Abstract
Increasing evidence has suggested that microRNA (miRNA) may play a role in the pathogenesis of cardiovascular disease, which has led to a greater understanding of the complex pathophysiological processes underlying unstable angina (UA). The present study aimed to investigate changes in the miRNA expression profiles of patients with UA using gene-chip analysis, in order to further elucidate the pathogenesis of UA. Total RNA was extracted and purified from plasma samples collected from patients with UA and healthy controls. The samples underwent microarray analysis using an Exiqon miRCURY LNA™ microRNA Array. Differentially expressed miRNAs were identified by volcano plot filtering, and were validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). In addition, functional annotation of the differentially expressed miRNAs involved gene ontology analyses. Among the 212 miRNAs differentially expressed between the two groups, 82 were upregulated and 130 were downregulated. Notably, the results of the RT-qPCR were consistent with the gene-chip results. The miRNAs identified in the present study may be potential novel biomarkers for the prevention and early diagnosis of UA. Furthermore, the results of the present study suggested that UA occurs as a result of complex and dynamic processes regulated by numerous factors, including multiple miRNAs.
Keywords: unstable angina, microRNA, gene chip, reverse transcription-quantitative polymerase chain reaction
Introduction
Coronary artery disease (CHD) is an umbrella term for a group of cardiovascular diseases, including unstable angina (UA) and acute myocardial infarction (MI) (1). CHD is a leading cause of mortality in developed and certain developing countries (2). UA, whose prevalence in China has been increasing in male patients over the age of 40, is associated with MI and an increased risk of cardiac-associated mortality (3). At present, there is an absence of effective therapeutic strategies for UA due to a limited understanding of the molecular mechanisms underlying the disease. Therefore, significant improvements in its diagnosis and treatment may be achieved if the underlying pathogenesis of UA is elucidated.
MicroRNAs (miRNAs) are a group of endogenous non-coding small RNAs that are typically 19–24 nucleotides in length (4). They have important roles in the post-transcriptional regulation of endogenous gene expression by degrading target mRNAs and inhibiting the translation of target gene transcripts, thereby altering the transcriptome and proteome (5). miRNAs are able to regulate gene expression at the post-transcriptional level by binding to sequences in the 3′-untranslated regions (3′UTR) of target mRNAs (6,7). Complete complementarity with the 3′UTR of the target mRNA typically initiates its degradation, whereas incomplete complementarity may inhibit its translation by the ribosome (8,9). It has been reported that ~30% of protein-coding genes are regulated by miRNAs (9), and miRNAs have been shown to be key regulators of various biological processes, including cell growth, proliferation, differentiation and apoptosis (10). Furthermore, previous studies have associated miRNAs with the pathogenesis of various human diseases, including a number of malignancies, and miRNA has been shown to serve as a critical biological marker in numerous human diseases. Therefore, the analysis of miRNA expression profiles is important for the elucidation of their roles in the regulation of gene expression in various diseases (11).
Gene-chips, which are also termed DNA microarrays or biological chips, are highly parallel, high-throughput, miniaturized and automated platforms based on the theory of hybridization (12). Microarray technology has previously been applied to the elucidation of molecular mechanisms, drug screening and medical diagnostics, and has wide application prospects (13). In addition, microarrays serve as a global tool for the identification and characterization of gene expression profiles (14), due to their ability to analyze the differentially expressed genes of a whole genome in a single experiment (15).
The present study applied high-throughput gene-chips in the analysis of differentially expressed genes between patients with UA and healthy controls, in order to identify miRNAs involved in the pathogenesis of UA and potential biomarkers. Furthermore, the results of the microarray were independently validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Functional annotation of the differentially expressed miRNAs involved gene ontology (GO) analyses. The aim of the present study was to identify potential factors involved in the pathogenesis of UA, as well as critical miRNAs that may serve as potential diagnostic biomarkers.
Materials and methods
Ethics statement
The experimental protocol of the present study was approved by the ethics committee of the Hebei Medical University (Shijiazhuang, China). In addition, the present study was approved by the Institutional Review Board for Human Studies, and written informed consent was obtained from all patients.
Patients and controls
Plasma samples were taken from 175 healthy subjects and 150 patients with UA at the Tangshan Workers' Hospital (Tangshan, China) between January 2012 and June 2013, in order to establish seven plasma pools for the healthy subjects and six pools for the patients with UA. Each plasma pool consisted of 25 cases (100 µl plasma/case). UA patients were eligible to participate in the present study if they met the American College of Cardiology/American Heart Association criteria for UA (16).
