of 40 nm of the EGFR-mEos2 distribution in cryo-arrested HeLa cells. Upper row: fraction of molecules in clusters of different diameters. Lower row: fraction of molecules shown as differential plot between the compared conditions. Left column: before (red) and 5 min after EGF stimulation (blue); middle column: unstimulated cells that were cryo-arrested twice with an interval of 5 min; right column: chemically fixed cells before (red) and 5 min after EGF stimulation (blue). (mean ± s.d.; n=8 cells per condition; *: p<0.05; **p<0.01 using two sample KS-test) (c) Mean fluorescence anisotropy of EGFR-QG-mCitrine in HeLa cells as a function of its bleached fraction. Data obtained at 37°C before (purple, n=7) and 5 min after EGF stimulation (green, n=12) or at -45°C for unstimulated cryo-arrested cells (red, n=10) (d) Mean fluorescence anisotropy of EGFR-QG-mCitrine in cryo-arrested HeLa cells as a function of its binned fluorescence intensity (n=12), before (purple) and 5 min after EGF stimulation (green). (e) Fluorescence anisotropy per cell before and 5 min after EGF stimulation at 37°C (n=16) and at -45°C (n=12). Data in (c)-(e): mean ± s.e.m.
After EGF stimulation for 5 min, the fraction of large EGFR-clusters (> 250 nm) was increased at the expense of smaller ones. These effects were observed to a similar extent in the periphery and center of the basal membrane (Supplementary Fig. 8), indicating a uniform activation of EGFR. Inter-cluster network analysis on a larger scale revealed no change upon stimulation, indicating that the observed effects were specific for the analyzed nanometer length scale (Supplementary Fig. 7b).
To assess if self-association of EGFR causes its clustering, we measured homo-FRET between mCitrines inserted into EGFR (EGFR-QG-mCitrine) by fluorescence anisotropy 23,37. Upon photo-bleaching, EGFR-QG-mCitrine showed no significant increase in anisotropy, indicating that EGFR before stimulation is mainly monomeric 38. This did not change during cryo-arrest showing that the receptor remained monomeric (Fig. 5c). Stimulated cells showed a lower anisotropy that increased upon bleaching to the value of unstimulated cells, reflecting EGFR-dimerization (Fig. 5c). The mCitrine intensity independent anisotropy decrease upon stimulation 23 was also detectable in the cryo-arrested state (Fig. 5d, e). These experiments show that cryo-arrest is not affecting EGFR self-association and that EGFR-dimerization is still inducible after, and measurable during the arrest.
Cryo-SMLM analysis in conjunction with anisotropy measurements thus revealed local accumulations of EGFR in unstimulated cells that are not due to self-association. Upon stimulation and dimerization of EGFR, its partitioning changed from relatively small clusters to large domains containing more molecules.
Mapping microscale EphA2 activity during cryo-arrest
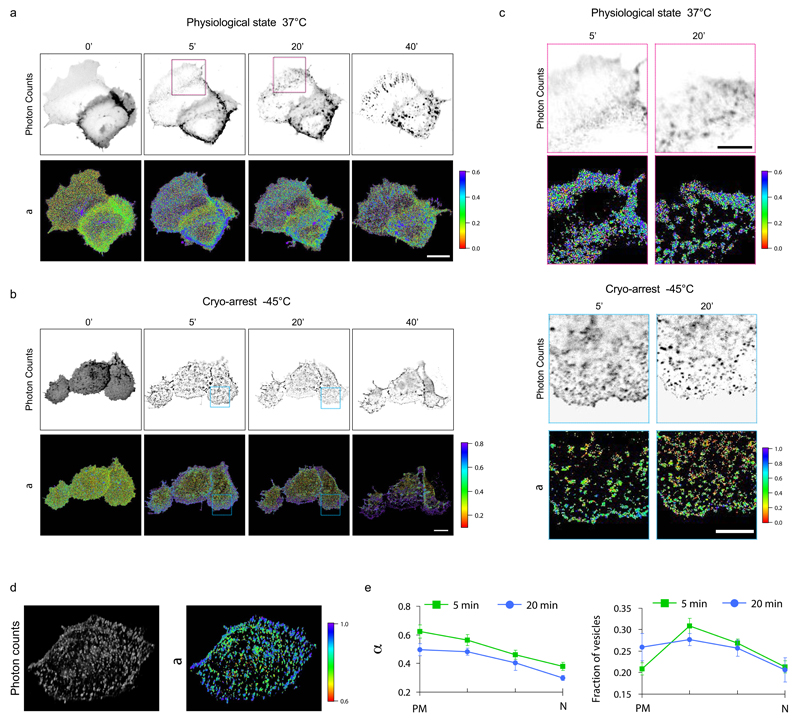
We measured the spatial distribution of the active conformation of EphA2 in consecutive cryo-cycles before and after stimulation with clustered ephrinA1-Fc in Cos-7 cells expressing LIFEA2 24. We quantified the spatially-resolved fraction of active LIFEA2 by global analysis of fluorescence decay profiles obtained by confocal FLIM 39. A FRET-FLIM time-series of LIFEA2 at 37°C showed an increase in the fraction of active LIFEA2 after 5 min stimulation, which was most pronounced at the cell periphery. Inspection of the corresponding fluorescence intensity micrographs showed a continuous distribution of LIFEA2 at the periphery, indicating that its active state resides at the plasma membrane. 20 and 40 min post-stimulation, blurred structures appeared in the cytoplasm with a similar fraction of active LIFEA2 as at the plasma membrane. Additionally, a minority of larger vesicular structures with active LIFEA2 were apparent (Fig. 6a). Based on these experiments, one would conclude that after stimulation EphA2 is mostly active at the plasma membrane and on stable endocytic vesicles at later time points. However, because the acquisition of enough photons for quantifying the fraction of active LIFEA2 took on average 3 min, rapidly moving early endocytic vesicles could not be detected in FRET-FLIM time-series. Indeed, reversible cryo-arrest enabled the detection of individual endosomal vesicles after ephrin stimulation in cryo-arrested cells (Fig. 6b, c and Supplementary Fig. 9a, c). This showed that massive endocytosis already ensued after 5 min stimulation where most of the active LIFEA2 resided in small peripheral endocytic vesicles. 3D confocal FRET-FLIM of cryo-arrested cells showed that these structures were indeed endocytosed receptors and not clusters at the plasma membrane (Fig. 6d, Supplementary Fig. 9d, Supplementary Video 3). We determined the active fraction of LIFEA2 on each individual endosome as well as the number of LIFEA2 containing vesicles and calculated the average in 4 concentric spatial bins with equal area (Fig. 6e and Supplementary Fig. 9b, c). This quantification revealed a descending gradient of active LIFEA2 in endosomes towards the interior of the cell that persisted for at least 20 min after stimulation.
