Abstract
The present study aimed to investigate the expression levels of components of the Hedgehog signaling pathway (HH) during the proliferation of a liver stem cell subgroup, namely small hepatocyte-like progenitor cells (SHPCs). Retrorsine-treated Fisher 344 rats underwent a partial hepatectomy (PH) to induce the proliferation of SHPCs, after which reverse transcription-polymerase chain reaction (PCR), quantitative PCR, immunohistochemistry and western blot analysis were performed to analyze the expression of various components of the HH in primary SHPCs at different times points post-PH. A number of components of the HH, including Indian hedgehog (IHH), patched (PTCH), smoothened and glioma-associated oncogene (GLI)1, 2 and 3, were continuously expressed and showed dynamic changes in proliferating SHPCs. In addition, the expression levels of IHH, PTCH and GLI1 were significantly different as compared with those of the control group at the same time point, and there were significant differences among the various time points in the experimental group (P<0.01). Furthermore, there was an association between the postoperative day and expression levels of HH components in the retrorsine-treated group. An immunohistochemical analysis demonstrated that PTCH was also expressed at the protein level. In conclusion, the results of the present study suggested that the HH was continuously activated during the proliferation of SHPCs, thus indicating that SHPCs may be a subgroup of stem cells that are regulated by the HH.
Keywords: Hedgehog signaling pathway, small hepatocyte-like progenitor cells, liver regeneration
Introduction
Retrorsine treatment combined with partial hepatecomy (PH) is a commonly used method to establish a model of liver regeneration (1). A subgroup of liver stem cells, termed small hepatocyte-like progenitor cells (SHPCs), has been previously shown to initiate liver regeneration in retrorsine-pretreated Fisher 344 rats that have undergone PH (2–3). The morphological characteristics and regeneration dynamics of SHPCs were found to be markedly different from the normal hepatic oval cells (1–3); however, the histological origin, cell biology characteristics and mechanisms that regulate the proliferation of SHPCs remain unclear.
In our previous study, it was demonstrated that the Hedgehog signaling pathway (HH) was activated during various liver injury processes, including liver regeneration (4). The HH regulates the development of the embryonic digestive tract and its subsidiary organs, including the liver and pancreas (5,6). The structures and functions of the various components of the HH have been reported in the literature (5,6). Briefly, the signaling molecules Indian hedgehog (IHH) and sonic hedgehog (SHH) bind to the Patched (PTCH) membrane receptor, relieving inhibition of the transmembrane transduction protein Smoothened (SMO) (5,6). This leads to the intracellular transduction of signals and eventual activation of the glioma-associated oncogene (GLI) family of zinc finger transcription factors, including GLI1, GLI2 and GLI3 (5,6). This in turn results in the expression of GLI target genes, including PTCH, GLI and cyclin D, ultimately leading to changes in the cell cycle and cell proliferation and differentiation. At present, there is no consensus regarding the role of the HH in the mature liver and liver regeneration process (7–10). Furthermore, to the best of our knowledge, there have been no previous studies regarding the expression and significance of the HH in the SHPC-mediated regeneration process in the liver. Therefore, the present study aimed to investigate the expression levels of various components of the HH, including IHH, SHH, PTCH, SMO, GLI1, GLI2 and GLI3, in primary SHPCs during liver regeneration, in order to elucidate the role of the HH in SHPC proliferation.
Materials and methods
Animals and liver regeneration model
A total of 140 male Fisher 344 rats (clean grade; average weight, 80 g; age, 5 weeks) were provided by the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China). After 1 week of adaptive feeding, the rats were randomly divided into four groups, as follows (35 rats/group): i) Normal control (N) group; ii) retrorsine-treated (R) group; iii) PH group; and iv) R plus PH group (R/PH or SHPC group). Animals were maintained in conditions described in a previous study (4). The present study was conducted in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (Bethesda, MA, USA). The animal use protocol has been reviewed and approved by the Institutional Animal Care and Use Committee of the China Academy of Chinese Medical Sciences (Beijing, China). All experimental procedures were approved by Animal Ethics Committee of Beijing University of Chinese Medicine under the guidelines issued by Regulations of Beijing Laboratory Animal Management (Beijing, China).
In order to induce SHPC proliferation, the rats in the R and R/PH groups were intraperitoneally injected with 30 mg/kg retrorsine (Sigma-Aldrich, St. Louis, MO, USA) once in week 6 and once in week 8, as previously described (1,2), The N and PH groups were injected with an equal volume of saline at these time points. At 5 weeks after the second dose (week 13), the PH and R/PH groups underwent a two-thirds PH (11), whereas the N and R groups underwent sham surgery to ensure that changes possibly caused by surgical trauma and narcotic drugs were accounted for in all groups (11). No mortalities caused by retrorsine administration and surgery occurred during the experiment. The rat-feeding and experiments were performed at the specific-pathogen-free Animal Experiment Center of the Dongfang Hospital (Beijing, China), and the surgical procedure was performed in accordance with the Regulations of Laboratory Animal Management (Beijing University of Chinese Medicine).