Exiqon miRNA chip
The 6th generation of the miRCURY LNA™ microRNA Array (Exiqon A/S, Vedbaek, Denmark) contains >1,891 capture probes, covering all human, mouse and rat microRNAs annotated in the miRBase 16.0 (http://www.mirbase.org/), as well as all viral miRNAs associated with these species. In addition, the array contains capture probes for 66 novel miRPlus™ human miRNAs. Exiqon gene-chips are able to detect specific hybridization and to perform a rigid statistical analysis in order to reduce the rate of false-positive signals (17).
miRNA isolation
All plasma samples were subjected to RNA extraction. Briefly, total RNA was isolated from the plasma samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and an miRNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocols. This RNA extraction step efficiently recovered 90% RNA species, including miRNAs. The concentration and purity of the total RNA samples were assessed by measuring the absorbance at 260, 280 and 230 nm in a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity was determined by agarose gel electrophoresis (using 1% gels). Briefly, agarose (0.5 g; Zhongshan Jinqiao Biology & Technology Co., Ltd., Beijing, China) was dissolved in electrophoresis buffer (50 ml, Zhongshan Jinqiao Biology & Technology Co., Ltd.), following mixing. The sample was then added to the wells, and the voltage was adjusted to 100 v, with the RNA electrophoresis from anode to cathode. The gel was dyed for 5 min with EB dyeing liquid (Applygen Technologies Inc. Beijing, China).
cDNA probe preparation
Following RNA isolation from the plasma samples, RNA was labeled using the miRCURY LNA™ microRNA Array Hy3™/Hy5™ Power labeling kit (Exiqon A/S). Briefly, 1 µg of each sample was 3′-labeled with Hy3 fluorescent dye, using the T4 RNA ligase (provided in the kit), according to the following procedure: RNA in 2.0 µl water was combined with 1.0 µl calf intestinal phosphatase buffer (Exiqon A/S), after which the mixture was incubated for 30 min at 37°C, followed by termination of the reaction by incubating for 5 min at 95°C. Subsequently, 3.0 µl labeling buffer, 1.5 µl Hy3 and 2.0 µl labeling enzyme (both Beijing Zhongshan Jinqiao Biotechnology Co., Ltd. Beijing, China) were added to the mixture. The labeling reaction was incubated for 1 h at 16°C, and terminated by incubation for 15 min at 65°C.
Array hybridization
Following termination of the labeling reaction, the Hy3-labeled samples were hybridized to the miRCURY LNA™ microRNA Array, according to the manufacturer's protocol. Briefly, the total 25 µl mixture consisting of 5 µl Hy3-labeled samples and 20 µl hybridization buffer were denatured for 2 min at 95°C, incubated on ice for 2 min and then hybridized to the microarray for 16–20 h at 56°C in a 12-Bay NimbleGen Hybridization System (Roche Diagnostics, Basel, Switzerland), which provides an active mixing action and constant incubation temperature in order to improve hybridization uniformity and enhance signals. Following hybridization, the slides were obtained, washed three times using Wash buffer kit (Roche Diagnostics GmbH, Mannheim, Germany; cat no. 5188–5327) and finally dried by centrifugation for 5 min at 400 × g and 4°C. Subsequently, the slides were scanned using the Axon GenePix® 4000B Microarray Scanner (Molecular Devices, LLC, Sunnyvale, CA, USA).
Data analysis
Microarray data analysis was performed using a series of models in the limma package (version 3.22.7; https://bioconductor.org/packages/release/bioc/html/limma.html). Scanned images were then imported into GenePix® Pro 6.0 Microarray Acquisition and Analysis software (http://axon-genepix-pro.software.informer.com/6.0/) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs with intensities ≥30 in all samples were selected for normalization. Data was normalized using the Median normalization method and ComBat software (version 1.1.4; http://www.bu.edu/jlab/wp-assets/ComBat/Download.html) was used to adjust the normalized intensity to eliminate batch effects. Following normalization, significantly differentially expressed miRNAs were identified via volcano plot filtering, with P-value value between single fluorescent chip data group and a fold change value established by Volcano plot using R Software (version 5.50; MathSoft, Windows). Hierarchical clustering was performed using Multiexperiment Viewer software, version 4.6 (http://www.tm4.org/mev.html).