Figure 6. Evolution of LIFEA2 activity patterns monitored during reversible cryo-arrest.
(a) Confocal FLIM measurements of LIFEA2 activation in Cos7 cells upon stimulation with pre-clustered ephrinA1-Fc (2 µg mL-1) for the indicated time (minutes) at 37°C. Upper row: representative images of LIFEA2 donor (mCitrine) photon count, lower row: fraction of LIFEA2 in the active conformation (α). (b) Same as above, but Cos7 cells were cryo-arrested at -45°C at the indicated time points following the scheme depicted in figure 3a. (c) Magnified insets from respective donor photon count and α-maps masked for defined structures (see Online Methods for details) at the indicated time (a, magenta boxes) and (b, blue boxes). (d) Volume representation of a deconvolved confocal FLIM z-scan (Imaging time: 37 min) through a representative HeLa cell cryo-arrested 5 min after stimulation with pre-clustered ephrinA1-Fc. Left: LIFEA2 donor (mCitrine) photon count; right: α (e) Quantification of α on vesicles (left) and fraction of vesicles (right) in 4 concentric radial bins of equivalent area (compare Supplementary Fig. 9b). PM: outermost bin close to the plasma membrane; N: innermost bin around the nucleus. Data: mean ± s.e.m. (n=9 cells from 4 time series). Scale bars: a,b = 20 µm; c = 10 µm.
Discussion
The presented cryo-arrest method enables physical fixation of living cells in situ in a reversible manner, thereby permitting fluorescence imaging with long exposure times of arrested but living cells at multiple points in time during a biological process. Moreover, fluorophores are more photostable and produce less cytotoxic radicals at lower temperatures 40,41. Cryo-fixation is frequently used for electron microscopy, avoiding ice formation by rapid cooling below -130°C 7. However, these approaches are not yet reversible 5 and application to SMLM showed that the resolution, though limited by non-immersion objectives 40,42,43, can be superior to ambient temperature due to better photon-economics 44. The reversible cryo-arrest presented here is compatible with immersion objectives by cooling to -45°C, a temperature where diffusion of proteins and lipids is so negligible that it enables the measurement of their patterns with high precision. As shown by high survival rates and preservation of overall morphology after three consecutive cryo-arrests, DMSO ensures cryo-tolerance without cytotoxicity at this temperature 12, DMSO can replace water in hydration shells 45 and when added at 37°C, it influences protein folding and interactions and thereby cellular functions 10. However, addition of DMSO at low temperatures did not disrupt inter- and intramolecular interactions, such as dimerization of EGFR, binding of EGFR to a PTB-domain and conformation of LIFEA2. EGF-induced downstream signaling was conserved after cryo-arrests, which shows that there was no deleterious effect on multiple protein reactions/interactions involved in these complex signaling processes. The proteome composition is likely also not affected by cryo-arrest, since stress proteins were not induced.
The reversible cryo-arrest performed on the microscope has the advantage that molecular dynamics at physiological temperature can be measured in conjunction with the precise determination of molecular patterns at cryo-temperatures in the same cells. The cooling procedure from 37°C to -45°C takes in our current setup about 10 min. This is fast compared to chemical fixation, which usually takes at least 30 min to arrest membrane bound proteins and cannot fix lipids 1. However, biochemical reactions are already dramatically reduced at 4°C, which is reached after ~50 s. That biochemical reactions are indeed arrested on the timescale of a minute is apparent from the unaltered ERK-activity profiles of cells that were stimulated with EGF before cryo-arrest (Fig. 4d). Nevertheless, the state of the molecules cannot be precisely measured at this temperature due to residual diffusion, for which cryo-arrest at -45°C was necessary. The observed molecular state at -45°C is thereby representative of ~1 min after the beginning of cryo-arrest. This is for example shown in the early vesicular structures in which EphA2 resides after 5 min stimulation. Thus, the method is applicable to study processes that evolve on the timescale of minutes.
