Abstract
The Asian soybean rust fungus, Phakopsora pachyrhizi, is an obligate biotrophic pathogen causing severe soybean disease epidemics. Molecular mechanisms by which P. pachyrhizi and other rust fungi interact with their host plants are poorly understood. The genomes of all rust fungi encode many small, secreted cysteine-rich proteins (SSCRP). While these proteins are thought to function within the host, their roles are completely unknown. Here, we present the characterization of P. pachyrhizi effector candidate 23 (PpEC23), a SSCRP that we show to suppress plant immunity. Furthermore, we show that PpEC23 interacts with soybean transcription factor GmSPL12l and that soybean plants in which GmSPL12l is silenced have constitutively active immunity, thereby identifying GmSPL12l as a negative regulator of soybean defenses. Collectively, our data present evidence for a virulence function of a rust SSCRP and suggest that PpEC23 is able to suppress soybean immune responses and physically interact with soybean transcription factor GmSPL12l, a negative immune regulator.
Author Summary
Asian soybean rust is a disease caused by the rust fungus, Phakopsora pachyrhizi, which was first described in Japan in 1902 and eventually reached North and South America in the early 2000s. Like other rust fungi, P. pachyrhizi, is capable of causing disease epidemics that severely reduce crop yields. Rust fungi require living host tissue in order to complete their life cycles, and thus, we expect that they produce and secrete effector proteins that are transferred into plant cells to interfere with immune responses. Small proteins that are rich in the amino acid cysteine and predicted to be secreted have been proposed to be likely candidates for effector proteins. Here, we report that a secreted, small cysteine-rich protein from P. pachyrhizi suppresses plant immunity, and furthermore, it interacts with a soybean transcription factor protein. Interestingly, silencing of this transcription factor results in plants that have increased expression of defense marker genes and enhanced disease resistance, which demonstrates that it is a negative regulator of soybean immune responses. Our work shows that, indeed, a small cysteine-rich protein from a rust fungus can suppress plant immunity, and furthermore, it identifies a new function for SQUAMOSA PROMOTER BINDING-LIKE (SPL) transcription factors as components of the defense regulatory networks that holds immune responses in check.
Introduction
Rust fungi comprise many pathogenic species in the order Pucciniales that cause devastating plant diseases [1–3]. They are notable for their obligate biotrophy and complex life cycles that include distinct spore-producing stages and host specialization. As rust fungi colonize their hosts, they form specialized hyphal structures, called haustoria, within their host plant cells, and these structures are critical for nutrient uptake, metabolic processes, and delivery of effector proteins that manipulate host immune systems [4,5]. Many rust candidate effector proteins have been predicted through whole-genome sequencing and in planta expression studies [3,6–8].
The most well-studied effector proteins are from four different rust species: AvrM, AvrL567, AvrP123, and AvrP4 from Melampsora lini (flax rust), Rust Transferred Protein (RTP1) from Uromyces fabae (broad bean rust), PGTAUSPE10-1 from Puccinia graminis f. sp. tritici (wheat stem rust), and most recently, PEC6 from Puccinia striiformis f. sp. tritici (wheat stripe rust). AvrM, AvrL567, AvrP123, AvrP4 and PGTAUSPE10-1 have been studied intensely for their avirulence properties defined by their abilities to trigger R-protein mediated immune responses [9–12]. RTP1 was the first rust protein proven to be directly transferred from haustoria to plant cells during infection [11], and moreover, it was shown to form fibrils and be a protease inhibitor [9], although its targets and detailed mechanisms still remain unclear. PEC6 suppresses basal defense responses in non-host and host plants and interacts with adenosine kinases [13]. Pathogen-free assays further demonstrate the autonomous entry of AvrP4, AvrM and AvrL567 into plant cells [14–16]. Additionally, AvrP4, AvrP123, and PEC6 all are small, secreted cysteine-rich proteins (SSCRP) [13,14].
Asian soybean rust (ASR), caused by Phakopsora pachyrhizi, is a serious threat to soybean production. The rapid lifecycle and the production of large numbers of infectious urediospores under optimal environmental conditions lead to high inoculum levels and potentially high yield losses [17]. To date, six P. pachyrhizi R genes have been identified [18], but monogenic resistance to ASR is rapidly overcome by virulent P. pachyrhizi isolates [19]. The variation in P. pachyrhizi virulence makes research on conserved effectors highly significant, because they may provide new targets for broad spectrum resistance to many virulent isolates of this pathogen as well as provide new insights into mechanisms by which rust pathogens manipulate host immune systems. Recently, haustoria of P. pachyrhizi were purified and the haustorial transcriptome was sequenced and analyzed, which provided a resource for predicting secreted P. pachyrhizi effector candidates (PpECs) [8]. Among the PpECs were several SSCRP, i.e., a class of proteins that had been hypothesized to be strong candidates for rust fungus effectors [20].
Suppression of soybean immunity is an expected function for some PpECs that promote P. pachyrhizi virulence. This is necessary because plant immune recognition systems activate defenses in response to conserved features or specific effector proteins produced by pathogens [21,22]. Pathogen-associated molecular patterns (PAMPs) are conserved features of pathogens that can elicit a type of basal defense known as PAMP-triggered immunity (PTI) when they are perceived by pattern recognition receptors (PRR) on the plant cell surface [23–25]. Basal defense responses include the deposition of callose in the cell walls, a reactive oxygen species (ROS) burst, and transcriptional up-regulation of immune-related genes, all of which restrict pathogen proliferation [26]. However, these defense responses are suppressed by successful pathogens through multiple means including secretion of effector proteins [22]. In return, host plants have evolved the more specific and much stronger effector-triggered immune (ETI) responses, such as the hypersensitive response (HR), which is a form of programmed cell death. HR is often mediated by nucleotide binding site leucine-rich repeat (NBS-LRR) proteins that recognize the presence of specific avirulence (former effector) proteins [27]. Even though basal defense and HR can be roughly discriminated based on the response strength and specificity, they share many signaling components [28].
We are interested in identifying PpECs that suppress immunity and characterizing the mechanisms by which they interfere with soybean immune signaling. To initiate this work, we performed functional screens designed to identify PpECs that suppress defense responses elicited by a bacterial pathogen, Pseudomonas syringae pv. tomato (Pst). One SSCRP, which we named PpEC23, was unique in its ability to suppress HR and basal defense responses. The major feature of PpEC23 is a tandem repeat of a ten-cysteine motif that appears in the predicted secretomes of other rust fungi [1,3,8], and it interacts with soybean transcription factor GmSPL12l (SQUAMOSA promoter-binding-like protein 12 like). RNA silencing of GmSPL12l indicates that this transcription factor functions as a negative regulator of soybean defense responses. While prior work documented avirulence functions for a few SSCRPs, i.e. their detection by the plant immune system leading to plant defenses, the current study establishes that SSCRP PpEC23 has functions consistent with a virulence factor that regulates host plant immunity.
Results
PpEC23 suppresses HR induced by Pst strain DC3000 in soybean, Nicotiana benthamiana, and tobacco
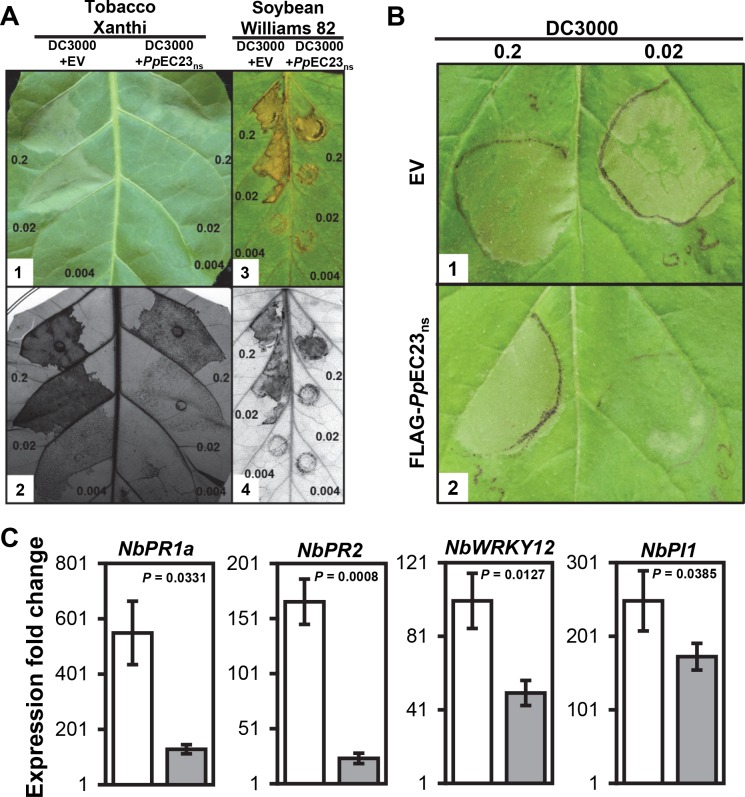
We wanted to determine if a PpEC could suppress plant immune responses. To test this, we cloned 82 of 156 PpECs identified by Link et al. [8] minus their predicted signal peptides (PpECns) into the bacterial type III secretion system (T3SS) vector, pEDV6 [29]. The pEDV6::PpECns constructs plus the empty pEDV6 were introduced into Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000), which elicits an HR on our experimental plants N. benthamiana, tobacco (N. tabacum cv. Xanthi), and soybean (Glycine max cv. Williams 82) [30,31]. As expected, infiltration of N. benthamiana, N. tabacum, and soybean leaves with Pst DC3000 carrying the empty vector (EV) at optical densities (OD) 600 nm of 0.2 and 0.02 caused strong macroscopic HR (Fig 1A1 and 1A3, S1A and S1B Fig, left side of leaves). Of the 82 PpECns screened for suppression of Pst DC3000-induced HR, pEDV6::PpEC23ns was unique in its ability to delay and reduce the HR (Fig 1A1 and 1A3, S1A and S1B Fig, right side of leaves). Infiltrated areas on soybean leaves are much smaller compared to those on Nicotiana leaves, and more force is required to introduce the inoculum, so there is an obvious wound at the infiltration site. Therefore, we only kept soybean leaves in which the inoculum had clearly expanded beyond the infiltration site and photographed them two days later. To confirm this HR suppression phenotype, trypan blue staining was performed to detect dead tobacco and soybean cells (Fig 1A2 and 1A4). For both tobacco and soybean, trypan blue staining was reduced in leaf areas inoculated with Pst DC3000 expressing PpEC23ns at each inoculum density compared to Pst DC3000 carrying empty pEDV6, which confirmed the phenotype of weakened HR.
Fig 1. PpEC23 suppresses HR induced by Pst DC3000.
A. HR on leaves of N. tabacum cv. Xanthi (A1, A2) and G. max cv. Williams 82 (A3, A4) infiltrated with Pst DC3000 carrying empty pEDV6 vector (EV) (left half) and Pst DC3000 carrying pEDV6::PpEC23ns (right half). Leaves were infiltrated with bacteria at OD 600 nm = 0.2, 0.02, and 0.004 (approximately equal to 108, 107 and 2x106 CFU (colony forming units) / mL, respectively [32]). Leaves were photographed at 18 h post infiltration (hpi) for tobacco (A1) and at 30 hpi for soybean (A3) and then stained with trypan blue to detect non-viable cells (A2, A4). B. HR on leaves of N. benthamiana stably transformed with EV (B1) and FLAG-PpEC23ns (B2) infiltrated with Pst DC3000. Leaves were infiltrated with bacteria at OD 600 nm = 0.2 (left), 0.02 (right). Leaves were photographed at 18 hpi. Representative images are shown (n ≥ 8). C. The expression fold change of immune marker genes PR1a, PR2, WRKY12, and PI1 in the leaves of N. benthamiana stably transformed with EV (white bars) and FLAG-PpEC23ns (gray bars) at 16 hpi with Pst DC3000. NbAct1 was used as the internal reference gene. A t-test was performed for each pair-wise comparison and the P value for each comparison is shown. Four biological and four technical replicates were performed.