The rats from each group were sacrificed on days 2, 3, 4, 7, 14, 21 and 30 following PH, with 5 rats from each group sacrificed at each time point, according to previous studies (1,2). Following ligation and removal of the caudate lobe, the liver was separated in situ for preparation of primary SHPCs. The obtained caudate lobe was cut into 3–5 sections with a size of 0.5×0.5×0.2 cm, fixed in 10% paraformaldehyde and embedded in paraffin 24 h later. Sections were then stained with hematoxylin and eosin and the proliferation of SHPCs were identified by three pathologists.
Separation and preparation of primary hepatocytes
The separation of hepatic tissues and the preparation of primary hepatocytes were performed according to previous reports (12,13). Briefly, following ligation and resection of the caudate lobe, the in situ-isolated liver was perfused with 350 ml EDTA solution at 37°C in Hanks' balanced salt solution (HBSS; Biyuntian Biological Technology Co., Ltd., Shanghai, China) via the abdominal aorta, at a flow rate of 35 ml/min; the lavage solution was naturally discharged via the resected vena cava. Subsequently, Ca2+-enriched HBSS-dissolved 0.05% collagenase IV (<38°C; Sigma-Aldrich), was perfused at the same rate for 10 min. Upon separation of the liver capsule from the liver parenchyma, 150 ml William's E medium (4°C; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was perfused at a flow rate of 150 ml/min for 1 min. Next, the liver tissues were placed into a sterile mortar, chopped and homogenized, and then filtered using a stainless steel mesh (thickness, 150 µm; pore size, 80 µm) to eliminate the components of the liver parenchyma. The obtained filtrate was washed with 4°C William's E medium and centrifuged through a speed gradient to obtain the enriched primary hepatocytes or SHPCs. The centrifugation conditions were as follows, according to a previous study (13): Three times centrifugation (4°C) at 50 × g for 1 min to obtain the enriched primary hepatocytes (as the control); subsequently, the lamellar sediment was discarded and the supernatant was centrifuged three times at 150 × g for 5 min to obtain the enriched primary SHPCs. The obtained primary SHPCs were placed into cell preservation solution (ABY; Aohua Medical Technology Co., Ltd., Xiaogan, China)for polymerase chain reaction (PCR) and western blotting, or placed into a low temperature refrigerator at −80°C for future use.
Reverse transcription (RT)-PCR
RT-PCR was performed to detect the expression of molecular markers, including oval cell marker 6, cytokeratin 19 (CK19, hepatic duct system marker), CK18 (hepatocyte marker) and α-smooth muscle actin (hepatic stellate cell marker), within the primary SHPCs at various time points following PH. The primer sequences are shown in Tables I and II. PCR was performed using the CFX96 Touch real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the manufacturer's protocol. Briefly, total RNA was extracted from the primary SHPCs using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), followed by purification using DNase (Invitrogen; Thermo Fisher Scientific, Inc.). The purity was assessed by spectrophotometry. Total RNA (3 µg) was added into 20 µl PrimeScript RT kit (Takara Biotechnology Co., Ltd., Dalian, China) for reverse transcription into cDNA. cDNA (5 µl) was then added into a 20 µl PCR reaction mixture, containing 4 µl PrimeScript Buffer (5X), 1 µl PrimeScript RT Enzyme Mix I, 1 µl Oligo Dt Primer (50 µmol/l), 1 µl Random 6 mers (100 µmol/l) and 13 µl total RNA. The PCR system (15 µl for each sample) was as follows: 7.5 µl Premix Ex Taq (2X), 0.25 µl forward primer (10 µmol/l), 0.25 µl reverse primer (10 µmol/l) and 4 µl dH2O. The cycling conditions were as follows: 93°C for 5 min, followed by 35 cycles of 94°C for 30 sec, 54°C for 45 sec and 72°C for 1 min, and a final extension step at 72°C for 10 min. The reaction products were subjected to 2% agarose gel electrophoresis. Ethidium bromide was used to visualize the DNA ladder. The remaining cDNA was stored at −20°C until further use.
Table I.
Primer sequences for reverse transcription-polymerase chain reaction.