We applied reversible cryo-arrest to study the evolution of spatial-temporal patterns of RTKs at different length-scales in living cells. The TIRF-PALM and fluorescence anisotropy microscopy results in cryo-arrested cells showed that non-stimulated EGFR in the plasma membrane is monomeric, yet locally clustered. EGFR-clustering in unstimulated cells is therefore not due to self-association, but caused by interactions with specialized membrane structures. This might prime the system for efficient activation by the formation of asymmetric dimers 46 upon EGF stimulation. After EGF-stimulation, EGFR localization shifted from mostly small clusters of 50-150 nm to clusters bigger than 250 nm. This indicates that activated and phosphorylated EGFR-dimers interact stronger with adaptor molecules in specialized membrane domains than non-activated EGFR. This could lead to the formation of signaling-clusters with high local concentrations of activated EGFR that can contribute to the autocatalytic amplification of the signal upon activation 19,47 and are likely the sites for receptor endocytosis 48.
Formaldehyde-fixed unstimulated cells exhibited mainly large clusters resulting in a smaller shift to large clusters upon stimulation. The cross-linking activity of formaldehyde takes more than 30 min to arrest transmembrane molecules 1, which might have been the cause for the observed large EGFR clusters.
Measuring FRET-FLIM of living, arrested cells allowed to obtain precise data on EphA2-activity in early endocytic vesicles. LIFEA2 was previously developed to quantitatively image the active state of EphA2 in living cells at 37°C 24. However, the long acquisition time required to collect enough photons to reconstruct a high resolution fluorescence lifetime image, prohibited the detection of LIFEA2 activity levels in dynamic subcellular structures. Under cryo-arrest, we observed a high fraction of activated LIFEA2 in peripheral endosomes that gradually decreased towards the perinuclear area. This gradient persisted over time despite the reduction in total receptor activity. These results suggest that EphA2-signaling response is controlled by the duration of endosomal trafficking to the perinuclear area, during which receptor dephosphorylation by an opposed PTP activity gradient occurs 24,49.
Cryo-arrest is compatible with different fluorescence microscopy techniques that extract structural and functional information on different length scales. Although the preservation of biological functionality after cryo-arrest will have to be verified on a case-by-case basis, our results demonstrate the applicability of reversible cryo-arrest to investigate molecular patterns with high precision during dynamic processes in cells.
Online Methods
Preparation of cover slides for cell culture
A 10 μm thick double-sided adhesive tape (Modulor GmbH, Berlin, Germany) was cut to the size of a 21 × 26 mm cover slide (Gerhard Menzel GmbH, Braunschweig, Germany). In the middle a rectangular area with the dimension of the cavity of the aluminum flow chamber was cut out. The adhesive tape was glued on the cover slides (keeping the upper side of the tape covered by the release liner), sterilized in ethanol and washed with sterile H2O. Cover slides with adhesive tape facing upwards were placed in sterile 6-well-plates for cell culture (Sarstedt AG & Co, Nümbrecht, Germany) (Supplementary Fig. 10a).
Cell Culture and Transfection
HeLa (ATCC No. CCL-185), MDCK (ATCC No. CCL-34), Cos7 (ATCC No. CRL-1651), and MCF7 cells (ATCC No. HTB-22) were obtained from ATCC without further authentication. HCT116 (ATCC No. CCL-247) were tested by short tandem repeat analysis (IFG, Münster, Germany), which indicated presence of chromosomal instability or polyploidy. All cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 µg mL-1 streptomycin, 100 U mL-1 penicillin, 1% L-Glutamine (200mM), and 1% nonessential amino acids (all PAN-Biotech, Aidenbach, Germany). Cells were regularly tested for mycoplasma infection with the Mycoalert™ Mycoplasma detection kit (Lonza, Basel, Switzerland) and cultured at 37°C with 95% air and 5% CO2. Cells were plated on the prepared cover slides in 6-well dishes at a concentration of 0.2-1x105 cells per well at least three days before the experiment. 24-48 h after seeding, HeLa, MCF7 and Cos7 cells were transfected using FuGENE™ 6 (Promega, Madison, WI, USA); MDCK cells were transfected with Effectene transfection reagent (Qiagen, Hilden, Germany) and HCT116 cells were transfected with lipofectamine 2000 (Fisher Scientific GmbH, Schwerte, Germany) following the standard protocols of the manufacturers.
Mounting of the cryo-stage and cryo-arrest
The cover slides with cells were removed from the 6-well plates. The release liner of the double-sided adhesive tape was removed and the cover slides were glued to the flow chamber. The rectangular part in the middle without adhesive tape was mounted right under the cavity of the flow chamber (Supplementary Fig. 1a, b and 10b). Care was taken to keep the cells moist by medium during this process. The cryo-stage was assembled as shown in supplementary figure 1. Solution exchanges during cooling and warming are realized with the help of a neMESYS low-pressure syringe-pump (Cetoni GmbH, Korbußen, Germany) that enables a constant flow rate of 3 µL s-1. For live-cell imaging, cells were flushed with HEPES buffered DMEM medium without phenol red and temperature was set to 37°C. For cryo-arrest, temperature and medium changes were performed as described in figure 1a. All experiments were performed using the maximum cooling and warming rates that within a cryo-cycle vary from −35°C min-1 to −50°C min-1 and 130°C min-1 to 75°C min-1, respectively.