To exclude the possibility that the expression of PpEC23ns had a negative effect on the general viability of Pst DC3000, a bacterial growth assay was performed. The growth curves for Pst DC3000 carrying PpEC23ns and Pst DC3000 carrying the empty vector were similar, indicating that the delayed and reduced HR was not due to the reduced viability in Hrp-inducing minimal medium (S2 Fig). To further investigate whether the reduced HR was a direct effect of expressing PpEC23 in Pst DC3000, we tested if pEDV6::PpEC23ns could suppress Pst DC3000-induced HR when expressed in trans from the non-pathogenic P. fluorescens strain EtHAn [33]. EtHAn carrying PpEC23ns was diluted to an OD 600 nm of 0.2 and then co-inoculated with serially diluted Pst DC3000 on N. benthamiana (S1C–S1F Fig). At 42 hpi, EtHAn expressing PpEC23ns had clearly suppressed HR caused by Pst DC3000 at an OD 600nm of 0.02, which was in contrast to EtHAn carrying empty pEDV6 that did not suppress the HR (S1E and S1F Fig). These data support the conclusion that the ability of PpEC23 to suppress HR was not due to general effects on Pst DC3000, and they further indicate that HR suppression was the result of PpEC23’s function in planta.
To directly test if PpEC23 suppresses HR in planta, stably transformed N. benthamiana plants expressing a FLAG-PpEC23ns fusion protein under control of the Cauliflower mosaic virus 35S promoter were generated. The accumulation of FLAG-PpEC23ns protein was confirmed in eight independent transgenic lines by Western blot using an anti-FLAG antibody (S3 Fig). We serially diluted Pst DC3000 and inoculated plants from four independent transgenic lines with the highest expression of FLAG-PpEC23ns, along with empty vector transformed controls. On control plants, Pst DC3000 caused strong HR when inoculated at an OD 600 nm of 0.02 and 0.2 (Fig 1B1). However, on FLAG-PpEC23ns transformants, HR was clearly suppressed in response to Pst DC3000 at an OD 600 nm of 0.02 (Fig 1B2). These results demonstrate that PpEC23 suppresses Pst DC3000-induced ETI through an activity in planta.
We hypothesized that suppression of HR would be accompanied by decreased mRNA accumulation of plant immune marker genes. To test this, quantitative reverse transcriptase-PCR (qRT-PCR) was performed to detect whether PpEC23 had an effect on the expression of four genes that were previously shown to represent immune responses in Nicotiana species: pathogenesis related genes PR1a and PR2 [34], WRKY12 transcription factor [35,36], and protease inhibitor 1 (PI1) [34,37,38]. N. benthamiana plants transformed with FLAG-PpEC23ns or empty vector were inoculated with Pst DC3000 at an OD 600 nm of 0.02 and samples were collected 16 hpi, which was prior to the appearance of macroscopic cell death. The expression of all four genes was induced in control and FLAG-PpEC23ns plants in response to Pst DC3000, but the levels of induction were much less in FLAG-PpEC23ns plants compared with plants transformed with the empty vector (Fig 1C). The lower expression was significant for all four genes, especially for PR1a and PR2, with 4.3 and 7 fold lower fold changes, respectively. These data show that PpEC23 suppresses plant immune responses and it dampens accompanying downstream transcriptional activation.
PpEC23 encodes a modular SSCRP that is present in diverse P. pachyrhizi isolates
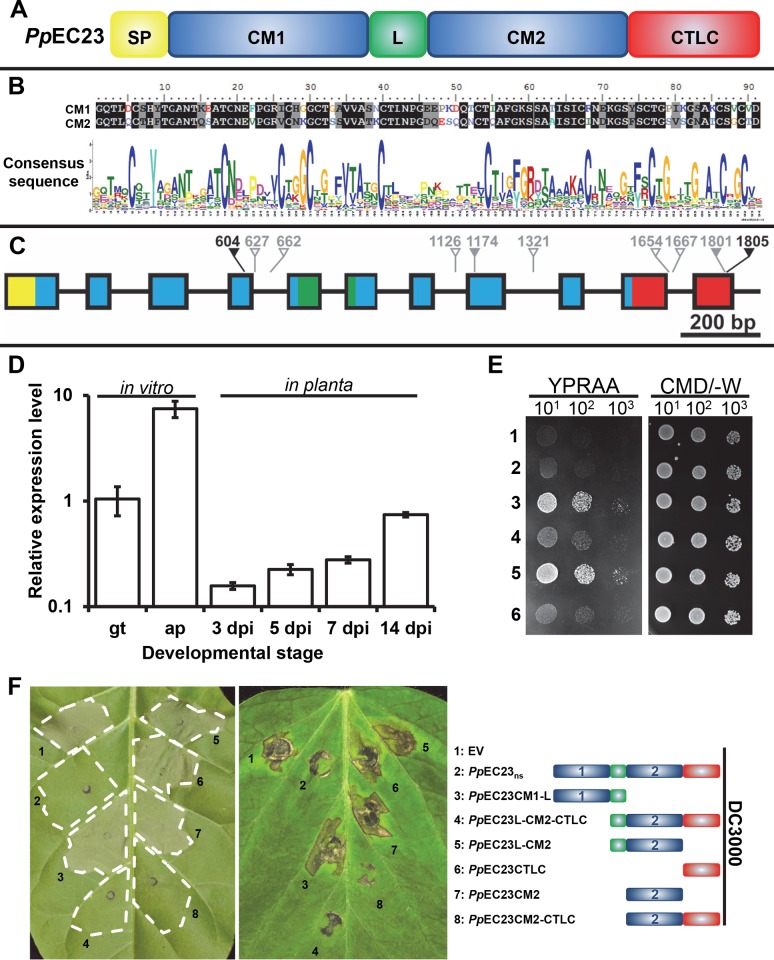
PpEC23 is a SSCRP containing a signal peptide (SP), two tandem ten-cysteine motifs (CMs) designated CM1 and CM2 separated by a linker region (L), and a C-terminal low complexity (CTLC) domain (Fig 2A). Alignment of the amino acid sequences of CM1 and CM2 showed that they share 65.9% identity (Fig 2B). PpEC23 belongs to a large family of putatively secreted proteins, previously designated as cluster 112 by Link et al. [8], comprised of many SSCRPs from various species of rust fungi. All members contain a single CM, with the exception of PpEC23 which contains two (Fig 2B). The unique structure of PpEC23 and its ability to suppress HR led us to further characterize its properties and functions.
Fig 2. PpEC23 is a modular SSCRP that is present in diverse P. pachyrhizi isolates.
A. Diagram depicting the different motifs of PpEC23. SP, signal peptide; CM1, ten-cysteine motif 1; L, linker; CM2, ten-cysteine motif 2; CTLC, C-terminal low complexity motif. B. Sequence alignment of the two CMs in PpEC23 (top) and the consensus sequence of CM for all members of cluster 112, generated by WebLogo (bottom). C. Genomic structure of the PpEC23 gene. Colors of the exons shown as blocks correspond with motif colors in panel A. Arrows show the SNPs, together with the nucleotide position, among ten geographically distinct isolates. Hollow gray, solid gray and black arrows show the intron SNPs, synonymous SNPs, and nonsynonymous SNPs, respectively. D. Transcript levels of PpEC23 at different developmental stages. Transcript levels were normalized to the expression of the fungal reference genes, RPS14 and PDK. Three independent biological and four technical replicates were performed. Error bars = standard deviation. E. The PpEC23 signal peptide directs protein secretion in yeast. Fusion constructs of PpEC23 in the secretion signal trap plasmid: 1) pSuc2t7M13ori::PpEC23, 2) pSuc2t7M13ori::PpEC23ns, 3) pSuc2t7M13ori::PpEC23Sp-CM1-L, 4) pSuc2t7M13ori::PpEC23L-CM2-CTLC, 5) pSuc2t7M13ori::PpEC23Sp, 6) pSuc2t7M13ori EV control. YPRAA and CMD/-W indicates the selective and non-selective medium, respectively. Numbers below indicate the fold dilution of the yeast strains. F. Symptoms on leaves of N. tabacum cv. Xanthi (left) and G. max cv. Williams 82 (middle) infiltrated with Pst DC3000 carrying EV, PpEC23ns, and various truncated constructs of PpEC23. The name and structural diagram of each construct is provided in the right panel. Leaves were infiltrated with bacteria at OD 600 nm = 0.02. Leaves were photographed at 18 hpi for tobacco (left) and at 30 hpi for Williams 82 soybean (middle). Representative images are shown (n ≥ 8).
To examine the variability of PpEC23, it was PCR-amplified, cloned, and sequenced from the genomic DNA of 11 P. pachyrhizi isolates collected from ten geographically distinct areas. Four independent clones of each isolate were sequenced and compared to the sequence of isolate LA04-1 [39]. Single nucleotide polymorphisms (SNPs) were found at 10 positions (Fig 2C, Table 1). Eight SNPs were either in introns (hollow gray arrows) or synonymous (solid gray arrows), neither of which altered the coding sequence. Two SNPs, SNP604 and SNP1805, were non-synonymous—generating amino acid changes (black arrows) I125V and A289P, respectively. Eight isolates, including LA04-1, are heterozygous at the SNP1801 and SNP1805 sites. However, isolates BZ01-1, PG01-3, and HW94-1 have more SNPs across 8 of the 10 sites. We calculated an overall SNP density of 5.3 SNPs/kbp, and a non-synonymous SNP density of 1.1 SNPs/kbp in the PpEC23 genomic region, which is dramatically lower than the SNP density of 15.4 SNPs/kbp in the housekeeping genes analyzed previously [40]. The low non-synonymous SNP density of the PpEC23 genomic region shows that its coding sequence is well maintained.
Table 1. SNPs of PpEC23 genes among P. pachyrhizi isolates.
| Isolate | Year | Origin | Source | SNPs* |
|---|---|---|---|---|
| IN73-1 | 1973 | India | D. N. Thapliyal, Pantnagar | 1801T or G; 1805C or G |
| TW72-1 | 1972 | Taiwan, China | Lung-Chi Wu, Taipei | 1801T or G; 1805C or G |
| TW80-2 | 1980 | Taiwan, China | AVRDC, Taiwan | 1801T or G; 1805C or G |
| BZ01-1 | 2001 | Brazil | J. T. Yorinori, Parana | 604A; 627T; 662A; 1126G; 1174A;1321T; 1654G; 1667T; 1801T or G; 1805C |
| PG01-3 | 2001 | Paraguay | W. M. Morel, Capitan Miranda | 604G or A; 627C or T; 662T or A; 1126A or G; 1174G or A; 1321C or T; 1654A or G; 1667C or T; 1801T or G; 1805C or G |
| HW94-1 | 1994 | Hawaii, US | E. Kilgore, Oahu | 604G or A; 627C or T; 662T or A; 1126A or G; 1174G or A; 1321C or T; 1654A or G; 1667C or T; 1805C or G |
| AU79-1 | 1979 | Australia | Unknown | 1801T or G; 1805C or G |
| SA01-1 | 2001 | South Africa | Z. A. Pretorius, Natal Province | 1801T or G; 1805C or G |
| ZM01-1 | 2001 | Zimbabwe | C. Levy, Harare | 1801T or G; 1805C or G |
| AL04-1 | 2004 | Alabama, US | R. Frederick, Mobile County | 1801T or G |
*: The genomic sequence of PpEC23 gene from LA04-1 (Louisiana, US) was taken as the reference sequence. Only variations are shown. SNP locations are shown schematically in Fig 2C.
To test if the amino acid differences caused by the two non-synonymous SNPs could affect suppression of Pst DC3000-induced HR, SNP604 and SNP1805 were introduced independently into the pEDV6::PpEC23ns sequence, which originated from the LA04-1 isolate. Pst DC3000 strains carrying the resulting constructs were infiltrated into soybean (S4 Fig). Both PpEC23 mutants maintained their ability to suppress HR demonstrating that the I125V and A289P amino acid changes did not affect this function.