| Gene | Primers | Product size (bp) |
|---|---|---|
| CD45 | F: 5′-TGTCGTGACGATCAATGACCA-3′ | 306 |
| R: 5′-ACATCCACTTTGCCCTCTGCT-3′ | ||
| CD133 | F: 5′-ACCAGTTGCCTGACGGAAATC-3′ | 304 |
| R: 5′-GCGTACAAGACCCATTGCAGAT-3′ | ||
| CD117 | F: 5′-GGCATCACCATCAAAAACGTG-3′ | 317 |
| R: 5′-TGGCGTTCGTAATTGAAGTCG-3′ | ||
| CD326 | F: 5′-GATGAAGGCGGAGATGACTCAC-3′ | 315 |
| R: 5′-CGAGATGCAAATGTGTCCTGAA-3′ | ||
| CK18 | F: 5′-GGATGCTCCCAAATCTCAGGA-3′ | 331 |
| R: 5′-TGATTCCAGATGCAGAAGGACC-3′ | ||
| CK19 | F: 5′-TCCACACCAGGCATTGACCTA-3′ | 403 |
| R: 5′-GCCTCGACTTGATGTCCATGA-3′ | ||
| CLDN3 | F: 5′-CGAGCTCTCATCGTGGTGTCTA-3′ | 256 |
| R: 5′-CGTACAACCCAGTTCCCATCTC-3′ | ||
| c-Met | F: 5′-TCAACAGCGGCAATTCTAGACA-3′ | 364 |
| R: 5′-GTGGTGCAGCAGATGATCTCTG-3′ | ||
| Cx43 | F: 5′-TTCATGCTGGTGGTGTCCTTG-3′ | 323 |
| R: 5′-CGATTTTGCTCTGCGCTGTAGT-3′ | ||
| HNF3β | F: 5′-CCAACAAGATGCTGACGCTGA-3′ | 331 |
| R: 5′-TGAACCTGAGAAGCCTGTGTCC-3′ | ||
| HNF4α | F: 5′-AGGCAGTGCGTGGTAGACAAA-3′ | 421 |
| R: 5′-TGAACACCATGGACCTCTTGG-3′ | ||
| OV-6 | F: 5′-ATGACATGAAGGTTGTCCTCGG-3′ | 263 |
| R: 5′-CCCCTTGGTCTTAACATGCTGA-3′ | ||
| Sca-1 | F: 5′-GCACGATGATCCCATTTGGT-3′ | 230 |
| R: 5′-TGCTGCGTTGCAAAGATCTG-3′ | ||
| Thy1 | F: 5′-TGCCGTCATGAGAATAACACCA-3′ | 201 |
| R: 5′-ACACATGTAGTCGCCCTCATCC-3′ | ||
| SHH | F: 5′-AACTCACCCCCAATTACAACCC-3′ | 321 |
| R: 5′-GGATGCGAGCTTTGGATTCA-3′ | ||
| IHH | F: 5′-ACCGCGACCGAAATAAGTACG-3′ | 301 |
| R: 5′-AAAGCTCTCAGCCTGTTTGGC-3′ | ||
| PTCH | F: 5′-TGTTGGTGTGGACGACGTCTT-3′ | 354 |
| R: 5′-GGTTCAACTTGAATCACCCGG-3′ | ||
| VIM | F: 5′-TACATCGACAAGGTGCGCTTC-3′ | 422 |
| R: 5′-TCGATCTGGACATGCTGTTCC-3′ | ||
| α-SMA | F: 5′-TGGAGAAGAGCTACGAACTGCC-3′ | 342 |
| R: 5′-ATAGAGAAGCCAGGATGGAGCC-3′ |
F, forward; R, reverse; CK, cytokeratin; CLDN3, claudin-3; Cx43, connexin 43; HNF, hepatocyte nuclear factor; OV-6, oval cell marker 6; Sca-1, stem cell antigen 1; Thy1, thymocyte antigen 1; SHH, sonic hedgehog; IHH, Indian hedgehog; PTCH, patched; VIM, vimentin; α-SMA, α-smooth muscle actin.
Table II.
Reverse transcription-polymerase chain reaction primer sequences for components of the Hedgehog signaling pathway.
| Gene | Primers | Product size (bp) |
|---|---|---|
| SHH | F: 5′-AACTCACCCCCAATTACAACCC-3′ | 321 |
| R: 5′-GGATGCGAGCTTTGGATTCA-3′ | ||
| IHH | F: 5′-ACCGCGACCGAAATAAGTACG-3′ | 301 |
| R: 5′-AAAGCTCTCAGCCTGTTTGGC-3′ | ||
| PTCH | F: 5′-TGTTGGTGTGGACGACGTCTT-3′ | 354 |
| R: 5′-GGTTCAACTTGAATCACCCGG-3′ | ||
| SMO | F: 5′-CAATGTGAAGCACCCTTGGTG-3′ | 323 |
| R: 5′-CCATCTGCTCGGCAAACAAT-3′ | ||
| GLI1 | F: 5′-CCAGTGTCCTCGACTTGAGCAT-3′ | 323 |
| R: 5′-ACAATTCCTGCTGCGACTGAAC-3′ | ||
| GLI2 | F: 5′-TGGATCTCTGAACCAGTTTGCC-3′ | 325 |
| R: 5′-TCGGTGACGACTAGCTGTGTTG-3′ | ||
| GLI3 | F: 5′-ATCAAAATGGAGGCACACGG-3′ | 303 |
| R: 5′-CCCTGACATTAGGCTGGTATGG-3′ |
SHH, sonic hedgehog; IHH, Indian hedgehog; PTCH, patched; SMO, smoothened; GLI, glioma-associated oncogene; F, forward; R, reverse.
Quantitative PCR (qPCR)
A semi-quantitative fluorescence probe method was used to analyze the expression levels of IHH, PTCH and GLI1 within the primary cell extract of each group at various time points. The primers and fluorescent probe sequences are shown in Table III. PCR was performed on the stored cDNA using the PE 7000 automated fluorescence qPCR instrument (Perkin Elmer Corp., Waltham, MA, USA), according to the manufacturer's protocol. Briefly, 5 ml cDNA, primers and probes were added to the PCR reaction solution (7.5 µl Premix Ex Taq (2X) and 4 µl dH2O) and the cycling conditions were as follows: 40 Cycles of 93°C for 3 min, 93°C for 45 sec and 55°C for 1 min. Following the reaction, the computer automatically analyzed the results. GAPDH was used as the internal control. The relative mRNA expression levels were calculated using the 2−∆∆Cq method (14). Each reaction was repeated three times, and the final result for a specific time point is presented as the mean of these three measurements.