Propidium iodide (PI) staining
For assessment of survival, cells transfected with eGFP were stained within the cryo-stage with 100 ng mL-1 PI for 5 min. Subsequently cells were imaged with filter sets for green (eGFP) and red (PI) fluorescence. The number of eGFP-transfected, PI-positive cells was normalized to the total number of eGFP-positive cells. The survival of each cell line was tested in three independent experiments with a total number of 64 (Cos7), 70 (HeLa), 100 (HCT116), 77 (MCF7) and 73 (MDCK) cells.
Quantification of stress response gene expression
The expression levels of the stress induced heat shock protein HSP70 and the cold-stress induced RNA binding protein CIRBP were tested in HeLa cells by quantitative real-time PCR 15 min after cryo-arrest (n=6 experiments for each condition). As a positive control for heat stress, cells were treated for 30 min at 42°C. As a positive control for mild cold stress, cells were incubated for 24 h at 32°C. Afterwards, the mRNA of cells was extracted directly out of the microscopy stage. For this, 300 µL RNA lysis buffer (Zymo Research, Freiburg, Germany) were conducted through the stage with a flow of 3 µL s-1 and collected directly after the stage in RNA free tubes (Eppendorf AG, Hamburg, Germany). Purification of the mRNA was done with the Quick-RNA™ MicroPrep kit (Zymo Research, Freiburg, Germany) according to the standard procedures of the manufacturer. Quality and amount of RNA were measured by measuring adsorption of 230, 260 and 280 nm in a spectrophotometer (Nanodrop products, Wilmington, DE, U.S.A). A one-step reverse transcription-quantitative PCR (RT-qPCR) protocol was performed using the GoTaq™ 1-Step RT-qPCR System (Promega Madison, WI, USA). Specifically, 10 ng input RNA was used in a 20 µL reaction in a 96-well thermal plate. All samples were tested in duplicate. The plates were sealed and run in an IQ5 real-time PCR system cycler (Bio-Rad, Munich, Germany). Cycling conditions: 40 cycles of 95°C for 10 sec, 57°C for 30 sec. Data were analyzed using the ΔΔCt method for determination of relative gene expression by normalization to an internal control gene (GAPDH) and fold expression change was determined compared to untreated control samples. The following primers were used for the qPCR: human GAPDH (sense, 5’-TTCATTGACCTCAACTACAT-3’; antisense, 5’-GTGGCAGTGATGGCATGGAC-3’); human CIRBP (cold inducible RNA binding protein, Gene ID: 1153) (sense, 5’-GGTCTTCTCAAAGTACGG-3’; antisense, 5’-CATCATGGCATCCTTAGC-3’); human HSPA1A (heat shock protein family A (Hsp70) member 1A, Gene ID: 3303) (sense, 5’-CCTACTTCAACGACTCG-3’; antisense, 5’-AGGTCAAAGATGAGCAC-3’).
Confocal imaging of EGFR and DiOC18
HeLa cells were transfected with SNAP-EGFR (SNAP-tag fused to the extracellular N-terminus of EGFR 20) one day before the experiment and starved over night. Cholesterol extraction was done by incubating the cells in 10 mM methyl-β-cyclodextrin in medium without FBS for 60 min (ref. 50). Cells were labeled for 15 min with 20 μM SNAP-Surface Alexa-647 (New England Biolabs GmbH, Frankfurt, Germany) in FBS-free medium and subsequently labeled with DiOC18 in FBS-free medium for 2 min (Vybrant-DiO 1:200; Thermo Fisher Scientific, Waltham, MA, USA). This lipid-dye prefers liquid-ordered over disordered domains 50. Cells were imaged on a confocal microscope (Leica SP8) with a 63x 1.4 NA objective (Leica Microsystems CMS GmbH, Mannheim, Germany). To create scatterplots, intensity values of both channels were plotted pixel-by-pixel against each other using ImageJ software (Rasband, ImageJ, National Institutes of Health, Bethesda, MD, USA). To quantify the extent of correlation the image correlation quotient (ICQ) 51 was computed using ImageJ. To determine the ICQ, the number of pixels in which the intensity values deviate in both fluorescence intensities in the same direction from their respective means is divided by the total number of pixels. Afterwards a value of 0.5 is subtracted. This results in a value of 0.5 for absolutely co-localized fluorescence intensities and -0.5 for completely separated fluorescence intensities 51. Three independent experiments were performed and a total of 12 cholesterol depleted and 6 control cells were imaged.
Measurement of kinase activities by kinase translocation reporters (KTR)
HeLa cells were transfected with ERK-KTR, JNK-KTR or p38-KTR 30 one day before the experiment and starved over night. These reporter construct translocate from the nucleus to the cytoplasm of cells, when the kinase is active 30. Cells were imaged using a 100x 1.3NA objective and filter settings for green fluorescence. The amount of translocation was quantified by dividing the mean fluorescence in the cell by the mean fluorescence in the nucleus. For each condition three independent experiments were performed. Cells: ERK-KTR: ncontrol=6; nEGF(±cryo-arrest)=6; ncryo-arrest=10, nEGF=12, JNK-KTR: ncryo-arrest=31; nanisomycin=23, p38-KTR ncryo-arrest=18; nanisomycin=13 (p38).