RT-qPCR
RT-qPCR was performed in order to validate the microarray data. RT-qPCR was conducted using the RNA samples used in the microarray analyses. Single-strand cDNA synthesis was performed using a PrimeScript II 1st strand Cdna Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's instructions using dNTPs (0.15 µl), enzyme mix (1 µl), primer mix (3 µl), RT buffer (1.5 µl), RNAase inhibitor (0.19 µl), RNase free H2O (4.16 µl), RNA (5 µl). Primer sequences targeting specific genes were designed using Primer Express® software (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to sequences published by GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The primers designed in the present study were as follows: Forward, 5′-CGCGGTATGGCACTGGTAGA-3′, and reverse, 5′-AGTGCAGGGTCCGAGGTATTC-3′ for miRNA-183-5p; forward, 5′-CCGGAATTCCCTCAACTCCACTCGTGTCC-3′, and reverse, 5′-ATTGCGGCCGCTGGGACTGTGACTCCTACCTG-3′ for miRNA-9-3p; forward, 5′-GGGAGCTGGTGTGTGAAT-3′, and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′ for miRNA-138-5p; forward, 5′-AACCUGAUCCCGUCUGAGAUUG-3′, and reverse, 5′-CCGGAUCAAGAUUAGUUCGGUU-3′ for miRNA-204-3p; forward, 5′-ACACTCCAGCTGGGTAAGGCACGCGGTGAAT-3′, and reverse, 5′-CTCAACTGGTGTCGTGGA-3′ for miRNA-124-3p; and sense, 5′-TCCACCACCCTGTTGCTGTA-3′ and antisense, 5′-ACCACAGTCCATGCCATCAC-3′ for GAPDH. GAPDH was used as an internal control. RT-qPCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions were as follows: i) 50°C for 2 min; ii) 95°C for 15 min; iii) 40 cycles of 95°C for 15 sec, 55°C for 30 sec and 72°C for 30 sec; and iv) final fluorescence detection at 95°C for 15 sec. miRNA expression levels were normalized against an endogenous reference gene (GAPDH) and relative to a control. The relative expression levels for each mRNA were determined using the 2−ΔΔCq method (18).
Statistical analysis
Statistical analyses were performed with the SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean ± standard error of the mean. Differences in the mean values were evaluated by Student's t-test (two means comparison), and one-way analysis of variance was conducted in order to compare the means among groups. Differences in the mean values were evaluated by Student's t-test (two means comparison). P<0.05 was considered to indicate a statistically significant difference.
Results
Sample RNA quality control
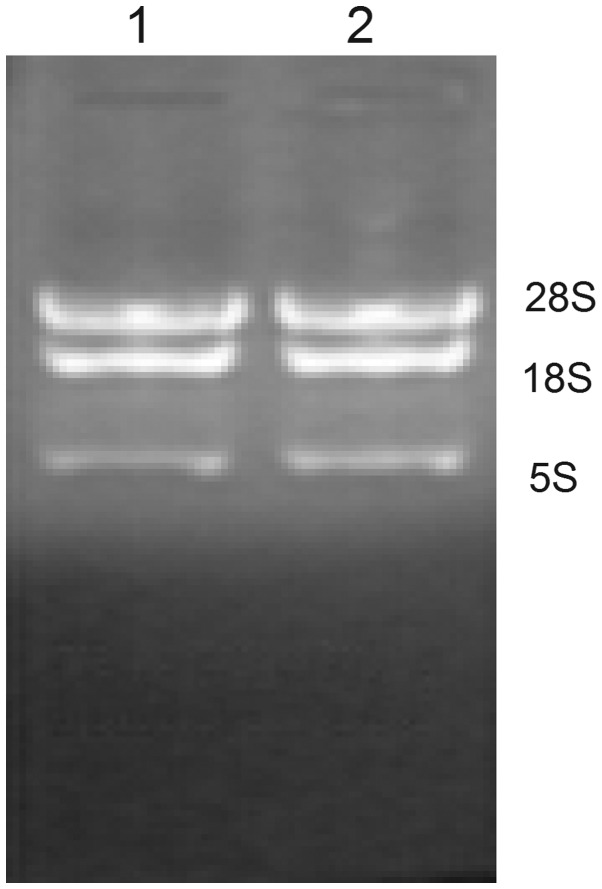
The concentration and purity of the RNA was determined using a NanoDrop ND-1000 spectrophotometer. The A260/A280 ratio was >2.0 and the A260/A230 ratio was >1.7. In addition, the integrity of RNA was assessed by denaturing agarose gel electrophoresis. Intact total RNA run on a denaturing gel will exhibit sharp 28S and 18S ribosomal RNA bands, as is shown in Fig. 1. Therefore, the concentration, purity and integrity of the RNA samples in the present study were suitable for microarray experiments.
Figure 1.
RNA electrophoresis on a denaturing agarose gel. A sharp distinction is evident at the small site of the 18S and 28S ribosomal RNA bands/peaks in total RNA electrophoresis. 1: control group; 2: UA group.