Table III.
Sequences of the primers and probes used for quantitative polymerase chain reaction.
| Gene | Primers and probes | Product size (bp) |
|---|---|---|
| IHH | F: 5′-CGGCCATCACTCAGAGGAAT-3′ | 104 |
| R: 5′-CTGCTAAGCGCGCCAGTAGT-3′ | ||
| P: 5′-FAM-TTTACACTATGAGGGCCGCGC-TAMRA-3′ | ||
| PTCH | F: 5′-GCCTTTCTGACAGCCATTGG-3′ | 103 |
| R: 5′-CACCCAGCAGAGTGGACACA-3′ | ||
| P: 5′-FAM-CAAGAACCACAGGGCTATGCTCGC-TAMRA-3′ | ||
| GLI1 | F: 5′-CCTGAAGTGGGCAGGTTAGG-3′ | 100 |
| R: 5′-GCTGAGTGTTGTCCAGGTCAAG-3′ | ||
| P: 5′-FAM-AGGGCAGGTGTGTAACCCTCTGG-TAMRA-3′ |
IHH, Indian hedgehog; PTCH, patched; GLI1, glioma-associated oncogene 1; F, forward; R, reverse; P, probe.
Immunohistochemical and western blot analyses
Immunohistochemical and western blot analyses were performed to detect the protein expression of IHH, PTCH and GLI1 within the liver tissue sections, according to previous studies (4,8,9,15). The following antibodies were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA) and used in the present study: Goat anti-IHH (C-15; immunohistochemistry) and anti-IHH (I-19; western blot) polyclonal antibodies (cat. nos. sc-1196 and sc-1782, respectively; 1:50); rabbit anti-PTCH polyclonal antibody (H-267; sc-9016; 1:50); goat anti-GLI1 polyclonal antibody (N-16; sc-6153; 1:50); and horseradish peroxidase-conjugated rabbit anti-goat (sc-2033; 1:1,000) and goat anti-rabbit (sc-2004; 1:1,000) polyclonal secondary antibodies. A negative control consisted of 0.01 mol/l phosphate-buffered saline instead of the primary antibody.
Statistical analysis
Data are presented as the mean ± standard deviation. Repeated measures analysis of variance was used to compare the expression levels of IHH, PTCH and GLI1 between the experimental and control groups at various time points. Statistical analyses were conducted with SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of HH components and other molecular markers
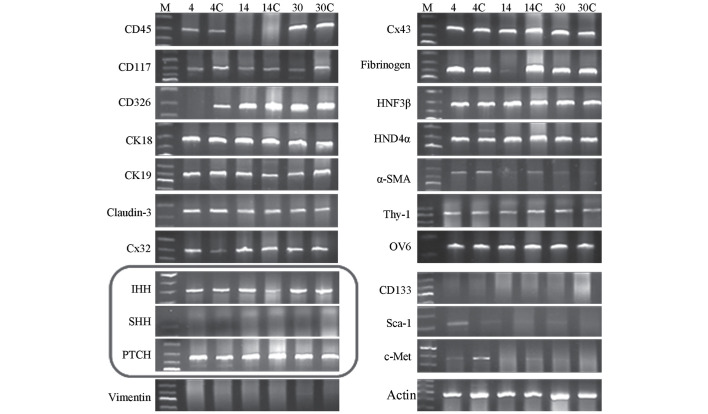
RT-PCR demonstrated that IHH and PTCH were continuously expressed in the early, middle and late stages of SHPC proliferation (postoperative days 4, 14 and 30, respectively), whereas SHH mRNA expression was negative. In addition, a number of other molecular markers were expressed in the primary SHPC extract, including oval cell marker 6, CK19, CK18 and α-smooth muscle actin. These results suggested there were a variety of cell components in the extract (Fig. 1), in agreement with previous findings (16–20).
Figure 1.
Expression of various molecular markers in the cell homogenate of primary SHPCs. Reverse transcription-polymerase chain reaction demonstrated that numerous molecular markers were expressed in primary SHPCs undergoing proliferation in the early, middle and late stages of liver regeneration (postoperative days 4, 14 and 30, respectively). Primary SHPCs from the retrorsine-treated group within the same period (postoperative days 4, 14 and 30) were set as the controls (4C, 14C and 30C, respectively). The circled pane shows that IHH and PTCH were continuously expressed, whereas SHH was not. SHPCs, small hepatocyte-like progenitor cells; IHH, Indian hedgehog; PTCH, patched; SHH, sonic hedgehog.
According to Gordon et al (1,2), SHPC regeneration presented a diffused distribution and continued proliferation. In the present study, RT-PCR detected the continuous expression of HH-associated genes on postoperative days 2, 3, 4, 7, 14, 21 and 30. IHH, PTCH, GLI1, GLI2 and GLI3 mRNA expression was consistently observed in the primary SHPCs, whereas SMO was expressed at a low level (Fig. 2) and the expression of SHH was negligible. These results confirm the continued activation of HH in the process of proliferation of SHPCs.
Figure 2.