Super-Resolution Microscopy
The microscope was built up on an Olympus IX-81 body, equipped with a total internal reflection fluorescence illuminator and a PLAPON60xO/TIRFM-SP objective (NA 1.45; Olympus GmbH, Hamburg, Germany). A diode laser emitting at 405 nm (Toptica Photonics, Graefelfing, Germany), the 476 nm laser line of an argon laser and a 561 nm solid-state laser (both Coherent GmbH, Dieburg, Germany) were coupled in using single-mode optical fibers. Emission light was spectrally separated with a 570 LP dichroic mirror and a 575-625 band pass filter (Olympus GmbH, Hamburg, Germany). Light detection was done using an EM-CCD camera with an effective pixel size of 107 nm (iXon EM-CCD; Andor Technology Ltd., Belfast, UK).
For PALM measurements, HeLa cells were transfected with EGFR-mEos2 (mEos2 fused to the intracellular C-terminus of EGFR) 24 h before the experiments and starved 12 h prior to the experiment in medium without FBS. 1000 frames of 100 ms exposure time were recorded for each super-resolution image. Super-resolution images were reconstructed using the ThunderSTORM Plugin (Ovesny, Bioinformatics, 2014) for ImageJ (Rasband, ImageJ, National Institutes of Health, Bethesda, MD, USA). Localization precision was calculated by ThunderSTORM as described 52.
For single-particle tracking, HeLa cells were transfected with SNAP-EGFR or EGFR-mCitrine 24 h before the experiments and starved 12 h prior to the experiment in medium without FBS. For EGF-Quantum Dot conjugation, 3 μL Quantum Dots (Qdot 705 with streptavidin; 1 uM; Thermo Fisher Scientific, Waltham, MA, USA) + 72 μL PBS with 0.1%BSA were mixed with 4.7 μL biotin-EGF (40 μg mL-1) + 70.3 μL PBS with 0.1% BSA. This solution was incubated for 30 min at 4°C with slow rotation on a spinning wheel and purified on a PD SpinTrap G-25 column according to the manufacturers instructions. HeLa cells were incubated with a 1:200 dilution in imaging medium for 5 min. For SNAP-tag labeling, cells were incubated with 500 nM benzylguanine-Cy3 in medium containing 0.5% BSA for 5 min at 37°C. Afterwards cells were washed three times in medium containing 0.5% BSA and incubated for 15 min at 37°C before the experiment. 1000 frames of 31.6 ms exposure time were recorded for each single-particle tracking.
Lateral drift was corrected by cross-correlation using ThunderSTORM 53.
Cluster analysis
Single molecule localization maps were analyzed using density-based spatial clustering of applications with noise (DBSCAN) algorithm 36. Localizations with neighbors closer than 40 nm were grouped together and identified as members of the same cluster (eps radius = 40 nm). Clusters with less than five localizations were not considered in the quantification of the results. Using custom scripts written in Python, detected clusters were categorized by area and number of molecules per cluster and then quantified. Cluster area was expressed by its equivalent diameter in the plots. The center of each cluster was calculated and used for further analysis of the inter-cluster arrangement by DBSCAN (Supplementary Fig. 7c). Clusters were considered as neighbors if they were closer than 1 μm. The number of neighbors per node (node degree) of the generated networks of clusters was calculated using the NetworkX Python package 54.
For single-molecule localization imaging and cluster analysis during cryo-arrest, three independent experiments were performed at -45°C for stimulated cells (8 cells were analyzed in total) and two independent experiments were performed at -45°C without stimulation (8 cells were analyzed in total). Statistical significance was estimated by the two-sample Kolmogorov-Smirnov test and significance levels are indicated as: p<0.05 (*) and p<0.01 (**).
Single particle tracking analysis
Single particle tracks were identified using TrackPy Python package 55. Further analysis of the tracks using custom written Python scripts was performed to analyze the tracks, group them into tracks of confined and free moving particles and generate the mean-squared displacement curves. Only trajectories with more than 10 consecutive frames were considered for the analysis. To identify mobile and confined molecules, trajectories were classified into two states based on their diffusion coefficient. The threshold value was set to D0=0.015 μm2 s-1. Molecules diffusion coefficient > D0 were classified as mobile and molecules with diffusion constant ≤ D0 were classified as confined. Diffusion coefficients were estimated using the first 5 time points of the tracks. MSD curves were corrected for localization error 56.
Anisotropy
Anisotropy microscopy was performed before and 5 min after EGF stimulation in HeLa cells ectopically expressing EGFR-QG-mCitrine 23. Cells were either cryo-arrested at -45°C before anisotropy measurements or anisotropy was measured at 37°C in the cryo-stage. Cells were starved in FBS-free medium over night and imaged 24 h post-transfection. The microscope consisted of an Olympus IX-81 body equipped with a MT20 illumination system and CellR software (Olympus GmbH, Hamburg, Germany). A linear dichroic polarizer (Meadowlark Optics, Frederick, CO, USA) was placed in the illumination path of the microscope, and two identical polarizers in an external filter wheel were used to measure parallel (I||) and perpendicular (I⊥) polarization of the excitation light in two separate images per measurement. For bleaching steps a depolarization filter was inserted into the excitation path. Steady state anisotropy (ri) was calculated in each pixel i by:
Whereby the G-factor (Gi) was determined by acquiring the ratio of the intensities at perpendicular and parallel orientations for a fluorophore in solution (fluorescein) that has a steady-state anisotropy close to zero due to its fast rotation. Anisotropy analysis was performed using custom written Python scripts.