The expression pattern of PpEC23
PpEC23 was originally identified by sequencing the P. pachyrhizi haustorial transcriptome [8], and we were interested in determining its mRNA expression profile throughout the fungal lifecycle. Therefore, we quantified the abundance of PpEC23 mRNA at early infection stages and throughout infection using qRT-PCR. We included in vitro produced germ tubes and appressoria, representing the structures that rust fungi form on the leaf surface, and infected leaves at four different time points [3, 5, 7, and 14 days post inoculation (dpi)] using P. pachyrhizi RPS14 and PDK as reference genes [41]. Our analysis shows that transcript levels of PpEC23 are highly elevated during appressoria formation and also continue to rise throughout the infection time course (Fig 2D). This expression profile shows that PpEC23 is not exclusively expressed in haustoria.
The signal peptide of PpEC23 is sufficient for secretion in a yeast assay
To test the functionality of the predicted signal peptide, we determined if it was able to mediate secretion when fused with invertase in Saccharomyces cerevisiae. Expression constructs for fusion proteins containing PpEC23, PpEC23SP, PpEC23ns (the PpEC23 open reading frame minus SP), PpEC23SP-CM1-L and PpEC23L-CM2-CTLC were generated using the yeast signal sequence trap vector pSuc2t7M13ori [42]. The resulting plasmids were transformed into yeast strain YTK12 that lacks a secreted invertase, and transformants were streaked on CMS/loD-W medium, which only supports growth of yeast capable of secreting invertase. As expected, yeast expressing PpEC23SP or PpEC23SP-CM1-L fusions grew well on the selective medium, whereas the transformants expressing the PpEC23ns or PpEC23L-CM2-CTLC fusions lacking the SP showed the same weak growth as the empty vector control (Fig 2E). Transformants expressing full length PpEC23 fused to invertase also grew poorly on selective media. Overall, these results support the functionality of the PpEC23 signal peptide.
PpEC23 CM2 and CTLC domains are necessary for suppression of HR
To determine the functional domains of PpEC23 required for HR suppression, six truncated constructs consisting of different domain combinations were generated and cloned into pEDV6 (Fig 2F and S5 Fig). Pst DC3000 expressing the truncated PpEC23 constructs were inoculated on N. tabacum and soybean leaves to test if any had the ability to suppress HR (Fig 2F left and middle panels, respectively). Only the PpEC23L-CM2-CTLC and PpEC23CM 2-CTLC constructs retained HR suppression function. These data demonstrate that CM2 and CTLC together are necessary for the ability of PpEC23 to suppress HR induced by Pst DC3000.
PpEC23 suppresses basal defense responses
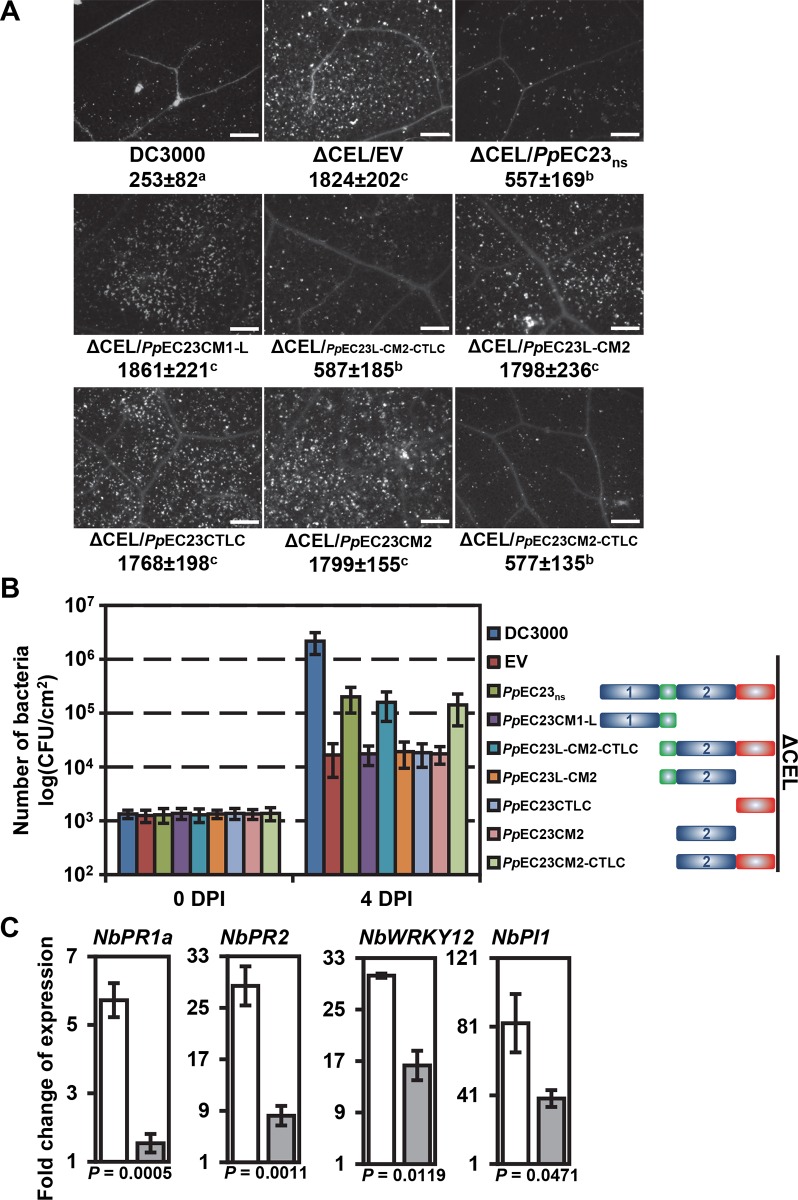
Because PpEC23 suppresses HR, we decided to further investigate its function by testing if it also suppresses basal defense responses associated with PTI. Arabidopsis thaliana ecotype Columbia-0 (Col-0) normally exhibits extensive callose deposition when inoculated with the Pst DC3000 ΔCEL strain CUCPB5115, whereas little callose deposition is observed in leaves inoculated with wild-type Pst DC3000 [43,44]. Callose deposition was quantified in A. thaliana leaves inoculated with Pst DC3000 and CUCPB5115 carrying empty pEDV6 or pEDV6 expressing PpEC23ns or its six truncated derivatives (Fig 3A). CUCPB5115 carrying PpEC23ns, PpEC23L-CM2-CTLC, or PpEC23CM 2-CTLC induced significantly less callose deposition than CUCPB5115 carrying empty pEDV6, or the other four truncated derivatives of PpEC23. These data show that PpEC23 also suppresses callose deposition and CM2 and CTLC together are necessary for this activity, which is consistent with their function in HR suppression.
Fig 3. PpEC23 suppresses basal defense responses.
A. Callose deposition in leaves of A. thaliana Col-0 induced by Pst DC3000, CUCPB5115 (ΔCEL)/EV, ΔCEL/PpEC23ns or ΔCEL/truncated constructs stained with aniline blue. The average number of callose spots ± standard deviation is listed under each representative image. Pair-wise t-tests were performed and a, b, c were designated groups with statistically significant difference. Bar = 50 μm. Representative images are shown (n ≥ 24). B. Bacterial growth in planta of Pst DC3000, ΔCEL/EV, ΔCEL/PpEC23ns or ΔCEL/truncated constructs. Diagrams of PpEC23ns or truncated constructs of PpEC23 are provided to the right of the graph. Initial inoculum was adjusted uniformly to 105 CFU/mL. Numbers of bacteria were evaluated at 0 dpi and 4 dpi. C. Transcript level fold change of immune marker genes PR1a, PR2, WRKY12 and PI1 in leaves of N. benthamiana stably transformed with EV (white bars) or FLAG-PpEC23ns (gray bars) at 6 hpi with P. fluorescens strain EtHAn. NbAct1 was used as the internal reference gene. T-tests were performed for each comparison. The corresponding P value is shown in the figure. Four biological and four technical replicates were performed.
Since PpEC23 suppressed basal defense, we expected that it would enhance growth of CUCPB5115 in planta. To test this, the same bacterial strains used for the callose deposition assays were inoculated on A. thaliana (Fig 3B). As expected, wild-type Pst DC3000 grew ~100-fold better than CUCPB5115 carrying the empty vector. CUCPB5115 carrying PpEC23ns, PpEC23L-CM2-CTLC or PpEC23CM 2-CTLC grew ~10-fold better than CUCPB5115 carrying the empty vector or the other truncated versions of PpEC23. These data show that the ability of PpEC23 to suppress callose deposition is associated with enhanced bacterial growth and that CM2 and CTLC together are necessary for this activity.
The four immune marker genes used in the HR suppression assay can also be used as basal defense markers [45]. Therefore, N. benthamiana plants transformed with FLAG-PpEC23ns or empty vector were inoculated with EtHAn at OD 600 nm of 0.2 to induce PTI. mRNA levels of the four markers genes were examined at 6 hpi using qRT-PCR (Fig 3C). All four marker genes were induced to lower levels in FLAG-PpEC23ns plants, especially PR1a and PR2, with 3.7 and 3.5 fold lower induction, respectively. The abilities of PpEC23 to suppress callose deposition, promote CUCPB5115 multiplication, and suppress defense gene expression are consistent with a basal defense suppression function.
PpEC23 interacts with itself
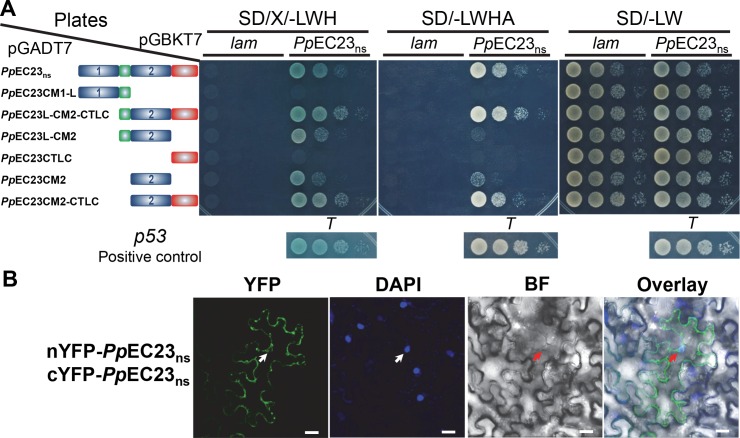
PpEC23 is a cysteine-rich protein and the cysteine residues in cysteine-rich proteins are reported to form inter- and intramolecular disulfide bridges, which could help to stabilize their ternary structures or to form homodimers or homopolymers [46]. Using a yeast two hybrid assay (Y2H), we found that PpEC23ns does interact with itself (Fig 4A and S6 Fig). To determine the motifs contributing to this self-interaction, we examined the interactions between PpEC23ns and six truncated PpEC23 derivatives (Fig 4A). Ten-fold serially diluted yeast strains were simultaneously cultured on SD (-Leu/-Trp) plates, where all the strains should grow well, and SD/X-α-gal (-Leu/-Trp/-His) and SD (-Leu/-Trp/-His/-Ade) plates, where only interaction-positive strains should grow. PpEC23ns strongly interacted with PpEC23L-CM2-CTLC and PpEC23 CM2-CTLC (Fig 4A). Surprisingly, we also found that PpEC23ns exhibits weaker interactions with PpEC23L-CM2 and PpEC23CM2. Collectively, these data show that CM2 is sufficient for dimerization, while CM2 and CTLC together are necessary for complete interaction.
Fig 4. PpEC23 interacts with itself.
A. Y2H assay showing that PpEC23 interacts through the C-terminal CM. SD/X/-LWH, SD/-LWHA and SD/-LW represent SD/X-α-gal (-Leu/-Trp/-His), SD (-Leu/-Trp/-His/-Ade) and SD (-Leu/-Trp), respectively. The structural diagrams of PpEC23ns or truncated constructs of PpEC23 are provided next to the relevant strain. The interaction of murine p53 (p53) and SV40 large T-antigen (T) was used as a positive control for the system, and human lamin C (lam) was used as a negative control. B. BiFC assay showing that PpEC23 interacts with itself in planta. YFP, yellow fluorescent protein epifluorescence. BF, bright field. DAPI signal was used as a nuclear marker. Arrows indicate nuclei. Representative images are shown (n ≥ 20). Bar = 20 μm.