Microarray hybridization analysis
A total of 1,891 miRNAs were detected in each group using the Exiqon gene-chip. In order to identify miRNAs that were significantly differentially expressed between the two groups, volcano plot filtering was performed. The cut-off threshold for significantly differentially expressed miRNAs was a fold-change ≥2.0 and P<0.05. A total of 212 miRNAs were shown to be differentially expressed between the UA and control groups. Of these, 82 were upregulated (Table I) and 130 were downregulated (Table II). Functional annotation using GO analyses suggested that the majority of the differentially expressed miRNAs were associated with cardiovascular development or cholesterol metabolism (Table III).
Table I.
Twenty-five most upregulated miRNAs.
| miRNA | logFC | AveExpr | t | P-value | adj. P-val |
|---|---|---|---|---|---|
| hsa-let-7f-5p | 6.412454 | 8.331218 | 9.437973 | 6.75E-08 | 7.31E-06 |
| hsa-miR-221-3p | 3.995805 | 10.306465 | 9.031834 | 1.22E-07 | 1.03E-05 |
| hsa-miR-20b-5p | 5.593950 | 6.231350 | 8.344389 | 3.48E-07 | 2.40E-05 |
| hsa-miR-599 | 6.517428 | 3.760962 | 8.195105 | 4.40E-07 | 2.67E-05 |
| hsa-miR-26b-5p | 4.451009 | 10.029735 | 7.844804 | 7.72E-07 | 3.90E-05 |
| hsa-miR-23a-3p | 3.148685 | 9.992869 | 7.701674 | 9.75E-07 | 4.70E-05 |
| hsa-miR-101-3p | 3.637871 | 10.415865 | 7.614987 | 1.13E-06 | 5.17E-05 |
| hsa-miR-223-3p | 5.569596 | 9.001532 | 7.439788 | 1.51E-06 | 6.18E-05 |
| hsa-miR-30e-5p | 3.550603 | 9.188997 | 7.371461 | 1.69E-06 | 6.56E-05 |
| hsa-miR-98 | 4.201956 | 9.808134 | 7.151605 | 2.46E-06 | 8.67E-05 |
| hsa-miR-130a-3p | 4.357032 | 4.928504 | 6.978797 | 3.32E-06 | 1.12E-0.4 |
| hsa-miR-1 | 7.004429 | 4.493636 | 6.882004 | 3.93E-06 | 1.25E-0.4 |
| hsa-miR-1297 | 4.975548 | 8.112952 | 6.876167 | 3.97E-06 | 1.25E-0.4 |
| hsa-let-7d-5p | 3.824436 | 10.538728 | 6.817833 | 4.40E-06 | 1.36E-0.4 |
| hsa-miR-126-5p | 3.921176 | 7.013740 | 6.779627 | 4.70E-06 | 1.40E-0.4 |
| hsa-miR-500a-3p | 4.878389 | 3.902607 | 6.709008 | 5.33E-06 | 1.55E-0.4 |
| hsa-miR-501-3p | 4.800957 | 5.033868 | 6.609521 | 6.36E-06 | 1.75E-0.4 |
| hsa-miR-27a-3p | 4.523209 | 6.709947 | 6.490825 | 7.88E-06 | 2.02E-0.4 |
| hsa-miR-3942-5p | 2.520883 | 2.727009 | 6.451306 | 8.46E-06 | 2.07E-0.4 |
| hsa-miRPlus-J98 | 5.575619 | 3.580042 | 6.250002 | 1.22E-05 | 2.77E-0.4 |
| hsa-miR-224-5p | 4.171181 | 5.815962 | 6.229069 | 1.27E-05 | 2.83E-0.4 |
| hsa-miR-664-5p | 3.777162 | 4.100721 | 6.193966 | 1.36E-05 | 2.98E-04 |
| hsa-let-7a-5p | 5.396839 | 9.215099 | 6.092876 | 1.64E-05 | 3.40E-04 |
| hsa-miR-421 | 5.960126 | 6.157626 | 6.004292 | 1.93E-05 | 3.95E-04 |
| hsa-miR-144-5p | 5.757770 | 4.995824 | 5.986946 | 1.99E-05 | 4.03E-04 |
miRNA, microRNA; logFC, log fold-change; AveExpr, average expression; adj. P-val; adjusted P-value.
Table II.