Fluorescence Recovery After Photobleaching (FRAP)
FRAP measurements were performed on a Leica SP5 confocal microscope (Leica Microsystems CMS GmbH, Mannheim, Germany). Cells transfected with EGFR-mCitrine were imaged by excitation with the 514-nm laser line of an argon laser at <10% power with 2.67 frames s-1. Cells transfected with eGFP were imaged by excitation with the 488-nm laser line with 5.2 frames s-1. Bleaching of a circular spot of 2 μm was performed by scanning with all laser lines of the argon laser at 80% intensity plus a 405-nm laser diode at 100% intensity. This resulted in a bleaching efficiency of ~80%. Bleaching without the 405-nm laser resulted in similar bleaching efficiencies but a fast recovery of fluorescence even in chemically fixated cells. This is presumably caused by a recovery of a reversible dark state. Fluorescence recovery was monitored for 100 frames (EGFR-mCitrine) or 150 frames (eGFP) after bleaching using the same settings as before bleaching. For representation, fluorescence intensity was normalized to pre-bleaching intensities and the first post-bleaching intensity was set to zero. Three independent experiments were performed for each condition and a total of 11 (EGFR 37°C), 19 (EGFR 4°C and -45°C) and 20 cells (eGFP) have been analyzed.
Fluorescence lifetime imaging microscopy (FLIM)
Time correlated single photon counting (TCSPC) FLIM of EGFR-mCitrine was performed on a microscope consisting of an Olympus IX-81 body (Olympus GmbH, Hamburg, Germany) and a time-correlated single-photon counting unit containing a picosecond-pulsed 515 nm diode laser (MicroTime 200; Picoquant, Berlin, Germany) as well as on a Leica SP8 microscope equipped with a white light laser that was set to 514 nm (Leica Microsystems CMS GmbH, Mannheim, Germany). HeLa cells (n=9 cells from 3 experiments) were co-transfected with EGFR-mCitrine and a phosphotyrosine binding domain (PTB) of Shc fused to mCherry in a ratio of 1:2, 24 h before the experiment. 12 h prior to the experiment, cells were starved in medium without FBS.
TCSPC-FLIM measurements of the genetically encoded conformational biosensor LIFEA2 24 were performed on a Olympus fluoview 1000 microscope (Olympus GmbH, Hamburg, Germany) and a time-correlated single-photon counting unit (LSM Upgrade Kit, Picoquant, Berlin, Germany) containing a picosecond-pulsed 470 nm diode laser (LDH 470, Picoquant, Berlin, Germany) as well as on a Leica SP8 microscope equipped with a white light laser that was set to 514 nm (Leica Microsystems CMS GmbH, Mannheim, Germany). One day prior to the experiment, Cos7 cells were transfected with LIFEA2 and BFP tagged c-Cbl in a ratio of 1:3. Two hours before the experiment the cells were starved in HEPES-buffered imaging medium containing 0.5% FBS. Clustered mouse ephrinA1-Fc (R&D Systems) fusion protein was used for LIFEA2 stimulation. Clustered ephrinA1-Fc was prepared by incubating with goat anti-human Fc at a ratio of 5:1 for 30 min at room temperature. Stimulations were carried out at 37°C for the indicated time (5 min, 20 min or 40 min) in HEPES buffered DMEM medium without phenol red. The recorded FLIM data was analyzed using custom written MATLAB and python codes for global analysis to reliably extract fluorescence lifetimes and quantify the fraction of LIFEA2 in the active conformation within each pixel in the image (α) 39. For this, we assumed a two state LIFEA2 conformation model, where the inactive and active state exhibit a defined FRET efficiency corresponding to two discrete, spatially invariant fluorescence lifetimes (τ1,τ2: global parameters). From the global analysis, an image of the fraction of active LIFEA2 (αi: local parameter at pixel i) was reconstructed.
3D confocal FLIM
FLIM images were acquired on the Leica SP8 microscope described in the previous section. For the cell presented in figure 6d and supplementary figure 9d, a total number of 36 frames were acquired in a total time of 37 min. The pixel size was 103 nm for the x-y dimensions and 300 nm for the z dimension. The 3D stack was deconvolved using the “Iterative Deconvolve 3D” plugin for ImageJ [http://www.optinav.info/Iterative-Deconvolve-3D.htm]. The PSF function was calculated based on the microscope configuration used for the acquisition of the stack by using “Deconvolve PSF 3D” plugin for ImageJ [http://www.optinav.info/Diffraction-PSF-3D.htm]. After deconvolution, an intensity threshold was applied in order to generate a mask to identify endosomes.
Spatial analysis of endosomes
For each cell, intensity images were Fourier transformed and high-pass filtered in the frequency domain. After transforming back the images, an intensity threshold was applied to generate the binary masks to identify endosomes. The masks were multiplied with the fluorescence intensity images and the following parameters were extracted for each punctum: position, area, intensity and α. Further spatial analysis was done by segmenting the cells in 4 concentric spatial bins with equal area (Supplementary Fig. 9b). For each area, mean α per endosome and the number of endosomes were quantified.
Code availability
Requests for the used python and MATLAB codes may be directed to the corresponding author.
Supplementary Material
Acknowledgments
The authors would like to thank M. Reichl, J. Luig and P. Glitz for excellent technical assistance and A. Krämer for help in writing the manuscript. This study was funded by the Fraunhofer Society and the Max-Planck Society for the Promotion of Science (CryoSystems grant to G.R.F. and P.I.H.B.) and the European Research Council (ERC AdG 322637 to P.I.H.B.).