To test if the PpEC23 self-interaction occurs in planta, a bimolecular fluorescence complementation (BiFC) assay was used in N. benthamiana leaves [47]. For the BiFC assay, the N-terminal half of YFP (nYFP) and the C-terminal half of YFP (cYFP) were fused to the N- or C-terminal end of PpEC23ns. Reciprocal combinations of fusion proteins were transiently expressed in N. benthamiana leaves infiltrated with Agrobacterium tumefaciens and were tested for interaction. Representative confocal microscopy images showing PpEC23 dimerization are presented in Fig 4B. We observed strong YFP signals in the cytoplasm, suggesting that PpEC23 protein can also form stable dimers in plant cells with a cytoplasmic subcellular localization (Fig 4B; positive and negative controls for BiFC assays are presented in S7 Fig). Fluorescence could only be observed when the nYFP and cYFP domains were fused to the N terminus of PpEC23ns, without disturbing the functional domains in the C terminus. We noticed some YFP signal appeared to be associated with the nucleus. However, based on overlay images, these appear to be surrounding the nucleus. A similar localization pattern was observed when PpEC23 was tagged with full-length GFP, although there also appears to be a significant amount of aggregation of the protein in the cytoplasm (S8 Fig).
PpEC23 interacts with a soybean transcription factor, GmSPL12l
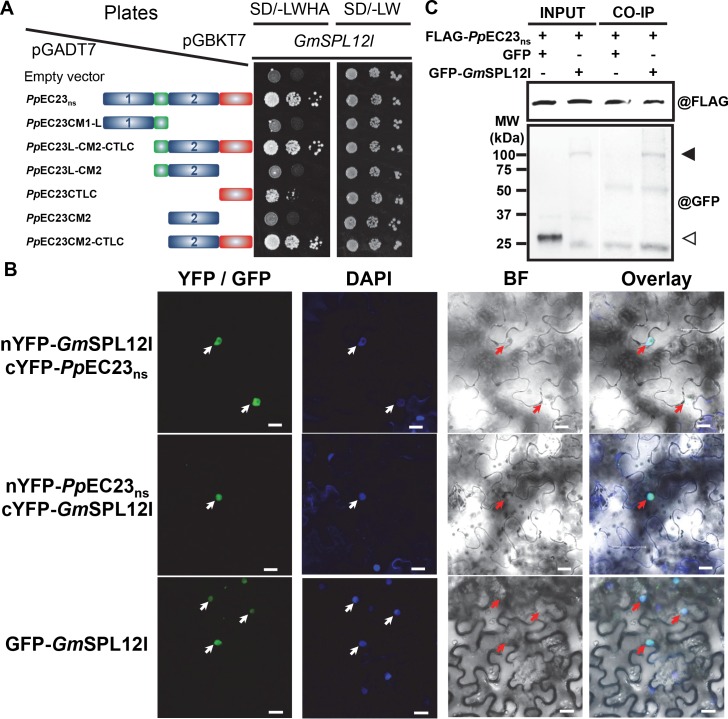
To further investigate the mechanism by which PpEC23ns interferes with plant immune responses, we performed an Y2H screen against a soybean cDNA library. A single interacting protein candidate was identified to be the C-terminal 899 amino acids (aa) of GmSPL12l (Glyma.10g009200), which is a 1,016 aa full-length protein. To confirm the protein-protein interaction observed in yeast, we cloned the full length GmSPL12l and performed Y2H against the PpEC23ns and 6 truncated constructs of PpEC23 using the GAL4 Y2H system (Fig 5A). All strains grew well on SD (-Leu/-Trp) plates. On SD (-Leu/-Trp/-His/-Ade) plates, we observed that PpEC23ns, together with PpEC23L-CM2-CTLC and PpEC23CM2-CTLC, exhibit strong protein-protein interactions with GmSPL12l. Moreover, we found that PpEC23CTLC weakly interacted with GmSPL12l. These results suggest that CTLC is sufficient for the interaction between PpEC23 and GmSPL12l, while the full interaction requires both CM2 and CTLC.
Fig 5. PpEC23 interacts with soybean transcription factor GmSPL12l.
A. PpEC23 and GmSPL12l interaction confirmed by Y2H. The empty vector, pGADT7, was included as a negative control. SD/-LWHA and SD/-LW represents SD (-Leu/-Trp/-His/-Ade) and SD (-Leu/-Trp), respectively. The structural diagrams of PpEC23ns or truncated constructs of PpEC23 are shown next to the corresponding strain. B. PpEC23 and GmSPL12l interaction detected in N. benthamiana nuclei by BiFC assay. YFP/GFP, yellow or green fluorescent protein epifluorescence. BF, bright field. DAPI signal was used as a nuclear marker. Arrows indicate nuclei. Representative images are shown (n ≥ 20). Bar = 20 μm. C. PpEC23 and GmSPL12l interaction confirmed by co-immunoprecipitation assay (CoIP). MW, molecular weight marker. The solid triangle indicates the band corresponding to the GFP-GmSPL12l fusion protein. The white triangle indicates the GFP protein band. @FLAG and @GFP indicate detection using anti-FLAG and anti-GFP antibodies, respectively.
We then used the BiFC assay to test if the PpEC23 and GmSPL12l interaction could occur in planta [47]. nYFP and cYFP were fused to the N or C-terminal ends of the test proteins, PpEC23ns and GmSPL12l, and these fusion proteins were expressed in N. benthamiana by Agrobacterium infiltration (S9 Fig). Representative confocal microscopy images show that PpEC23 and GmSPL12l can interact with each other (Fig 5B). Co-expression of PpEC23ns and GmSPL12l resulted in bright fluorescence in the nuclei, similar to the localization pattern of GFP-GmSPL12l alone (Fig 5B). Fluorescence was only observed when the nYFP and cYFP domains were fused to the N terminus of PpEC23ns and GmSPL12l, indicating the importance of the C termini of both proteins for the interaction. These data suggest that PpEC23 interacts with GmSPL12l in planta and the interaction occurs in nuclei. One Cluster 112 family member from P. pachyrhizi, PpC112-7, which contains a 10-cysteine-motif [8], and one SPL family member from A. thaliana, AtSPL6, which is a positive immune regulator [48], were included to test the specificity of the interaction (S7 Fig). Neither PpC112-7 and GmSPL12l, nor AtSPL6 and PpEC23ns resulted in detectable fluorescence, indicating that the interaction between PpEC23ns and GmSPL12l is specific.
To further confirm this interaction, we tested if PpEC23 interacts with GmSPL12l in N. benthamiana extracts using a pull-down assay. GFP or GFP-GmSPL12l was transiently expressed in leaves of the N. benthamiana lines expressing FLAG-PpEC23ns. Total soluble proteins were extracted from leaves at 48 hpi, and FLAG-PpEC23ns was pulled down using anti-FLAG beads. Western blot analysis showed that GFP-GmSPL12l was pulled down together with FLAG-PpEC23ns, while GFP, as a negative control, was not pulled down under the same conditions (Fig 5C and S10 Fig). The results demonstrate that FLAG-PpEC23ns physically associates with GFP-GmSPL12l in plant extracts.
Because SPL transcription factors exist widely in many plant species and PpEC23 also suppresses defense in A. thaliana and N. benthamiana, we investigated the hypothesis that PpEC23 could interact with SPL homologs in these species by using the BiFC assay. Co-expression of NbSPL1-1 (Niben101Scf01773g04003.1) or AtSPL1 (AT2G47070) with PpEC23ns resulted in bright fluorescence in the nuclei, similar to the localization pattern of the PpEC23-GmSPL12l interaction (S11 Fig). The interaction specificity between PpEC23 and SPL12 homologs in other non-host plants was confirmed by including the same set of negative control combinations. These data suggested that PpEC23 can also similarly interact with homologs of GmSPL12l in A. thaliana and N. benthamiana, suggesting that PpEC23 may use similar mechanisms to suppress the defense responses in non-host and host plants.
GmSPL12l can suppress the soybean defense responses
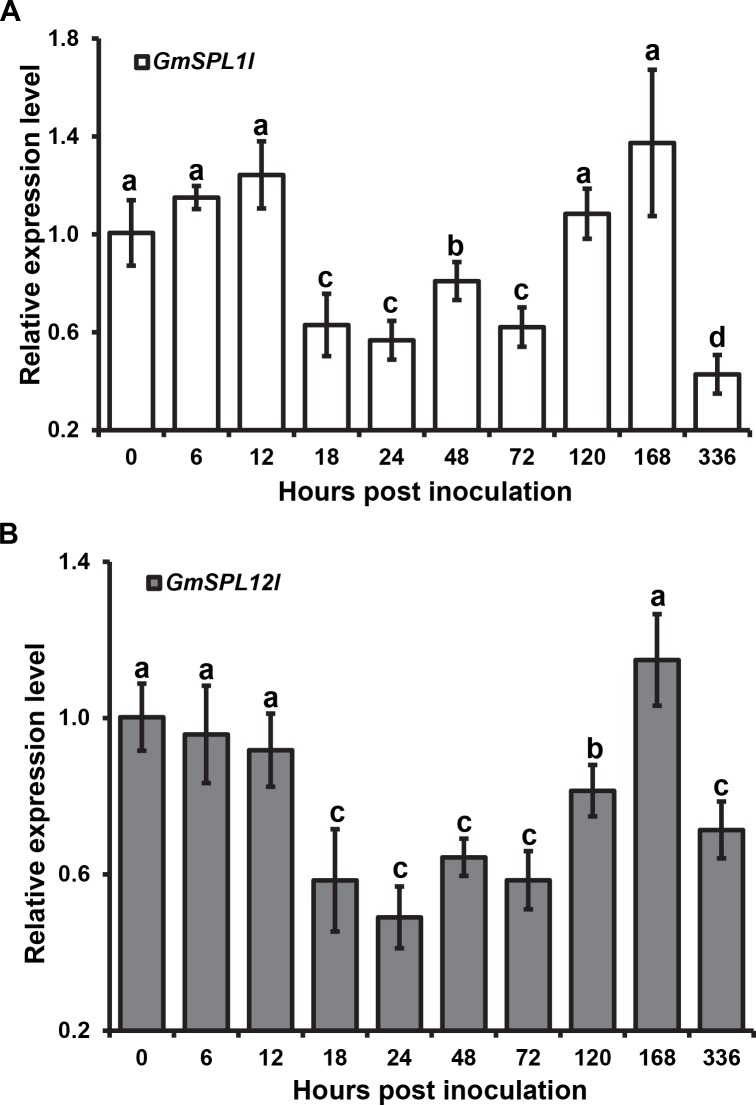
GmSPL12l belongs to the large SQUAMOSA promoter binding-like (SPL) transcription factor family that is involved in many biological processes [49]. In the soybean genome, GmSPL12l has a paralogue, GmSPL1l (Glyma.02g008600), that differs by only 57 aa. By using qRT-PCR, we found that expression profiles of these two transcription factors are similar during infection (Fig 6). The mRNA transcripts of both genes are down regulated in the time range of 18 to 72 hpi. Afterwards their mRNA levels return to levels comparable to the 0 hpi time point before decreasing again at 336 hpi.
Fig 6. The expression profile of GmSPL1l (A) and GmSPL12l (B) mRNA during P. pachyrhizi infection.
Expression levels are relative to the expression in soybean without P. pachyrhizi infection. Three independent biological and four technical replicates were performed. Error bars = standard deviation. Single factor ANOVA and pair-wise two-tailed t-test analyses were performed. The letters above the error bars indicate the different groups with statistical significance (P < 0.05). The soybean Ukn2 gene [41] was used as the internal reference gene.