Twenty-five most downregulated miRNAs.
| miRNA | logFC | AveExpr | t | P-value | adj. P-val |
|---|---|---|---|---|---|
| hsa-miR-124-3p | −9.086063 | 9.471658 | −13.177200 | 1.40E-09 | 2.98E-07 |
| hsa-miR-522-3p | −4.028076 | 5.529201 | −11.382700 | 1.02E-08 | 1.18E-06 |
| hsa-miR-125b-5p | −4.837694 | 9.099674 | −9.766860 | 7.63E-08 | 5.26E-06 |
| hsa-miR-874 | −4.054182 | 6.042472 | −9.521790 | 1.06E-07 | 6.42E-06 |
| hsa-miR-183-3p | −4.486712 | 7.526373 | −9.243540 | 1.55E-07 | 8.27E-06 |
| hsa-miR-363-5p | −5.495176 | 5.372322 | −9.304010 | 1.42E-07 | 8.27E-06 |
| hsa-miR-376b | −3.416201 | 5.919605 | −8.796220 | 2.90E-07 | 1.26E-05 |
| hsa-miR-668 | −3.881453 | 5.132914 | −8.601630 | 3.84E-07 | 1.49E-05 |
| hsa-miR-433 | −3.968384 | 6.703331 | −8.583870 | 3.94E-07 | 1.49E-05 |
| hsa-miR-3622b-3p | −4.405204 | 4.583577 | −8.570250 | 4.02E-07 | 1.49E-05 |
| hsa-miR-204-5p | −4.039169 | 5.750276 | −8.538700 | 4.21E-07 | 1.52E-05 |
| hsa-miR-9-5p | −5.371110 | 6.675159 | −8.503100 | 4.43E-07 | 1.56E-05 |
| hsa-miR-3144-5p | −4.400906 | 4.194498 | −8.374680 | 5.35E-07 | 1.74E-05 |
| hsa-miR-1253 | −4.005080 | 6.183476 | −8.256420 | 6.38E-07 | 1.90E-05 |
| hsa-miR-124-5p | −3.563842 | 4.473439 | −8.165870 | 7.30E-07 | 2.09E-05 |
| hsa-miR-1275 | −5.232220 | 8.268528 | −8.110010 | 7.94E-07 | 2.19E-05 |
| hsa-miR-933 | −4.227328 | 3.658511 | −8.064700 | 8.51E-07 | 2.28E-05 |
| hsa-miR-938 | −4.524226 | 5.752147 | −8.037610 | 8.87E-07 | 2.28E-05 |
| hsa-miR-138-5p | −4.796323 | 7.314068 | −8.051380 | 8.68E-07 | 2.28E-05 |
| hsa-miR-642b-3p | −3.479234 | 8.897743 | −7.982010 | 9.65E-07 | 2.36E-05 |
| hsa-miR-487b | −4.319478 | 7.409457 | −7.820470 | 1.24E-06 | 2.82E-05 |
| hsa-miRPlus-C1076 | −3.634989 | 5.587049 | −7.701560 | 1.49E-06 | 3.32E-05 |
| hsa-miR-296-5p | −3.156563 | 6.051010 | −7.597880 | 1.75E-06 | 3.59E-05 |
| hsa-miR-490-5p | −2.747711 | 5.614571 | −7.444480 | 2.23E-06 | 4.23E-05 |
| hsa-miR-9-3p | −4.097505 | 7.552229 | −7.355800 | 2.57E-06 | 4.69E-05 |
miRNA, microRNA; AveExpr, average expression; logFC, log fold-change; adj. P-val, adjusted P-value.
Table III.
Circulating miRNAs associated with cardiovascular development or cholesterol metabolism.
| miRNA | logFC | AveExpr | t | P-value | adj. P-val |
|---|---|---|---|---|---|
| hsa-miR-1 | 7.004429 | 4.493636 | 6.882004 | 0.000004 | 0.000125 |
| hsa-miR-144-5p | 5.757770 | 4.995824 | 5.986946 | 0.000020 | 0.000403 |
| hsa-miR-144-3p | 1.983646 | 5.655513 | 2.859536 | 0.011455 | 0.039290 |
| hsa-miR-122-3p | 1.839073 | 5.463513 | 2.875871 | 0.011076 | 0.038335 |
| hsa-miR-133b | 1.619471 | 7.419827 | 2.282713 | 0.036636 | 0.090144 |
| hsa-miR-133a | 1.054411 | 5.824366 | 1.872975 | 0.079676 | 0.158932 |
| hsa-miR-378a-3p | 1.645260 | 9.811212 | 2.174933 | 0.045163 | 0.104850 |
| hsa-miR-33a-3p | −0.313460 | 3.832990 | −0.644450 | 0.528523 | 0.649785 |
| hsa-miR-33b-5p | −0.053875 | 3.835549 | −0.109040 | 0.914544 | 0.941881 |
| hsa-miR-9-3p | −4.097505 | 7.552229 | −7.355800 | 0.000003 | 0.000047 |
miRNA, microRNA; AveExpr, average expression; logFC, log fold-change; adj. P-val, adjusted P-value.