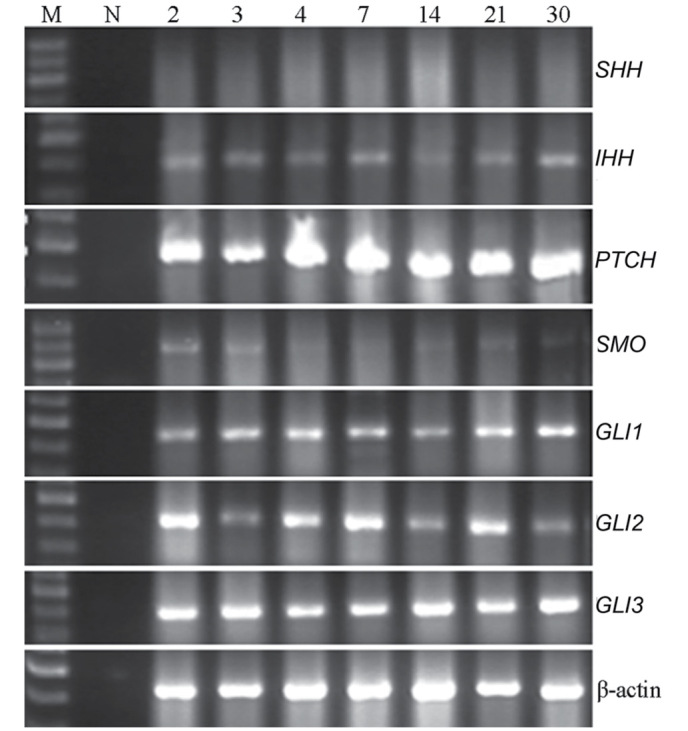
Expression levels of various Hedgehog signaling pathway components in proliferating SHPCs. On postoperative days 2, 3, 4, 7, 14, 21 and 30, IHH, PTCH, GLI1, GLI2 and GLI were continuously expressed in the cell homogenate of primary SHPCs, whereas SMO was only weakly expressed and SHH expression was negligible. SHPCs, small hepatocyte-like progenitor cells; M, DNA marker; N, negative control; SHH, sonic hedgehog; PTCH, patched; IHH, Indian hedgehog; SMO, smoothened; GLI, glioma-associated oncogene.
Activation status of the HH during the proliferation of SHPCs
In order to further clarify the role of HH activation in the process of liver regeneration, qPCR was performed to detect the expression levels of IHH, PTCH and GLI1 in the SHPC extract at various time points. IHH is a signaling molecule that is synthesized and released into the microenvironment. The synthesis of GLI1 is entirely dependent on the stimulation of the HH, such that GLI1 is considered a direct marker of HH activation (8,21,22). In addition, PTCH is not only a membrane receptor for the HH, but also a target protein of HH activation, which may establish a negative feedback mechanism that helps to avoid the abnormal or excessive activation of HH (8,21). Therefore, upregulation of PTCH expression has been regarded as a sign of the constitutive activation of the HH (8,21).
In the present study, the expression levels of IHH, PTCH and GLI1 were significantly different in proliferating SHPCs, as compared with the control group at the same time points, as well as among the different time points within the R/PH group (P<0.01). Furthermore, there was an association between the postoperative time point and the expression levels of the HH components in the R/PH group. However, the same association was not observed for the N and R groups (P>0.05; Tables IV–VI). In contrast to the expression in the N, R and PH groups, significantly higher expression levels of IHH and GLI1 were observed during the early stage of SHPC proliferation in the R/PH group (the earliest detection point in this experiment was day 2 post-PH). Conversely, the expression levels of IHH and GLI1 gradually decreased after reaching a peak on day 3 post-PH. Subsequently, IHH and GLI1 expression levels approached similar levels as in the control group on days 4–7 post-PH, although the difference between the control and R/PH groups remained significant (P<0.05), with the exception of IHH expression on the days 4 and 21 post-PH (Tables IV–VI). The expression of PTCH exhibited the opposite pattern, since it was lower compared with that in the N group at the early stages, but gradually increased and reached a peak on day 14 post-PH. In addition, the expression levels of PTCH remained elevated until day 30 post-PH.
Table IV.
Expression of Indian hedgehog during the regeneration of small hepatocyte-like progenitor cells.
| Timea | n | N | R | PH | R/PH | P-value |
|---|---|---|---|---|---|---|
| 2 | 5 | 0.461±0.014 | 0.544±0.032 | 0.765±0.009 | 0.651±0.022 | 0.014 |
| 3 | 5 | 0.463±0.005 | 0.546±0.016 | 0.610±0.100 | 0.987±0.100 | <0.001 |
| 4 | 5 | 0.456±0.020 | 0.552±0.025 | 0.523±0.010 | 0.554±0.070 | 0.060 |
| 7 | 5 | 0.493±0.015 | 0.560±0.013 | 0.300±0.047 | 0.543±0.010 | 0.020 |
| 14 | 5 | 0.556±0.009 | 0.539±0.060 | 0.443±0.013 | 0.534±0.080 | 0.041 |
| 21 | 5 | 0.562±0.030 | 0.566±0.010 | 0.553±0.050 | 0.507±0.010 | 0.120 |
| 30 | 5 | 0.555±0.060 | 0.560±0.015 | 0.672±0.020 | 0.536±0.030 | 0.016 |
| P-value | 0.54 | 0.39 | 0.002 | <0.001 |
Days post-PH. Data are presented as the mean ± standard deviation. N, normal control group; R, retrorsine-treated group; PH, partial hepatectomy; R/PH, retrorsine-treated plus PH group.