To investigate its role in plant immunity, GmSPL12l was silenced using Bean pod mottle virus (BPMV)-mediated gene silencing [50]. At 21 dpi, shorter plants with smaller leaves were observed when GmSPL12l was silenced compared to the BPMV empty-vector controls (Fig 7A, 7B and 7D). Phenotypes were consistent in three independent replicates. To confirm the silencing of GmSPL12l, qRT-PCR was performed to demonstrate that GmSPL12l mRNA levels were decreased (Fig 7C). Since GmSPL1l is nearly identical in sequence with GmSPL12l, we also detected GmSPL1l mRNA levels in GmSPL12l silencing plants. GmSPL1l mRNA levels were also significantly decreased as expected (S12 Fig). We reasoned that the GmSPL12l silencing phenotype may have two causes: 1) GmSPL12l silencing disturbs plant developmental processes, yielding stunted plants; or 2) GmSPL12l-silenced plants have induced defense responses that antagonistically suppress plant growth and development.
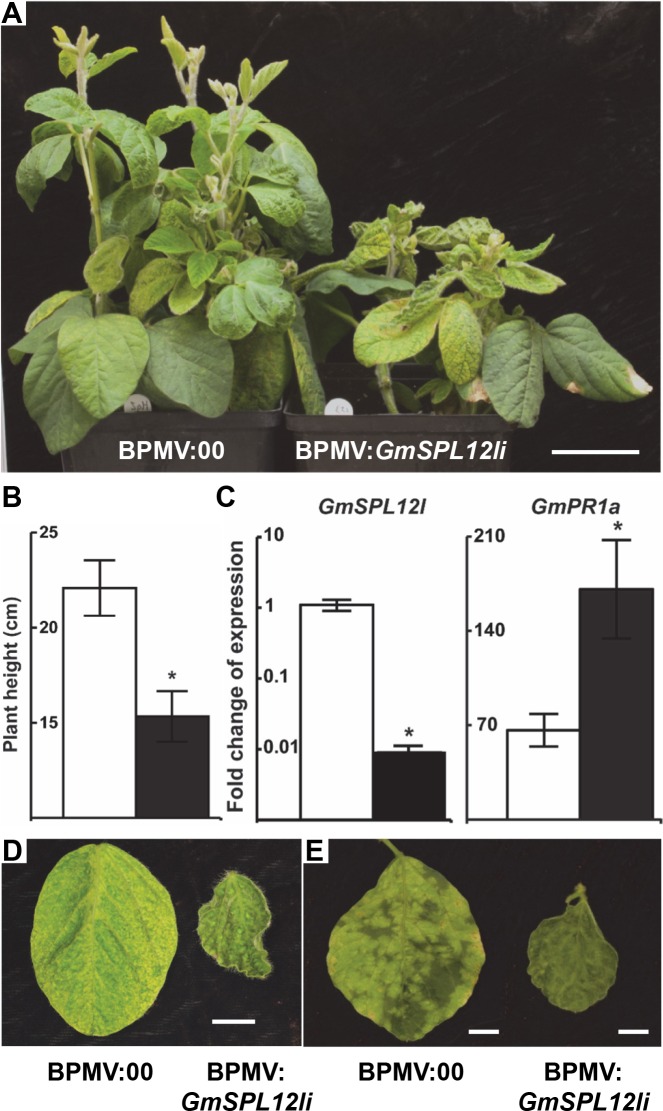
Fig 7. GmSPL12l is a negative regulator of plant immunity.
A. Phenotypes of soybean plants 21 dpi with BPMV. A. Soybean plants infected with the BPMV empty vector (BPMV:00) (left) and BPMV:GmSPL12li (right). Bar = 5 cm. Representative images are shown (n ≥ 6). B. Heights of plants at 21 dpi with BPMV:00 (white bars) and BPMV:GmSPL12li (black bars). A t-test was performed for the pair-wise comparison. The * indicates significant difference (P < 0.05). Six biological replicates were performed. C. Fold change of GmSPL12l (left panel) and GmPR1a (right panel) mRNA in BPMV:00 (white bars) and BPMV:GmSPL12li (black bars) plants. GmAct1 was used as the internal reference gene. A t-test was performed for each comparison. The * indicates significant difference (P < 0.05). Four biological and four technical replicates were performed. D. Representative leaves from BPMV:00 (left) and BPMV:GmSPL12li (right) plants (n ≥ 18). Bar = 1 cm. E. Downy mildew symptoms on leaves of BPMV:00 (left) and BPMV:GmSPL12li (right) plants inoculated with P. manshurica. Leaves were photographed at 28 days post BPMV inoculation (7 days post P. manshurica inoculation). Representative third trifoliate leaves are shown (n ≥ 12). The assay was repeated three times. Bar = 1 cm.
To test these hypotheses, GmSPL12l-silenced and BPMV empty-vector control plants at 21 dpi were inoculated with the biotrophic pathogen Peronospora manshurica (downy mildew). Seven days later, strong chlorotic lesions caused by P. manshurica infection were observed on leaves of BPMV empty-vector control plants, while no lesions could be observed on the leaves of GmSPL12l-silenced plants (Fig 7E). The lack of disease indicated that GmSPL12l silencing decreased the susceptibility of soybean to P. manshurica. We then examined the expression level of GmPR1a, a marker gene for soybean defense responses [51], in both BPMV empty-vector control and GmSPL12l-silenced plants at 21 dpi using qRT-PCR to determine if defense responses were activated in GmSPL12l-silenced plants (Fig 7C). Expression GmPR1a was induced 2.6 fold in GmSPL12l-silenced plants compared to that in empty-vector control plants, suggesting that GmSPL12l silencing significantly activates soybean defense responses. Taken together, these results indicate that GmSPL12l functions as a negative regulator of soybean defense responses.
To test if N. benthamiana homologs of GmSPL12l might also function as negative regulators of plant immune responses NbSPL1-1 (Niben101Scf01773g04003.1), NbSPL1-2 (Niben101Scf09781g00016.1), NbSPL1-3 (Niben101Scf00687g04018.1) and NbSPL12 (Niben101Scf02558g00030.1) were silenced using Tobacco rattle virus (TRV)-mediated gene silencing [52,53]. Three silencing constructs were designed: NbSPL1-1/NbSPL1-2i for silencing both NbSPL1-1 and NbSPL1-2, NbSPL1-3/SPL12i for silencing both NbSPL1-3 and NbSPL12, and NbSPL1si for silencing all four NbSPL1s. At 10 dpi, the TRV-infected plants started to show viral symptoms. There were no obvious differences in morphological or growth phenotypes in the NbSPL1s silenced plants, compared to the plants infected with TRV empty vector control. To assess the silencing of NbSPL1 target genes, qRT-PCR was performed to demonstrate that NbSPL1 mRNA levels were decreased (S13 Fig). The expression level of NbSPL1-1/SPL1-2 was decreased in NbSPL1-1/SPL1-2i and NbSPL1si plants by 64% and 18%, respectively, compared to that of the plants with the TRV empty vector control. The expression level of NbSPL1-3/SPL12 was decreased in NbSPL1-3/SPL12i and NbSPL1si plants by 49% and 67%, respectively. These data confirmed the silencing of NbSPL1s. To determine the impact of silencing NbSPL1s on N. benthamiana defense responses, the expression levels of the same set of four immune marker genes used in the HR suppression assay were detected by performing qRT-PCR (S13 Fig). The expression levels of all four marker genes were increased in the NbSPL1 silenced plants, except NbPI1 in NbSPL1-1/SPL1-2i plants. These data indicated that NbSPL1s, the homologs of GmSPL12l in N. benthamiana, also act as negative regulators of defense responses, similar to GmSPL12l in soybean.
Discussion
In this study, we determined that PpEC23 has the ability to suppress HR and basal defense when delivered into plant cells via the T3SS or expressed in transgenic plants. PpEC23 is a modular cysteine-rich protein, in which CM2 and the CTLC domains mediate self-interaction and interaction with a soybean protein. The interacting soybean protein identified is GmSPL12l, a member of the large SPL family of transcription factors. Silencing of GmSPL12l activates mRNA expression of soybean defense genes and enhances resistance to the downy mildew pathogen, P. manshurica, suggesting that PpEC23 may promote P. pachyrhizi infection by interacting with a negative regulator of soybean immune responses. The most likely hypothesis postulates that PpEC23 binding alters the stability or activity of GmSPL121, thereby weakening the soybean defenses.
PpEC23, a SSCRP
Known and predicted effectors from filamentous plant pathogens, including fungi and oomycetes, are often SSCRPs, which are major components of the secretome [54,55]. SSCRPs are normally defined as small proteins (less than 300 aa) with high cysteine content (the total number of cysteine residues representing more than 5% of the mature protein and/or greater than 4 cysteine residues in total), following the predicted secretion signal at the N terminus [55]. SSCRPs from plant pathogens play very important and even decisive roles in the processes of suppression of signal transduction or gene expression in plant cells, protection against antifungal compounds or enzymes. In rust fungi, hundreds of SSCRPs have been predicted, but few of them have been characterized for their roles in virulence [1,8,13,56].
The N-terminal 25 aa of PpEC23 were predicted to encode a putative secretion signal [8], and it was proven functional in a yeast secretion assay (Fig 2E). Excluding the secretion signal, mature PpEC23 is predicted to be 267 aa in total and contains 21 cysteine residues (7.9%), qualifying it as an SSCRP. Furthermore, the cysteine content is as high as 11% within its two tandem repeats of the highly conserved ten-cysteine-motif (Fig 2A). The higher cysteine content significantly increases the number of possibilities for disulfide bridges, which makes the ternary structure of PpEC23 difficult to predict.
Previous work suggested that PpEC23 belongs to a large fungal protein family, Cluster 112, whose members all contain the ten-cysteine motif and most also possess a predicted secretion signal [8]. Sequence alignment showed that the number and positions of the 10 cysteine residues are conserved in all family members, while the intervening amino acid sequences are highly variable, suggesting that the family-specific structure is essentially determined by the number and positions of cysteine residues. The abundance of family members in each rust fungal species implies their importance for these fungi. Notably, all of the family members contain only one copy of the 10-cysteine-motif except for PpEC23, which contains two.
C-terminal low complexity motif
The C-terminal 59 aa of PpEC23 was predicted as a CTLC motif, which has no reference structures in the structural prediction databases. Thus, the CTLC is expected to have structural flexibility. Surprisingly, we found that the CTLC was sufficient to mediate PpEC23’s interaction with GmSPL12l in the Y2H assay, and this interaction was strengthened by the CM2. Based on this observation, we concluded that the CTLC, together with the CM2, can acquire and maintain the proper structure to interact with GmSPL12l. It was reported that proteins with a terminal low complexity region (LCR) tend to interact with more proteins than those with central LCRs or those without an LCR [57]. Other than in translation and in transport processes, terminal LCR-containing proteins are also enriched in stress-related processes. The N terminal LCR of XopD from Xanthomonas campestris pv. vesicatoria was demonstrated to enhance bacterial virulence on host plants by forming a coiled-coil-like structure and dimerization [58]. Thus, it is reasonable to expect that the CTLC of PpEC23 interferes with the host plant immune system by interacting with host proteins.
The genomic sequence of PpEC23 is well maintained
SNP analyses indicated that nucleotide and aa sequences encoded by PpEC23 are maintained in diverse P. pachyrhizi isolates (Fig 2C, Table 1). These isolates were collected from Asia, Africa, South America, and the U.S.A. over a period of approximately 32 years. Most of the isolates, with the exception of BZ01-1, carry two alleles at most of their SNP sites indicating that they are heterozygous. The SNP604, which is only present in BZ01-1, PG01-3, and HW94-1, causes a conservative aa change from isoleucine to valine at position 125, which is in CM1. Based on our analysis of the six PpEC23 truncation mutants, a mutation at this position was not expected to affect ability of PpEC23 to suppress plant immunity or interact with the host target GmSPL12l. SNP1805 is within the CTLC and causes an alanine to proline change, which could potentially have interfered with the function of PpEC23. The introduction of the aa differences in the PpEC23 sequence did not affect its ability to suppress plant immunity.