RT-qPCR analysis
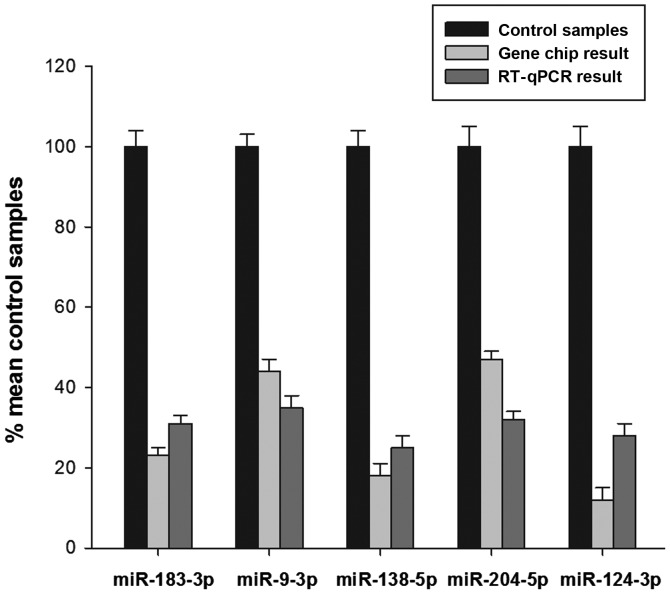
In order to validate the microarray results, RT-qPCR was performed. A total of 5 miRNAs from the differential gene expression profile were selected to validate the gene-chip results. As is shown in Fig. 2, RT-qPCR demonstrated the expression levels of these miRNAs were markedly downregulated, which was consistent with the gene-chip results. In addition, statistical analyses identified a conformity between the RT-qPCR analysis and microarray results.
Figure 2.
Gene-chip results were verified by RT-qPCR. Gene expression levels were normalized to the glyceraldehyde-3-phosphate dehydrogenase reference gene. RT-qPCR analysis demonstrated that the change tendency of the expression of miR-183-3p, miR-9-3p, miR-138-5p, miR-204-3p and miR-124-3p was consistent with the microarray data. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
The mechanisms underlying UA are currently unknown, and the timely biomarker-based diagnosis of UA remains a major clinical challenge. In order to address this problem, the present study assessed the levels of circulating miRNAs as potential novel biomarkers of UA using gene microarray analysis to compare the expression levels of miRNAs in the plasma of UA patients with those of healthy controls. A total of 212 differentially expressed miRNAs were detected in the present study, of which 82 were upregulated and 130 were downregulated. These results suggested that the progression of UA is a complex process affected by numerous factors and signaling pathways. Notably, downregulated miRNAs were less prevalent, as compared with upregulated miRNAs; suggesting that certain cellular functions may be inhibited and simplified in the process of UA. Due to inherent shortcomings in the microarray technique, including limitations in sensitivity, quantification and location, the present study employed RT-qPCR to validate the gene-chip results. Importantly, the results of the RT-qPCR were consistent with the microarray results.
GO analyses are commonly used to assign functions to differentially expressed genes and their downstream target genes (19). In the present study, a number of the differentially expressed miRNAs were closely associated with cardiovascular development, endothelial cell functions and cholesterol metabolism. Previous studies have suggested that miRNAs are involved in the pathogenesis of various cardiovascular diseases, including MI, heart failure, atherosclerosis and acute coronary syndromes (20,21). In addition, miRNAs have been shown to be key regulators of cardiac development, angiogenesis and vascular development (22). A previous study reported that miRNAs circulated in the blood of patients with cardiovascular diseases in a stable form (23); therefore, due to this stability, circulating miRNAs may serve as novel biomarkers for the diagnosis of cardiovascular diseases.
Previous studies have identified the mechanisms underlying the regulation of miRNA expression levels (24). Of the differentially expressed miRNAs identified in the present study, miRNA-1, miRNA-133 and miRNA-128 have been associated with the development of the cardiovascular system in the embryo (25). Previous studies reported that miRNA-1 and miRNA-133a were associated with cardiovascular diseases, including CHD, myocardial fibrosis, heart failure and cardiac functional insufficiency (26–28), which is consistent with the present results. Furthermore, the upregulation of miRNA-150, miRNA-186 and miRNA-210 in the peripheral blood of patients with MI has been extensively reported (29). The present study demonstrated that miRNA-19a and miRNA-19b were significantly differentially expressed between the UA and control groups. miRNA-19b has been shown to have an anti-thrombotic role by inhibiting the expression of tissue factors in patients with UA (30). In addition, a previous study reported that downregulation of miRNA-19a and miRNA-19b expression was involved in age-associated remodelling in the heart (31). In the present study, the expression levels of miRNA-126b-5p, miRNA-27a-3p and miRNA-26b-5p were increased 4–6 times (P<0.001). miRNA-26 and miRNA-27 are specifically expressed in endothelial cells, and have been shown to participate in the regulation of endothelial cell function (32). Thus, the results of the present study suggested that circulating miRNAs may serve as candidate molecules for the monitoring of endothelial cell injury in patients with UA.