Table VI.
Expression of patched during the regeneration of small hepatocyte-like progenitor cells.
| Timea | n | N | R | PH | R/PH | P-value |
|---|---|---|---|---|---|---|
| 2 | 5 | 0.161±0.014 | 0.185±0.060 | 0.166±0.080 | 0.150±0.022 | 0.032 |
| 3 | 5 | 0.164±0.004 | 0.178±0.090 | 0.219±0.040 | 0.186±0.110 | 0.019 |
| 4 | 5 | 0.155±0.090 | 0.193±0.040 | 0.323±0.010 | 0.254±0.040 | 0.006 |
| 7 | 5 | 0.182±0.012 | 0.189±0.030 | 0.290±0.037 | 0.343±0.010 | <0.001 |
| 14 | 5 | 0.156±0.010 | 0.168±0.015 | 0.241±0.020 | 0.534±0.050 | <0.001 |
| 21 | 5 | 0.161±0.030 | 0.175±0.011 | 0.154±0.045 | 0.507±0.010 | <0.001 |
| 30 | 5 | 0.155±0.060 | 0.180±0.020 | 0.152±0.020 | 0.536±0.028 | <0.001 |
| P-value | 0.48 | 0.52 | <0.001 | <0.001 |
Days post-PH. Data are presented as the mean ± standard deviation. There was no significant difference among the time points of the N group (P>0.05). N, normal control group; R, retrorsine-treated group; PH, partial hepatectomy; R/PH, retrorsine-treated plus PH group.
Expression of HH components at the protein level
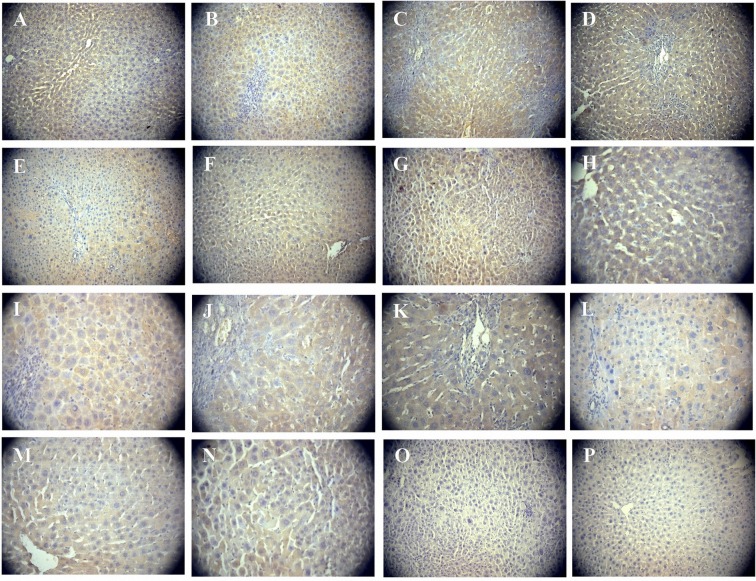
The protein expression levels of IHH, PTCH and GLI1 were detected by immunohistochemical and western blot analyses. In proliferating SHPCs, PTCH was constitutively expressed at a high level and, in the post-PH day 30 liver tissue sections, a large number of cells with brown membranes and a stained cytoplasm were diffused within the liver parenchyma (Fig. 3). By contrast, the expression of IHH was negative (Fig. 3) and GLI1 exhibited weak non-specific staining, and was thus difficult to confirm its expression (data not shown). Western blot analysis was unable to detect the expression of the three indicators.
Figure 3.
Immunohistochemical detection of PTCH and IHH in proliferating SHPCs. Immunohistochemistry detected the expression levels of PTCH and IHH in regenerated liver tissues during SHPC proliferation. (A)-(G), magnification ×200; (H)-(N), magnification ×400; (O) and (P), magnification ×200. PTCH was continuously strongly expressed on postoperative days (A and H) 2, (B and I) 3, (C and J) 4, (D and K) 7, (E and L) 14, (F and M) 21 and (G and N) 30, as demonstrated by cells with large brown membranes or cytoplasmic staining, diffused inside the liver parenchyma. (O) Negative control without the anti-PTCH antibody. (P) Regenerated liver tissue sections showed no IHH expression. PTCH, patched; IHH, Indian hedgehog; SHPC, small hepatocyte-like progenitor cells.
Discussion
Retrorsine-treated Fisher 344 rats may be able to complete the compensation of hepatic structural defects and functional loss by SHPCs in one time- and space-specific way after PH (1–3). Previous studies have demonstrated that SHPCs are a group of non-oval, non-bile duct cells that may be a stem cell subgroup derived from mature hepatocytes (23–25). However, the following features of SHPCs have yet to be fully elucidated: i) The histological origin and anatomical location of SHPCs; ii) the characteristic cytobiological features of SHPCs, in particular specific molecular markers and gene/protein expression profiles; iii) the mechanisms and signaling pathways underlying the regulation of SHPC proliferation; and iv) evidence that SHPCs exist in other mammalian species.