PpEC23 suppresses plant immunity
The obligate biotrophic nature of P. pachyrhizi accompanied by the inability to perform sexual crossings currently limits our ability to directly determine the functions of its genes. To overcome these limitations, we utilized the bacterial secretion vector pEDV6, bacterial strain Pst DC3000, and the non-pathogenic P. fluorescens strain EtHAn to explore the functions of PpEC23 [6,13,29,59,60]. In bacterial inoculation assays, PpEC23 could suppress both HR and basal defense (Figs 1 and 3). PpEC23 was not inherently antimicrobial based on the bacterial growth curve assay (S2 Fig), and its ability to suppress HR was not limited to a single NBS-LRR protein-effector interaction. Pst DC3000 causes HR on tobacco and soybean through the recognition of different effectors, HopQ1-1 and AvrD, respectively [30,31]. This observation indicates that PpEC23 functions at a point of convergence downstream of Avr-R recognition that is conserved between legumes and solanaceous plants. The possibility remained that PpEC23 has a direct effect on Pst DC3000. However, supplying PpEC23 in trans by co-inoculating Pst DC3000 with EtHAn strains, or inoculating Pst DC3000 on FLAG-PpEC23ns transgenic plants confirmed that HR suppression occurs in planta and is not due to an effect that occurred within Pst DC3000 (Fig 1B and S1 Fig). Subsequently, we demonstrated that PpEC23 also suppresses basal defense, suggesting that it may interfere with a common node of basal defense and HR signaling. There is increasing evidence that basal defense and HR are not entirely distinct processes [28]. Because PpEC23 affected the expression of the defense marker genes PR1a, PR2, WRKY12, and PI1 in response to HR and basal defense cues, we expect that it affects regulatory components that are upstream of activation of the expression of these genes. Our data suggests a function for PpEC23 that occurs within cells, which is consistent with its expression in haustoria. However, the finding that it is highly expressed beginning with the formation of appressoria suggests that it could have functions in the apoplast and/or earlier in infection. One of the peculiarities of P. pachyrhizi is that it does not use stomata to enter into the host, but instead it directly penetrates epidermal cells, which subsequently die [61]. It may be necessary for the fungus to express immune suppressing proteins early to slow cell death or to prevent it from spreading to neighboring cells. It will be interesting to investigate if PpEC23 may have functions in suppressing host immunity throughout the biotrophic interaction of P. pachyrhizi and soybean.
GmSPL12l as a target of PpEC23
SPL proteins constitute a plant-specific family of transcription factors, that are present in single cell algae to higher plants [49]. The SPL family is defined by the conserved SQUAMOSA promoter-binding protein (SBP) domain, consisting of a bipartite nuclear localization signal (NLS) and two, non-interleaved Zn-finger-like structures [62]. SPL proteins are conserved in the SBP domain, which is normally located in the N terminus, but they have diverse C-terminal domains. In the model plant A. thaliana, SPLs have been well characterized and most are involved in developmental and adaptive programs, including leaf morphogenesis [63], plastochron determination [64], vegetative phase transition [65], flowering [66], anther and gynoecium development [67] and copper homeostasis [68]. AtSPL6 was recently reported to positively regulate the defense transcriptome following its association with nuclear-localized R proteins [48]. Although the functions of SPL proteins have been analyzed in other plant species, this is the first report of a function for an SPL protein in soybean [69,70]. There are a total of 48 SPL family members predicted in the soybean genome. Sequence analyses of SBP domains suggested that they belong to eight clades along with the 16 AtSPLs [71]. GmSPL12l and its paralogue, GmSPL1l, are most closely related to AtSPL1/12/14/16 according to phylogenetic analyses. However, these AtSPLs are known to play roles in development but not defense.
We determined that GmSPL12l and PpEC23 interact using three independent assays: Y2H, BiFC, and Co-IP (Fig 5). In these assays, GmSPL12l and PpEC23 interacted only when the epitope tags were fused to the N terminus of both proteins, suggesting the C termini of both proteins are important for protein-protein interaction. Sequence analyses of the C-terminus of GmSPL12l shows that it contains ankyrin-repeats, which normally mediate protein-protein interactions [63]. In the BiFC assay, the interaction of GmSPL12l and PpEC23 was found to localize to the nuclei of plant cells (Fig 5B). Taking into account the cytoplasmic localization of PpEC23 and the nuclear localization of GmSPL12l, the BiFC results show that GmSPL12l can recruit PpEC23 into the nuclei. Furthermore, the observation that the PpEC23 self-interaction was not present in nuclei suggests that does not bind to GmSPL12l as a multimer. The ability of PpEC23 to also interact with AtSPL1 and NbSPL1-1 in the BiFC assay (S11 Fig) provides a potential mechanism by which it suppresses host immunity in legume, brassica, and solanaceous plants.
GmSPL12l and its paralogue GmSPL1l are highly conserved with only 57 aa differences. The mRNA sequences of these homologous genes, including the 5’- and 3’-UTRs, are also nearly identical. Therefore, our BPMV:GmPL12li gene silencing construct silenced both genes, although GmSPL12l was silenced ~10 fold more than GmSPL1l (Fig 7C and S11 Fig). The similar sequences and expression profiles of the two genes strongly suggest that they may have redundant function, but we have been unable to establish that GmSPL1 can interact with PpEC23 using Y2H. Silencing of GmSPL12l and its paralogue resulted in plants that were more resistant to downy mildew and exhibiting a stunted phenotype, suggesting that immunity was activated and plant growth was suppressed (Fig 7). The inverse relationship between immunity and growth is commonly observed in mutants that have elevated or constitutive defense responses, including in soybean [51]. We have performed P. pachyrhizi infection assays on the GmSPL12l-silenced and PpEC23-silenced soybeans, however, no change in macroscopic disease phenotypes was observed to date. This negative result is possibly due to the functional redundancy of P. pachyrhizi effector proteins that suppress immunity. Collectively, our data show that GmSPL12l acts as an immune suppressor possibly to ensure normal plant growth in the absence of infection. However, we postulate that during defense responses, the immune suppression by GmSPL12l could be overcome or dampened to protect the plants from infection.
Transcript levels of GmSPL12l and its paralogue decreased after P. pachyrhizi infection about two fold between 18 and 72 hpi compared to 0 hpi (Fig 6). After 72 hpi, their levels returned to that in non-infected plants suggesting that transcription of GmSPL12l is down regulated during early stages of P. pachyrhizi infection. Because silencing of GmSPL12l activated soybean immunity, we expect that it is a negative regulator of plant defenses. Therefore, decreased levels of its transcripts may also lead to activation of soybean defenses during early stages of P. pachyrhizi infection. The interaction of PpEC23 with GmSPL12l suggests that PpEC23 may manipulate post-translationally or modify GmSPL12l to suppress soybean immunity, which would potentially promote P. pachyrhizi infection. PpEC23 mRNA was originally observed in haustoria, but our analyses here showed that it is expressed in pre-infection structures as well as during the infection time course. These data suggest that PpEC23 functions early and throughout the biotrophic interaction between P. pachyrhizi and soybean, and one of its activities may be to interfere directly with the function or post-translational regulation of GmSPL12l. These will be interesting hypotheses to investigate in the future after we have developed the necessary tools in soybean.
The elevated expression of PR1a in GmSPL12l-silenced and NbSPL1-silenced plants is consistent with our conclusion that PpEC23 targets a protein upstream of defense gene expression at a point of convergence between HR and basal defense, which is conserved between non-host and host plants. However, we do not know if GmSPL12l or NbSPL1 directly or indirectly regulate the expression of defense marker genes such as PR1a. In general, the genes that are direct transcriptional targets of the SPLs are not yet known. Studies to investigate this will be aided by stable mutations that knock out GmSPL12l function and development of resources needed for performing chromatin immunoprecipitation coupled with RNA-seq. On the rust side of the interaction, the cluster 112 protein family characterized by the signature ten-cysteine motif is widely present in rust fungal species for which sequence information is available [8]. The presence of this motif suggests that PpEC23 could be a representative of a conserved protein-protein interaction platform that rust fungi use to manipulate host immune responses. It will be interesting to further explore the functions of this family of SSCRPs as virulence factors in rust pathogen-plant interactions.
Materials and Methods
Bacterial strains and plasmids
Bacterial strains used in this study are listed in Table 2. E. coli and A. tumefaciens were grown in Luria-Bertani (LB) broth at 37°C (E. coli) or 28°C (A. tumefaciens) using either liquid or solid media. Pseudomonas strains were grown in either LB or King’s B (KB) medium at 28°C. Plasmids were mobilized from E. coli to Pseudomonas strains by standard triparental mating using E. coli HB101 (pRK2013) as a helper strain.
Table 2. Strains and plasmids.
| Strain or plasmid | Genotype or relevant phenotype * | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK - mK +) λ– | Invitrogen |
| TOP10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ- | Invitrogen |
| P. syringae pv. tomato | ||
| DC3000 | Wild type, Rifr | |
| CUCPB5115 | ∆CEL::ΩSp/Smr, Rifr Spr | [72] |
| P. fluorescens | ||
| EtHAn | P. fluorescens Pf0-1 carrying a working TTSS from P. syringae pv. Syringae, Cmr | [33] |
| Agrobacterium tumefaciens | ||
| GV3101 | Carries Vir plasmid encoding T-DNA transfer machinery, Rifr, Gmr | |
| Saccharomyces cerevisiae | ||
| EGY48 | MATa, his3, trp1, ura3, LexA op(x6) -LEU2 | Clontech |
| AH109 | MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4∆, gal80∆, LYS2::GAL1 UAS -GAL1 TATA -HIS3, GAL2 UAS -GAL2 TATA -ADE2,URA3::MEL1 UAS -MEL1 TATA -lacZ | Clontech |
| YTK12 | suc2Δ9 trp1Δ ade2-101 ura3-52 | [42] |
| Plasmids | ||
| pCR8⁄GW⁄TOPO | Gateway-compatible entry vector, Spr | Invitrogen |
| pEDV6 | Gateway-compatible version of pEDV3, Gmr | [29] |
| pSITEII-3C1 | Gateway-compatible binary vector for transiently over-expression of EGFP-fused protein in planta, Smr | [73] |
| pBI121 | Binary vector for transformation in plant, Kmr in bacteria, Kmr in plants | [74] |
| pSuc2t7M13ori | Yeast signal sequence trap vector, Ampr in bacteria, -Trp in yeast | [42] |
| pLexA | Bait plasmid for yeast LexA two-hybrid system containing BD domain, Ampr in bacteria, -His in yeast | Clontech |
| pB42AD | Library plasmid for yeast LexA two-hybrid system containing AD domain, Ampr in bacteria, -Trp in yeast | Clontech |
| P8op-lacZ | LacZ reporter plasmid for yeast LexA two-hybrid system, Ampr in bacteria, -Ura in yeast | Clontech |
| pGBKT7 | Protein expression plasmid for yeast Gal4 two-hybrid system containing BD domain, Kmr in bacteria, -Trp in yeast | Clontech |
| pGADT7 | Protein expression plasmid for yeast Gal4 two-hybrid system containing AD domain, Ampr in bacteria, -Leu in yeast | Clontech |
| pSPYNE-35S | BiFC plasmid containing YFP N-terminal fragment fused to the C-terminus of insertion, Kmr in bacteria, Kmr in plants | [47] |
| pSPYCE-35S | BiFC plasmid containing YFP C-terminal fragment fused to the C-terminus of insertion, Kmr in bacteria, Barr in plants | [47] |
| phygII-SPYNE(R)155 | BiFC plasmid containing eYFP N-terminal fragment fused to the N-terminus of insertion, Kmr in bacteria, Hygr in plants | [75] |
| pkanII-VYCE(R) | BiFC plasmid containing Venus C-terminal fragment fused to the N-terminus of insertion, Kmr in bacteria, Kmr in plants | [75] |
| pBPMV-IA-V2 | BPMV-based gene silencing vector, Ampr | [50] |
| pYL192 | TRV-based gene silencing vector, containing TRV RNA1, Kmr in bacteria, Kmr in plants | [52] |
| pYL279 | TRV-based gene silencing vector, containing TRV RNA2-Gateway cassette, Kmr in bacteria, Kmr in plants | [53] |
| pYL124 | TRV-based gene silencing positive control, containing TRV RNA2-NtPDS, Kmr in bacteria, Kmr in plants | [52] |
*: Antibiotics concentrations (μg/ml) were used as follows: Rifampicin (Rif) 100, Kanamycin (Km) 75, Gentamycin (Gm) 50, Spectinomycin (Sp) 50, Chloramphenicol (Cm) 30, Ampicillin (Amp) 100, Streptomycin (Sm) 100, Hygromycin B (Hyg) 30 and Basta (Bas) 100.