Disorders of cholesterol metabolism are an important risk factor and serve a critical function in the pathological processes of UA disease (33). The regulation of cholesterol metabolism involves complex regulatory networks, including miRNA-122, miRNA-33, miRNA-144, miRNA-378 and miRNA-9-3p (34). Among these miRNAs, miRNA-33 may inhibit cholesterol metabolism to maintain cholesterol homeostasis (35,36). miRNA-22, which is specifically expressed in liver tissue, accounts for 70% of the total volume of miRNA in the liver tissue and serves to maintain the liver cell phenotype (37). miRNA-144 has been reported to regulate the level of high-density lipoprotein, promote myocardial cell survival, and reduce ischemia/reperfusion-induced cardiomyocyte injury (38). In the present study, the expression levels of miRNA-122, miRNA-33, miRNA-144 and miRNA-378 were not significantly different between the UA patients and controls, and thus may not be considered molecular markers of UA. Conversely, miRNA-9-3p, which was significantly differentially expressed between the two groups in the present study, targets the ATP-binding cassette subfamily A member 1 gene, which is a key regulator of cholesterol metabolism, resulting in translational repression or gene silencing at the post-transcriptional level. Therefore, miRNA-9-3p may emerge as a potential novel candidate in the development of miRNA-based therapeutic strategies for UA.
In conclusion, the results of the present study have highlighted a number of circulating miRNAs that may serve as potential candidate biomarkers for the diagnosis and treatment of UA. In addition, the present study provides an experimental basis for further functional studies to investigate the regulatory mechanisms underlying the differential expression of specific miRNAs in UA. The functions and significance of the differentially expressed miRNAs identified in the present study require further investigation.
Acknowledgements
The present study was supported by a grant from the Scientific Research Fund Project of Hebei Province (grant no. 20130310).
Glossary
Abbreviations
- UA
unstable angina
- miRNA
microRNA
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- CHD
coronary artery disease
- MI
myocardial infarction
- GO
gene ontology
References
- 1.Wiviott SD, Braunwald E. Unstable angina and non-ST-segment elevation myocardial infarction: Part I. Initial evaluation, and management and hospital care. Am Fam Physician. 2004;70:525–532. [PubMed] [Google Scholar]
- 2.Braunwald E, Morrow DA. Unstable angina: Is it time for a requiem? Circulation. 2013;127:2452–2457. doi: 10.1161/CIRCULATIONAHA.113.001258. [DOI] [PubMed] [Google Scholar]
- 3.Braunwald E. Unstable angina and non-ST elevation myocardial infarction. Am J Respir Crit Care Med. 2012;185:924–932. doi: 10.1164/rccm.201109-1745CI. [DOI] [PubMed] [Google Scholar]
- 4.Piscopo P, Albani D, Castellano AE, Forloni G, Confaloni A. Frontotemporal Lobar Degeneration and MicroRNAs. Front Aging Neurosci. 2016;8:17. doi: 10.3389/fnagi.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao J, Zhong H, Zhou Y, Yu F, Gao Y, Luo Y, Tang Z, Guo Z, Guo E, Gan X, et al. Identification and characterization of microRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus) by using solexa sequencing technology. PloS One. 2014;9:e86821. doi: 10.1371/journal.pone.0086821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Pal MK, Jaiswar SP, Dwivedi VN, Tripathi AK, Dwivedi A, Sankhwar P. MicroRNA: A new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol Med. 2015;12:328–341. doi: 10.7497/j.issn.2095-3941.2015.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JC, Hilsenbeck SG, Fuqua SA. Genomic approaches in the management and treatment of breast cancer. Br J Cancer. 2005;92:618–624. doi: 10.1038/sj.bjc.6602410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batzir NA, Shohat M, Maya I. Chromosomal Microarray Analysis (CMA) a Clinical Diagnostic Tool in the Prenatal and Postnatal Settings. Pediatr Endocrinol Rev. 2015;13:448–454. [PubMed] [Google Scholar]
- 14.Cobb JP, Brownstein BH, Watson MA, Shannon WD, Laramie JM, Qiu Y, Stormo GD, Morrissey JJ, Buchman TG, Karl IE, Hotchkiss RS. Injury in the era of genomics. Shock. 2001;15:165–170. doi: 10.1097/00024382-200115030-00001. [DOI] [PubMed] [Google Scholar]