Footnotes
Author Contributions
P.I.H.B., F.W. and G.R.F. conceived the project; M.E.M., J.H. and J.C. developed and tested the cryo-stage, performed and analyzed cell survival assay of HeLa cells, anisotropy and FLIM of EGFR-PTB; O.S., J.H. and M.E.M. performed and analyzed LIFEA2 FLIM measurements; J.H. developed the cryo-arrest protocol, performed and analyzed FRAP, confocal imaging of lipids, cell survival assay of additional cell lines and activity measurements of MAPKs; A.K., J.H. and M.E.M. performed and analyzed RNA extraction and qRT-PCR experiments; J.C., J.H. and M.E.M. performed and analyzed single particle tracking and PALM; P.I.H.B., J.H., J.C. and M.E.M. wrote the manuscript.
Competing Financial Interest
The authors declare no competing financial interest.
References
- 1.Tanaka KA, et al. Membrane molecules mobile even after chemical fixation. Nat Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 2.Saffarian S, Li Y, Elson EL, Pike LJ. Oligomerization of the EGF Receptor Investigated by Live Cell Fluorescence Intensity Distribution Analysis. Biophys J. 2007;93:1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnell U, Dijk F, Sjollema Ka, Giepmans BNG. Immunolabeling artifacts and the need for live-cell imaging. Nat Methods. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- 4.Pegg DE. In: Methods in Molecular Biology. Wolkers WF, Oldenhof H, editors. Vol. 1257. Springer; New York: 2015. pp. 3–19. [DOI] [PubMed] [Google Scholar]
- 5.Huebinger J, et al. Direct Measurement of Water States in Cryopreserved Cells Reveals Tolerance toward Ice Crystallization. Biophys J. 2015;110:1–10. doi: 10.1016/j.bpj.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 7.Dubochet J. Cryo-EM-the first thirty years. J Microsc. 2012;245:221–224. doi: 10.1111/j.1365-2818.2011.03569.x. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen DH, Mackenzie AP. Phase Diagram for the System Water–Dimethylsulphoxide. Nature. 1968;220:1315–1317. doi: 10.1038/2201315a0. [DOI] [PubMed] [Google Scholar]
- 9.Ott JB, Goates JR, Lamb JD. Solid-liquid phase equilibria in water + ethylene glycol. J Chem Thermodyn. 1972;4:123–126. [Google Scholar]
- 10.Santos NC, Figueira-Coelho J, Martins-Silva J, Saldanha C. Multidisciplinary utilization of dimethyl sulfoxide: Pharmacological, cellular, and molecular aspects. Biochem Pharmacol. 2003;65:1035–1041. doi: 10.1016/s0006-2952(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 11.Karow AM, Webb WR. Toxicity of various solute moderators used in hypothermia. Cryobiology. 1965;1:270–273. doi: 10.1016/0011-2240(65)90033-7. [DOI] [PubMed] [Google Scholar]
- 12.Farrant J. Mechanism of Cell Damage During Freezing and Thawing and its Prevention. Nature. 1965;205:1284–1287. [Google Scholar]
- 13.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014;24:959–976. doi: 10.1038/cr.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichinose J, Murata M, Yanagida T, Sako Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem Biophys Res Commun. 2004;324:1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]
- 16.Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008;26:316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- 17.Ariotti N, et al. Epidermal Growth Factor Receptor Activation Remodels the Plasma Membrane Lipid Environment To Induce Nanocluster Formation. Mol Cell Biol. 2010;30:3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peckys DB, Baudoin J-P, Eder M, Werner U, de Jonge N. Epidermal growth factor receptor subunit locations determined in hydrated cells with environmental scanning electron microscopy. Sci Rep. 2013;3:2626. doi: 10.1038/srep02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 2000;290:1567–1570. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- 20.Ibach J, et al. Single Particle Tracking Reveals that EGFR Signaling Activity Is Amplified in Clathrin-Coated Pits. PLoS One. 2015;10:e0143162. doi: 10.1371/journal.pone.0143162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Z, Zhang W, Yang Y, Xu L, Fang X. Single-molecule diffusion study of activated EGFR implicates its endocytic pathway. Biochem Biophys Res Commun. 2008;369:730–734. doi: 10.1016/j.bbrc.2008.02.084. [DOI] [PubMed] [Google Scholar]
- 22.Orr G, et al. Cholesterol Dictates the Freedom of EGF Receptors and HER2 in the Plane of the Membrane. Biophys J. 2005;89:1362–1373. doi: 10.1529/biophysj.104.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumdick M, et al. EGF-dependent re-routing of vesicular recycling switches spontaneous phosphorylation suppression to EGFR signaling. Elife. 2015;4 doi: 10.7554/eLife.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabet O, et al. Ubiquitination switches EphA2 vesicular traffic from a continuous safeguard to a finite signalling mode. Nat Commun. 2015;6:8047. doi: 10.1038/ncomms9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein R. Eph/ephrin signalling during development. Development. 2012;139:4105–9. doi: 10.1242/dev.074997. [DOI] [PubMed] [Google Scholar]
- 26.Kriete A, Papazoglou E, Edrissi B, Pais H, Pourrezaei K. Automated quantification of quantum-dot-labelled epidermal growth factor receptor internalization via multiscale image segmentation. J Microsc. 2006;222:22–27. doi: 10.1111/j.1365-2818.2006.01564.x. [DOI] [PubMed] [Google Scholar]