Plant and fungal material, inoculation procedure, and production of germ tubes and appressoria
N. benthamiana, N. tabacum, and soybean plants were grown in controlled environment chambers at an average temperature of 24°C (range 20°C—26°C), with 45% - 65% relative humidity under long-day conditions (16 h light). A. thaliana plants were grown in controlled environment chambers at an average temperature of 22°C (range 18°C—24°C), with 45% - 65% relative humidity under short-day conditions (10 h light). For P. pachyrhizi inoculation, 21 days old soybean plants (cultivar Thorne) were sprayed with a watery suspension containing 0.02% Tween 20, 0.2% milk powder and 0.02% P. pachyrhizi (isolate Thai1, laboratory collection Universität Hohenheim) urediospores. Plants were kept in the dark over night at 95% humidity and 20°C and then transferred to a greenhouse chamber (day/night 16 h/ 8 h, 22°C). To obtain the germ tube stage, 100 μg urediospores were spread on water in petri dishes and incubated for 12 h at room temperature in the dark. For appressoria, a watery suspension containing 0.02% Tween 20, 0.2% milk powder and 0.02% urediospores was sprayed on a polyethylene membrane and incubated for 16 h in 100% humidity at room temperature in the dark.
SNP identification
Genomic DNA was extracted from P. pachyrhizi urediospores using the QIAGEN DNeasy Plant Mini Kit. The PpEC23 genomic sequence was PCR-amplified with Q5 High-Fidelity DNA Polymerase (New England Biolabs) and primers KP680 and KP713 (S1 Table). PCR products were gel purified, modified with 3’-A overhangs using Taq polymerase, and cloned into pCR2.1-TOPO. Clones were sequenced and analyzed using BioEdit (Tom Hall, Ibis Biosciences) to identify SNPs. The SNP density was calculated as the number of SNPs/kb of genomic sequence.
Bacterial inoculation and growth in planta
Nicotiana plants used in this study were between 5 and 6 weeks old, A. thaliana plants were between 4 and 5 weeks old, and soybean plants were 14 days old. All plant assays were performed by infiltrating a bacterial suspension into plant leaves with a needleless syringe. A. tumefaciens strains were re-suspended in infiltration buffer (100 μM acetosyringone, 10 mM MES, pH 5.6, and 10 mM MgCl2) and kept at room temperature for 3 h before infiltration. All other strains were re-suspended in 10 mM MgCl2. Areas of bacterial infiltration were marked lightly with a Sharpie permanent marker. Levels of bacterial inoculum used in experiments are noted in the figures. Bacterial levels in planta were determined by cutting leaf disks with a cork borer (inner diameter 0.5 cm) and homogenizing them in 500 μl of the inoculation buffer. The resulting suspension containing the bacteria was diluted and plated on KB plates with the appropriate antibiotics.
Trypan blue staining
Trypan blue staining was performed as described [76] with some minor modifications. Briefly, N. tabacum and soybean leaves were harvested and incubated in an alcoholic trypan blue lactophenol solution (0.02% trypan blue in the mixture of phenol:glycerol:lactic acid:water:ethanol (1:1:1:1:8 (v/v/v/v))) at 95°C for 5 min, followed by incubation at room temperature for 24 h. Leaf samples were destained using 2.5 g/ml chloral hydrate. Pictures were taken using a Nikon D70 digital camera.
Callose staining and microscopic analysis
A. thaliana Col-0 leaves were harvested 12 h after bacterial infiltration, cleared, and stained with aniline blue for callose as described [77]. Leaves were examined with a Zeiss Axioplan II microscope with an A4 fluorescence cube. Numbers of callose depositions were determined with ImageJ software (NIH). Six adjacent fields along the length of the leaf (avoiding the midvein, leaf edge or the syringe-damaged area) were analyzed and averaged. Values in Fig 3 are the average and standard deviation of more than 6 independent leaves for each treatment.
Yeast secretion assay
Full length or truncated versions of PpEC23 were cloned into plasmid pSuc2t7M13ori for the yeast signal sequence trap assay [42]. The resulting plasmids were transformed into yeast strain YTK12 using the lithium acetate method [78] without additional carrier DNA. Transformants were plated on CMD/-W medium (0.17% yeast nitrogen base w/o aa, 0.13% dropout mix without tryptophan, 37.8 mM (NH4)2SO4, 2% glucose) [79]. The positive colonies were liquid cultured, adjusted to OD 600 nm = 1, serially diluted and plated on YPRAA medium plates (1% yeast extract, 2% bacto peptone, 2% raffinose, 2 ppm antimycin a) [42]. Plates were photographed after 4 days of incubation at 28°C.
Yeast constructs and two-hybrid screen
The Matchmaker LexA two-hybrid system (Clontech) was used for Y2H screening of a pathogen-infected soybean cDNA library. PpEC23 ns was cloned into the pLexA vector to create a fusion with the DNA BD. Approximately 2.6 × 106 yeast transformants were screened on 2% SD/Gal/Raf/X-β-gal (-Ura/-His/-Trp/-Leu) following the manufacturer’s instructions (Clontech). Direct protein-protein interaction was confirmed by co-transformation of the respective plasmids into the yeast strain AH109 using the Matchmaker GAL4 Two-hybrid System (Clontech), followed by selection of transformants on 2% SD (-Leu/-Trp) at 30°C for 3 days and subsequent transfer to 2% SD/X-α-gal (-Leu/-Trp/-His) and 2% SD (-Leu/-Trp/-His/-Ade) to select for growth of interacting clones. All the relevant constructs were listed in Supplemental Table 1.
Transient expression of GFP fusion and BiFC constructs in N. benthamiana
To create GFP-GmSPL12l, the GmSPL12l open reading frame was PCR amplified and cloned into the Gateway entry vector pCR8⁄GW⁄TOPO (Invitrogen) and then recombined into the Gateway binary destination vector pSITEII-3C1 (Table 2) using LR clonase II (Invitrogen). For BiFC constructs, GmSPL12l and PpEC23 ns were PCR amplified and cloned into the BiFC vectors pSPYNE-35S, pSPYCE-35S, phygII-SPYNE(R)155 and pkanII-VYCE(R) (Table 2). Each protein was independently tagged with either cYFP or nYFP at either the N or C terminus. All the binary vectors were introduced into A. tumefaciens GV3101 using the freeze-thaw method [80]. GFP or BiFC was detected in N. benthamiana leaves using a Leica SP5 X MP confocal/multiphoton microscope system at 48 h after infiltration.
Co-immunoprecipitation assay
For co-immunoprecipitation of PpEC23 and GmSPL12l, A. tumefaciens containing pSITEII-3C1 (empty vector) or pSITEII-3C1-GmSPL12l were infiltrated into the transgenic N. benthamiana leaves containing pBI121-FLAG-PpEC23ns (generated at the University of Nebraska Plant Transformation Core Research Facility). Total proteins were extracted as described previously [48]. The pre-cleared protein samples were incubated with anti-FLAG beads (Sigma-Aldrich) at 4°C for 3 h on a rotating shaker and washed 3 times with the buffer. The beads were boiled with 5 × loading buffer (250 mM Tris-Cl, pH 6.8, 10% SDS, 30% glycerol, 5% β-mercaptoethanol and 0.02% bromophenol blue) and samples were separated on an SDS-PAGE gel (4–15%) followed by western blotting using colorimetric alkaline phosphatase (AP) or horseradish peroxidase (HRP) systems.
RNA isolation and real-time PCR
Approximately 100 mg starting material (leaves, germ tubes, or appressoria) were used for RNA isolation using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Two μg of RNA and SuperScript III First Strand kit (Invitrogen) were used for cDNA synthesis. Quantitative real-time PCR (qRT-PCR) was performed using the cDNA and gene-specific primers listed in S1 Table. Each cDNA was amplified by quantitative PCR using iQ SYBR Green Supermix (Bio-Rad) and iCycler real-time PCR system (Bio-Rad). Plant ACTIN expression was used to normalize the expression value in each sample in most cases, and the relative expression values were determined against mock samples using the comparative Ct method (2-∆∆Ct). For normalization of P. pachyrhizi genes, RPS14 and PDK [41,81] were used, and in G. max, Ukn2 [41,82] was used for normalizing expression of the GmSPL genes.
BPMV-mediated gene silencing
BPMV vector constructs and inoculation of soybean seedlings via biolistic particle bombardment using a Biolistic PDS-1000/He system (Bio-Rad Laboratories, Hercules, CA, USA) have been previously described [50]. A 287 bp fragment from near the 3’ end of the GmSPL12l (Glyma.10g009200) open reading frame was amplified by PCR using the primer pair listed in S1 Table, and cloned into pBPMV-V2.