- 15.Liu WM, Laux H, Henry JY, Bolton TB, Dalgleish AG, Galustian C. A microarray study of altered gene expression in colorectal cancer cells after treatment with immunomodulatory drugs: Differences in action in vivo and in vitro. Mol Biol Rep. 2010;37:1801–1814. doi: 10.1007/s11033-009-9614-3. [DOI] [PubMed] [Google Scholar]
- 16.Cannon CP. Strategies and Therapies for Reducing Ischemic and Vascular Events (STRIVE): Updated standing orders for ST-segment elevation myocardial infarction and unstable angina/non-ST-segment elevation myocardial infarction. Crit Pathw Cardiol. 2010;9:55–81. doi: 10.1097/HPC.0b013e3181dfd6f8. [DOI] [PubMed] [Google Scholar]
- 17.Andreasen D, Fog JU, Biggs W, Salomon J, Dahslveen IK, Baker A, Mouritzen P. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010;50:S6–S9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time rt-PCR in four lepidopteran insects. J Insect Sci. 2012;12:60. doi: 10.1673/031.012.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papageorgiou N, Tousoulis D, Androulakis E, Siasos G, Briasoulis A, Vogiatzi G, Kampoli AM, Tsiamis E, Tentolouris C, Stefanadis C. The role of microRNAs in cardiovascular disease. Curr Med Chem. 2012;19:2605–2610. doi: 10.2174/092986712800493048. [DOI] [PubMed] [Google Scholar]
- 21.Romaine SP, Charchar FJ, Samani NJ, Tomaszewski M. Circulating microRNAs and hypertension—from new insights into blood pressure regulation to biomarkers of cardiovascular risk. Curr Opin Pharmacol. 2016;27:1–7. doi: 10.1016/j.coph.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Rognoni A, Cavallino C, Lupi A, Secco GG, Veia A, Bacchini S, Rosso R, Rametta F, Bongo AS. Novel biomarkers in the diagnosis of acute coronary syndromes: The role of circulating miRNAs. Expert Rev Cardiovasc Ther. 2014;12:1119–1124. doi: 10.1586/14779072.2014.953483. [DOI] [PubMed] [Google Scholar]
- 23.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 24.Latsios G, Tousoulis D, Androulakis E, Papageorgiou N, Synetos A, Toutouzas K, Siasos G, Oikonomou E, Stefanadis C. MicroRNAs in the diagnosis and treatment of unstable angina. Curr Top Med Chem. 2013;13:1596–1604. doi: 10.2174/15680266113139990108. [DOI] [PubMed] [Google Scholar]
- 25.Lee S, Choi E, Cha MJ, Park AJ, Yoon C, Hwang KC. Impact of miRNAs on cardiovascular aging. J Geriatr Cardiol. 2015;12:569–574. doi: 10.11909/j.issn.1671-5411.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond-the transcriptome of human monocytes and disease susceptibility. PloS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, Watanabe S, Baba O, Kojima Y, Shizuta S, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446–454. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 29.Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27:255–268. doi: 10.1155/2009/641082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Ren J, Xu N, Zhang J, Geng Q, Cao C, Lee C, Song J, Li J, Chen H. MicroRNA-19b functions as potential anti-thrombotic protector in patients with unstable angina by targeting tissue factor. J Mol Cell Cardiol. 2014;75:49–57. doi: 10.1016/j.yjmcc.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 31.van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, Cleutjens JP, van Zandvoort MA, Heymans S, Schroen B. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urbich C, Kaluza D, Frömel T, Knau A, Bennewitz K, Boon RA, Bonauer A, Doebele C, Boeckel JN, Hergenreider E, et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood. 2012;119:1607–1616. doi: 10.1182/blood-2011-08-373886. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien EC, Simon DN, Roe MT, Wang TY, Peterson ED, Alexander KP. Statin Treatment by Low-Density Lipoprotein Cholesterol Levels in Patients With Non-ST-Segment Elevation Myocardial Infarction/Unstable Angina Pectoris (from the CRUSADE Registry) Am J Cardiol. 2015;115:1655–1660. doi: 10.1016/j.amjcard.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT, Jegga AG, Fan GC. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49:841–850. doi: 10.1016/j.yjmcc.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]