In previous studies, SHPC regeneration presented a diffused distribution and continued proliferation (1,2). Therefore, the present study aimed to identify the signaling pathways involved in SHPC proliferation. RT-PCR demonstrated that numerous components of the HH were continuously expressed in primary SHPCs undergoing proliferation, and a quantitative analysis suggested that alterations in the expression levels of IHH, PTCH and GLI1 were indicative of HH activation. These results suggested that, during the process of SHPC proliferation, IHH is secreted and binds to the membrane receptor PTCH in order to reduce the expression of PTCH, and relieve the inhibition of the HH, thus upregulating the expression of the GLI1 target gene. However, following sustained activation of the pathway, the expression of PTCH was increased and ultimately sustained at a high level, thereby providing a negative feedback mechanism that inhibited excessive activation of the HH. This was accompanied by a gradual decrease in the expression levels of IHH and GLI1, reflecting the continuous activation of HH and the PTCH-mediated negative feedback mechanism (5,6). Notably, the results of the present study were consistent with those of Ochoa et al (10) and Agarwal et al (26), who also analyzed the expression of PTCH, SMO, GLI1, GLI2 and GLI3 in liver tissues. However, in contrast to these experiments, the present study was able to detect the expression of IHH, but not SHH, in the experimental and control groups. These results suggested that, in proliferating SHPCs, the HH may exert its physiological role via IHH rather than SHH, although these discrepancies may be associated with differences in the modeling method or differences in the genus/species used, and thus further verification is required.
The present study used immunohistochemistry to analyze the expression of HH components at the protein level and demonstrated that PTCH was continuously expressed in proliferating SHPCs. By contrast, western blot analysis was unable to further validate the results of the immunohistochemical analysis, which may have been due to a lack of reliable antibodies for detection of the mammalian liver HH molecules (4,9,10,15). Another reason for the unsuccessful detection may have been that, during the activation of the HH, the binding of the IHH signaling molecule to the PTCH membrane receptor may have altered the structure and location of the proteins, which in turn may have affected their extraction, identification and reaction with the antibodies (5,6,21).
In conclusion, to the best of our knowledge, the present study is the first to report the constitutive activation of a signaling pathway, in particular the HH, in proliferating SHPCs. The results of the present study suggested that the HH may serve an important role in the proliferation of SHPCs, and that SHPCs may be an HH-regulated stem cell subgroup. Further studies are required to validate these results, elucidate the histological position of SHPCs within the liver and observe the impacts of pathway blockage on SHPC proliferation, in order to delineate the regulatory mechanisms underlying SHPC proliferation and identify the role of the HH in liver regeneration.
Table V.
Expression of glioma-associated oncogene 1 during the regeneration of small hepatocyte-like progenitor cells.
| Timea | n | N | R | PH | R/PH | P-value |
|---|---|---|---|---|---|---|
| 2 | 5 | 0.261±0.014 | 0.280±0.007 | 0.566±0.001 | 0.651±0.022 | 0.021 |
| 3 | 5 | 0.263±0.005 | 0.277±0.004 | 0.610±0.100 | 0.780±0.100 | 0.034 |
| 4 | 5 | 0.255±0.090 | 0.290±0.005 | 0.523±0.010 | 0.354±0.040 | 0.027 |
| 7 | 5 | 0.262±0.015 | 0.287±0.030 | 0.270±0.047 | 0.343±0.010 | 0.004 |
| 14 | 5 | 0.256±0.020 | 0.274±0.014 | 0.241±0.013 | 0.311±0.005 | 0.017 |
| 21 | 5 | 0.261±0.030 | 0.277±0.010 | 0.154±0.050 | 0.287±0.010 | <0.001 |
| 30 | 5 | 0.255±0.060 | 0.280±0.040 | 0.172±0.020 | 0.290±0.030 | <0.001 |
| P-value | 0.245 | 0.43 | <0.01 | <0.01 |
Days post-PH. Data are presented as the mean ± standard deviation. N, normal control group; R, retrorsine-treated group; PH, partial hepatectomy; R/PH, retrorsine-treated plus PH group.