Downy mildew infection assay
Downy mildew infection was performed as described previously [83]. The field isolate of P. manshurica was maintained on soybean plants in the greenhouse. BPMV-infected soybean plants were spray-inoculated with the sporangia suspension (104 sporangia/ ml) of P. manshurica. Plants were kept overnight in the dark with high humidity and then moved to the growth chamber for 7 days. Symptoms of the top leaves were observed and recorded with a digital camera.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Acknowledgments
We thank S. P. Dinesh-Kumar and John McDowell for kindly providing materials and Matthias Kohlndorfer for technical assistance with the yeast secretion assay. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Iowa Soybean Association (SAW TJB), USDA National Institute of Food and Agriculture Hatch project 3708, and the Deutsche Forschungsgemeinschaft (VO595/3-1 and VO595/3-2 to RTV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Duplessis S, Cuomo CA, Lin Y-C, Aerts A, Tisserant E, et al. (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A 108: 9166–9171. 10.1073/pnas.1019315108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petre B, Joly DL, Duplessis S (2014) Effector proteins of rust fungi. Front Plant Sci 5 10.3389/fpls.2014.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nemri A, Saunders DGO, Anderson C, Upadhyaya NM, Win J, et al. (2014) The genome sequence and effector complement of the flax rust pathogen Melampsora lini . Front Plant Sci 5: 98 10.3389/fpls.2014.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Voegele RT, Mendgen KW (2011) Nutrient uptake in rust fungi: how sweet is parasitic life? Euphytica 179: 41–55. 10.1007/s10681-011-0358-5 [DOI] [Google Scholar]
- 5. Rafiqi M, Ellis JG, Ludowici VA, Hardham AR, Dodds PN (2012) Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr Opin Plant Biol 15: 477–482. 10.1016/j.pbi.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 6. Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, et al. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana . PLoS Biol 9: e1001094 10.1371/journal.pbio.1001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantu D, Segovia V, MacLean D, Bayles R, Chen X, et al. (2013) Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14: 270 10.1186/1471-2164-14-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Link TI, Lang P, Scheffler BE, Duke M V, Graham MA, et al. (2014) The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Mol Plant Pathol 15: 379–393. 10.1111/mpp.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pretsch K, Kemen A, Kemen E, Geiger M, Mendgen K, et al. (2013) The rust transferred proteins-a new family of effector proteins exhibiting protease inhibitor function. Mol Plant Pathol 14: 96–107. 10.1111/j.1364-3703.2012.00832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ellis JG, Dodds PN, Lawrence GJ (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45: 289–306. 10.1146/annurev.phyto.45.062806.094331 [DOI] [PubMed] [Google Scholar]
- 11. Kemen E, Kemen AC, Rafiqi M, Hempel U, Mendgen K, et al. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol Plant Microbe Interact 18: 1130–1139. 10.1094/MPMI-18-1130 [DOI] [PubMed] [Google Scholar]
- 12. Upadhyaya NM, Mago R, Staskawicz BJ, Ayliffe MA, Ellis JG, et al. (2014) A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol Plant Microbe Interact 27: 255–264. 10.1094/MPMI-07-13-0187-FI [DOI] [PubMed] [Google Scholar]
- 13. Liu C, Pedersen C, Schultz-Larsen T, Aguilar GB, Madriz-Ordeñana K, et al. (2016) The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol. 10.1111/nph.14034 [DOI] [PubMed] [Google Scholar]
- 14. Catanzariti A-M, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18: 243–256. 10.1105/tpc.105.035980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kale SD, Gu B, Capelluto DGS, Dou D, Feldman E, et al. (2010) External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 981–983. 10.1016/j.cell.2010.08.035 [DOI] [PubMed] [Google Scholar]
- 16. Rafiqi M, Gan PHP, Ravensdale M, Lawrence GJ, Ellis JG, et al. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22: 2017–2032. 10.1105/tpc.109.072983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, et al. (2010) Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol 11: 169–177. 10.1111/j.1364-3703.2009.00589.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harris DK, Kendrick MD, King ZR, Pedley KF, Walker DR, et al. (2015) Identification of unique genetic sources of soybean rust resistance from the USDA soybean germplasm collection. Crop Sci. [Google Scholar]
- 19. Hartman GL, Miles MR, Frederick RD (2005) Breeding for resistance to soybean rust. Plant Dis 89: 664–666. 10.1094/PD-89-0664 [DOI] [PubMed] [Google Scholar]
- 20. Duplessis S, Joly DL, Dodds PN (2012) Rust effectors In Effectors in plant-microbe interactions. Martin F, Kamoun S, editors West Sussex, UK: John Wiley & Sons; 155–193 p. 10.1002/9781119949138.ch7 [DOI] [Google Scholar]
- 21. Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 22. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 23. Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
- 24. Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 25. Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A 104: 2531–2536. 10.1073/pnas.0608779104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwessinger B, Zipfel C (2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol 11: 389–395. 10.1016/j.pbi.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 27. Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomma BPHJ, Nürnberger T, Joosten MHAJ (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. 10.1105/tpc.110.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sohn KH, Lei R, Nemri A, Jones JDG (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . Plant Cell 19: 4077–4090. 10.1105/tpc.107.054262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobayashi DY, Tamaki SJ, Keen NT (1989) Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc Natl Acad Sci U S A 86: 157–161. 10.1073/pnas.86.1.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei C-F, Kvitko BH, Shimizu R, Crabill E, Alfano JR, et al. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J 51: 32–46. 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- 32. Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 1: e0039 10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas WJ, Thireault CA, Kimbrel JA, Chang JH (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J 60: 919–928. 10.1111/j.1365-313X.2009.03998.x [DOI] [PubMed] [Google Scholar]
- 34. Sels J, Mathys J, De Coninck BMA, Cammue BPA, De Bolle MFC (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46: 941–950. 10.1016/j.plaphy.2008.06.011 [DOI] [PubMed] [Google Scholar]
- 35. Wang H, Avci U, Nakashima J, Hahn MG, Chen F, et al. (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci U S A 107: 22338–22343. 10.1073/pnas.1016436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim HS, Park YH, Nam H, Lee YM, Song K, et al. (2014) Overexpression of the Brassica rapa transcription factor WRKY12 results in reduced soft rot symptoms caused by Pectobacterium carotovorum in Arabidopsis and Chinese cabbage. Plant Biol 16: 973–981. 10.1111/plb.12149 [DOI] [PubMed] [Google Scholar]
- 37. Haq SK, Atif SM, Khan RH (2004) Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch Biochem Biophys 431: 145–159. 10.1016/j.abb.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 38. Valueva TA, Mosolov V V. (2004) Role of inhibitors of proteolytic enzymes in plant defense against phytopathogenic microorganisms. Biochem 69: 1305–1309. 10.1007/s10541-005-0015-5 [DOI] [PubMed] [Google Scholar]
- 39. Baysal-Gurel F, Ivey MLL, Dorrance A, Luster D, Frederick R, et al. (2008) An immunofluorescence assay to detect urediniospores of Phakopsora pachyrhizi . Plant Dis 92: 1387–1393. 10.1094/PDIS-92-10-1387 [DOI] [PubMed] [Google Scholar]
- 40. Zhang XC, Freire MCM, Le MH, De Oliveira LO, Pitkin JW, et al. (2012) Genetic diversity and origins of Phakopsora pachyrhizi isolates in the United States. Asian J Plant Pathol 6: 52–65. 10.3923/ajppaj.2012.52.65 [DOI] [Google Scholar]
- 41. Hirschburger D, Müller M, Voegele RT, Link T (2015) Reference genes in the pathosystem Phakopsora pachyrhizi/soybean suitable for normalization in transcript profiling. Int J Mol Sci 16: 23057–23075. 10.3390/ijms160923057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jacobs KA, Collins-Racie LA, Colbert M, Duckett M, Golden-Fleet M, et al. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198: 289–296. 10.1016/S0378-1119(97)00330-2 [DOI] [PubMed] [Google Scholar]
- 43. Alfano JR, Charkowski AO, Deng W-L, Badel JL, Petnicki-Ocwieja T, et al. (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci 97: 4856–4861. 10.1073/pnas.97.9.4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DebRoy S, Thilmony R, Kwack Y-B, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A 101: 9927–9932. 10.1073/pnas.0401601101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Wang L, Cai G, Jiang S, Sun L, et al. (2013) Response of tobacco to the Pseudomonas syringae pv. tomato DC3000 is mainly dependent on salicylic acid signaling pathway. FEMS Microbiol Lett 344: 77–85. 10.1111/1574-6968.12157 [DOI] [PubMed] [Google Scholar]
- 46. Rep M (2005) Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol Lett 253: 19–27. 10.1016/j.femsle.2005.09.014 [DOI] [PubMed] [Google Scholar]
- 47. Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- 48. Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, et al. (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 9: e1003235 10.1371/journal.ppat.1003235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen X, Zhang Z, Liu D, Zhang K, Li A, et al. (2010) SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J Integr Plant Biol 52: 946–951. 10.1111/j.1744-7909.2010.00987.x [DOI] [PubMed] [Google Scholar]
- 50. Zhang C, Bradshaw JD, Whitham SA, Hill JH (2010) The development of an efficient multipurpose Bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol 153: 52–65. 10.1104/pp.109.151639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu J-Z, Braun E, Qiu W-L, Shi Y-F, Marcelino-Guimarães FC, et al. (2014) Positive and negative roles for soybean MPK6 in regulating defense responses. Mol Plant Microbe Interact 27: 824–834. 10.1094/MPMI-11-13-0350-R [DOI] [PubMed] [Google Scholar]
- 52. Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to Tobacco mosaic virus . Plant J 30: 415–429. 10.1046/j.1365-313X.2002.01297.x [DOI] [PubMed] [Google Scholar]
- 53. Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786. 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- 54. Stergiopoulos I, de Wit PJGM (2009) Fungal effector proteins. Annu Rev Phytopathol 47: 233–263. 10.1146/annurev.phyto.112408.132637 [DOI] [PubMed] [Google Scholar]
- 55. Krijger J-J, Thon MR, Deising HB, Wirsel SGR (2014) Compositions of fungal secretomes indicate a greater impact of phylogenetic history than lifestyle adaptation. BMC Genomics 15: 722 10.1186/1471-2164-15-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saunders DGO, Win J, Cano LM, Szabo LJ, Kamoun S, et al. (2012) Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One 7: e29847 10.1371/journal.pone.0029847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coletta A, Pinney JW, Solís DYW, Marsh J, Pettifer SR, et al. (2010) Low-complexity regions within protein sequences have position-dependent roles. BMC Syst Biol 4: 43 10.1186/1752-0509-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Canonne J, Marino D, Noël LD, Arechaga I, Pichereaux C, et al. (2010) Detection and functional characterization of a 215 amino acid N-terminal extension in the Xanthomonas type III effector XopD. PLoS One 5: e15773 10.1371/journal.pone.0015773 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Cabral A, Stassen JHM, Seidl MF, Bautor J, Parker JE, et al. (2011) Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS One 6: e19328 10.1371/journal.pone.0019328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7: e1002348 10.1371/journal.ppat.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koch E, Ebrahim Nesbat F, Hoppe HH (1983) Light and electron microscopic studies on the development of soybean rust (Phakopsora pachyrhizi Syd.) in susceptible soybean leaves [Glycine max]. Phytopathol Zeitschrift 106: 302–320. [Google Scholar]
- 62. Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, et al. (2004) A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol 337: 49–63. 10.1016/j.jmb.2004.01.015 [DOI] [PubMed] [Google Scholar]
- 63. Stone JM, Liang X, Nekl ER, Stiers JJ (2005) Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J 41: 744–754. 10.1111/j.1365-313X.2005.02334.x [DOI] [PubMed] [Google Scholar]
- 64. Wang J-W, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana . Plant Cell 20: 1231–1243. 10.1105/tpc.108.058180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol 50: 2133–2145. 10.1093/pcp/pcp148 [DOI] [PubMed] [Google Scholar]
- 66. Wang J-W, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell 138: 738–749. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 67. Xing S, Salinas M, Garcia-Molina A, Höhmann S, Berndtgen R, et al. (2013) SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J 75: 566–577. 10.1111/tpj.12221 [DOI] [PubMed] [Google Scholar]
- 68. Zhang H, Zhao X, Li J, Cai H, Deng XW, et al. (2014) MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell. 10.1105/tpc.114.127340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Manning K, Tör M, Poole M, Hong Y, Thompson AJ, et al. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952. 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- 70. Miura K, Ikeda M, Matsubara A, Song X-J, Ito M, et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545–549. 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- 71. Wang L, Cao C, Ma Q, Zeng Q, Wang H, et al. (2014) RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol 14: 169 10.1186/1471-2229-14-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, et al. (2003) Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol 49: 1239–1251. 10.1046/j.1365-2958.2003.03647.x [DOI] [PubMed] [Google Scholar]
- 73. Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, et al. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta . Plant J 59: 150–162. 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
- 74. Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, et al. (2008) Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell 20: 3080–3093. 10.1105/tpc.108.063065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, et al. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta . Plant J 56: 505–516. 10.1111/j.1365-313X.2008.03612.x [DOI] [PubMed] [Google Scholar]
- 76. Keogh RC, Deverall BJ, McLeod S (1980) Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans Br Mycol Soc 74: 329–333. 10.1016/S0007-1536(80)80163-X [DOI] [Google Scholar]
- 77. Adam L, Somerville SC (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . Plant J 9: 341–356. 10.1046/j.1365-313X.1996.09030341.x [DOI] [PubMed] [Google Scholar]
- 78. Becker DM, Lundblad V (2001) Introduction of DNA into yeast cells. Curr Protoc Mol Biol Chapter 13: Unit13.7. 10.1002/0471142727.mb1307s27 [DOI] [PubMed] [Google Scholar]
- 79. Treco DA, Lundblad V (2001) Preparation of yeast media. Curr Protoc Mol Biol Chapter 13: Unit13.1. 10.1002/0471142727.mb1301s23 [DOI] [PubMed] [Google Scholar]
- 80. An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors in “Plant Molecular Biology Manual.” Gelvin SB, Schilperoort RA, Verma DPS, editors Dordrecht: Springer Netherlands; A3: 1–19 p. 10.1007/978-94-009-0951-9 [DOI] [Google Scholar]
- 81. Vieira A, Talhinhas P, Loureiro A, Duplessis S, Fernandez D, et al. (2011) Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biol 115: 891–901. 10.1016/j.funbio.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 82. Hu R, Fan C, Li H, Zhang Q, Fu Y-F (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10: 93 10.1186/1471-2199-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu J-Z, Horstman HD, Braun E, Graham MA, Zhang C, et al. (2011) Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol 157: 1363–1378. 10.1104/pp.111.185686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.