- 27.Morisaki T, McNally JG. Photoswitching-Free FRAP Analysis with a Genetically Encoded Fluorescent Tag. PLoS One. 2014;9:e107730. doi: 10.1371/journal.pone.0107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Offterdinger M, Georget V, Girod A, Bastiaens PIH. Imaging Phosphorylation Dynamics of the Epidermal Growth Factor Receptor. J Biol Chem. 2004;279:36972–36981. doi: 10.1074/jbc.M405830200. [DOI] [PubMed] [Google Scholar]
- 29.Haj FG. Imaging Sites of Receptor Dephosphorylation by PTP1B on the Surface of the Endoplasmic Reticulum. Science. 2002;295:1708–1711. doi: 10.1126/science.1067566. [DOI] [PubMed] [Google Scholar]
- 30.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gon Y, et al. Cooling and rewarming-induced IL-8 expression in human bronchial epithelial cells through p38 MAP kinase-dependent pathway. Biochem Biophys Res Commun. 1998;249:156–160. doi: 10.1006/bbrc.1998.9115. [DOI] [PubMed] [Google Scholar]
- 32.Al-Fageeh MB, Smales CM. Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochem J. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levental I, Grzybek M, Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc Natl Acad Sci U S A. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee I-H, et al. Live Cell Plasma Membranes Do Not Exhibit a Miscibility Phase Transition over a Wide Range of Temperatures. J Phys Chem B. 2015;119:4450–9. doi: 10.1021/jp512839q. [DOI] [PubMed] [Google Scholar]
- 35.Lambert S, Vind-Kezunovic D, Karvinen S, Gniadecki R. Ligand-independent activation of the EGFR by lipid raft disruption. J Invest Dermatol. 2006;126:954–962. doi: 10.1038/sj.jid.5700168. [DOI] [PubMed] [Google Scholar]
- 36.Ester M, Kriegel HP, Sander J, Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Second Int Conf Knowl Discov Data Min. 1996:226–231. doi:10.1.1.71.1980. [Google Scholar]
- 37.Squire A, Verveer PJ, Rocks O, Bastiaens PIH. Red-edge anisotropy microscopy enables dynamic imaging of homo-FRET between green fluorescent proteins in cells. J Struct Biol. 2004;147:62–69. doi: 10.1016/j.jsb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 39.Grecco HE, Roda-Navarro P, Verveer PJ. Global analysis of time correlated single photon counting FRET-FLIM data. Opt Express. 2009;17:6493. doi: 10.1364/oe.17.006493. [DOI] [PubMed] [Google Scholar]
- 40.Kaufmann R, Hagen C, Grünewald K. Fluorescence cryo-microscopy: current challenges and prospects. Curr Opin Chem Biol. 2014;20:86–91. doi: 10.1016/j.cbpa.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wäldchen S, Lehmann J, Klein T, van de Linde S, Sauer M. Light-induced cell damage in live-cell super-resolution microscopy. Sci Rep. 2015;5:15348. doi: 10.1038/srep15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufmann R, et al. Super-Resolution Microscopy Using Standard Fluorescent Proteins in Intact Cells under Cryo-Conditions. Nano Lett. 2014;14:4171–4175. doi: 10.1021/nl501870p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang Y-W, et al. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, et al. Three-dimensional super-resolution protein localization correlated with vitrified cellular context. Sci Rep. 2015;5:13017. doi: 10.1038/srep13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson ME, Malardier-Jugroot C, Head-Gordon T. Effects of co-solvents on peptide hydration water structure and dynamics. Phys Chem Chem Phys. 2010;12:393–405. doi: 10.1039/b915888j. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Gureasko J, Shen K, Cole Pa, Kuriyan J. An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Endres NF, et al. Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puri C, et al. Relationships between EGFR signaling-competent and endocytosis-competent membrane microdomains. Mol Biol Cell. 2005;16:2704–18. doi: 10.1091/mbc.E04-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yudushkin IA, et al. Live-Cell Imaging of Enzyme-Substrate Interaction Reveals Spatial Regulation of PTP1B. Science. 2007;315:115–119. doi: 10.1126/science.1134966. [DOI] [PubMed] [Google Scholar]
- 50.Hao M, Mukherjee S, Maxfield FR. Cholesterol depletion induces large scale domain segregation in living cell membranes. Proc Natl Acad Sci U S A. 2001;98:13072–7. doi: 10.1073/pnas.231377398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Q, et al. A Syntaxin 1, G o, and N-Type Calcium Channel Complex at a Presynaptic Nerve Terminal: Analysis by Quantitative Immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quan T, Zeng S, Huang Z-L. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging. J Biomed Opt. 2010;15:066005. doi: 10.1117/1.3505017. [DOI] [PubMed] [Google Scholar]
- 53.Mlodzianoski MJ, et al. Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt Express. 2011;19:15009. doi: 10.1364/OE.19.015009. [DOI] [PubMed] [Google Scholar]
- 54.Hagberg AA, Schult DA, Swart PJ. Exploring network structure, dynamics, and function using NetworkX. Proc 7th Python Sci Conf (SciPy 2008) 2008:11–15. [Google Scholar]
- 55.Allan D, et al. trackpy: Trackpy v0.2.4. 2014 doi: 10.5281/zenodo.12255. [DOI] [Google Scholar]
- 56.Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys Rev E - Stat Nonlinear Soft Matter Phys. 2010;82:1–13. doi: 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.