References
- 1.Gordon GJ, Coleman WB, Hixson DC, Grisham JW. Liver regeneration in rats with retrorsine-induced hepatocellular injury proceeds through a novel cellular response. Am J Pathol. 2000;156:607–619. doi: 10.1016/S0002-9440(10)64765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon GJ, Coleman WB, Grisham JW. Temporal analysis of hepatocyte differentiation by small hepatocyte-like progenitor cells during liver regeneration in retrorsine-exposed rats. Am J Pathol. 2000;157:771–786. doi: 10.1016/S0002-9440(10)64591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best D Hunter, Coleman WB. Cells of origin of small hepatocyte-like progenitor cells in the retrorsine model of rat liver injury and regeneration. J Hepatol. 2008;48:369–371. doi: 10.1016/j.jhep.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y, Zheng H, Gong W, Che Y, Jiang B. The role of hedgehog signaling pathway in liver regeneration. Hepatogastroenterology. 2011;58:2071–2076. doi: 10.5754/hge11155. [DOI] [PubMed] [Google Scholar]
- 5.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 2014;141:3445–3457. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merchant JL. Hedgehog signalling in gut development, physiology and cancer. J Physiol. 2012;590:421–432. doi: 10.1113/jphysiol.2011.220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMillan R, Matsui W. Molecular pathways: The hedgehog signaling pathway in cancer. Clin Cancer Res. 2012;18:4883–4888. doi: 10.1158/1078-0432.CCR-11-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Büller NV, Rosekrans SL, Westerlund J, van den Brink GR. Hedgehog signaling and maintenance of homeostasis in the intestinal epithelium. Physiology (Bethesda) 2012;27:148–155. doi: 10.1152/physiol.00003.2012. [DOI] [PubMed] [Google Scholar]
- 9.Tang L, Tan YX, Jiang BG, Pan YF, Li SX, Yang GZ, Wang M, Wang Q, Zhang J, Zhou WP, et al. The prognostic significance and therapeutic potential of hedgehog signaling in intrahepatic cholangiocellular carcinoma. Clin Cancer Res. 2013;19:2014–2024. doi: 10.1158/1078-0432.CCR-12-0349. [DOI] [PubMed] [Google Scholar]
- 10.Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, Zubiaga AM, Fresnedo O, Omenetti A, Zdanowicz M, et al. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 2010;51:1712–1723. doi: 10.1002/hep.23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmes D, Spiegel HU. Animal models of liver regeneration. Biomaterials. 2004;25:1601–1611. doi: 10.1016/S0142-9612(03)00508-8. [DOI] [PubMed] [Google Scholar]
- 12.Papeleu P, Vanhaecke T, Henkens T, Elaut G, Vinken M, Snykers S, Rogiers V. Isolation of rat hepatocytes. Methods Mol Biol. 2006;320:229–237. doi: 10.1385/1-59259-998-2:229. [DOI] [PubMed] [Google Scholar]
- 13.Gordon GJ, Butz GM, Grisham JW, Coleman WB. Isolation, short-term culture and transplantation of small hepatocyte-like progenitor cells from retrorsine-exposed rats. Transplantation. 2002;73:1236–1243. doi: 10.1097/00007890-200204270-00008. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Furmanski AL, Saldana JI, Ono M, Sahni H, Paschalidis N, D'Acquisto F, Crompton T. Tissue-derived hedgehog proteins modulate the differentiation and disease. J Immunol. 2013;190:2641–2649. doi: 10.4049/jimmunol.1202541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Than NN, Newsome PN. Stem cells for liver regeneration. QJM. 2014;107:417–421. doi: 10.1093/qjmed/hcu013. [DOI] [PubMed] [Google Scholar]
- 17.Snykers S, De Kock J, Vanhaecke T, Rogiers V. Differentiation of neonatal rat epithelial cells from biliary origin into immature hepatic cells by sequential exposure to hepatogenic cytokines and growth factors reflecting liver development. Toxicol In Vitro. 2007;21:1325–1331. doi: 10.1016/j.tiv.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: Their characteristics and regulatory mechanisms. J Biochem. 2011;149:231–239. doi: 10.1093/jb/mvr001. [DOI] [PubMed] [Google Scholar]
- 19.Koenig S, Krause P, Drabent B, Schaeffner I, Christ B, Schwartz P, Unthan-Fechner K, Probst I. The expression of mesenchymal, neural and haematopoietic stem cell markers in adult hepatocytes proliferating in vitro. J Hepatol. 2006;44:1115–1124. doi: 10.1016/j.jhep.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Böhm F, Köhler UA, Speicher T, Werner S. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol Med. 2010;2:294–305. doi: 10.1002/emmm.201000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Sci Signal. 2012;5:re6. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machado MV, Michelotti GA, Tde A Pereira, Boursier J, Kruger L, Swiderska-Syn M, Karaca G, Xie G, Guy CD, Bohinc B, et al. Reduced lipoapoptosis, hedgehog pathway activation and fibrosis in caspase-2 deficient mice with non-alcoholic steatohepatitis. Gut. 2015;64:1148–1157. doi: 10.1136/gutjnl-2014-307362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitaka T, Ichinohe N, Kon J. Thy1-positive cell transplantation activates the growth of small hepatocyte-like progenitor cells in rat livers treated with retrorsine and PH. FASEB J. 2013;27:257. [Google Scholar]
- 24.Best DH, Coleman WB. Bile duct destruction by 4, 4′-diaminodiphenylmethane does not block the small hepatocyte-like progenitor cell response in retrorsine-exposed rats. Hepatology. 2007;46:1611–1619. doi: 10.1002/hep.21876. [DOI] [PubMed] [Google Scholar]
- 25.Best DH, Coleman WB. Treatment with 2-AAF blocks the small hepatocyte-like progenitor cell response in retrorsine-exposed rats. J Hepatol. 2007;46:1055–1063. doi: 10.1016/j.jhep.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal JR, Wang Q, Tanno T, Rasheed Z, Merchant A, Ghosh N, Borrello I, Huff CA, Parhami F, Matsui W. Activation of liver X receptors inhibits hedgehog signaling, clonogenic growth and self-renewal in multiple myeloma. Mol Cancer Ther. 2014;13:1873–1881. doi: 10.1158/1535-7163.MCT-13-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]