ABSTRACT
Chromatin can function as an integrator of DNA-related processes, allowing communication, for example, between DNA replication and gene transcription. Such communication is needed to overcome the gene-dosage imbalance introduced during DNA replication, when certain genes are replicated prior to others. Increased transcription of early replicating genes could alter regulatory balances. This does not occur, suggesting a mechanism that suppresses expression from newly replicated DNA. Critical to this buffering is Rtt109, which acetylates the internal K56 residue of newly synthesized histone H3 prior to incorporation onto DNA. H3K56ac distinguishes replicated from non-replicated DNA, communicating this information to the transcription machinery to ensure expression homeostasis during S phase.
KEYWORDS: Chromatin, DNA replication, gene expression, H3K56ac, RTT109
Chromatin as an integrator of differentDNA-Related processes
DNA serves as a template for multiple processes, including gene transcription, DNA replication and DNA repair. As such, it needs to comply with different, often competing, requirements. For example, gene transcription is tightly regulated, requiring easy access of RNA polymerase to specific genomic regions, while preventing polymerase binding to others. DNA replication, on the other hand, requires that DNA polymerase accesses origins of replication, and proceeds to access each and every part of the genome. The repair machinery of DNA damage requires localized access to random regions. The genome is therefore co-optimized for multiple DNA-dependent processes, which interact and influence each other through the use of the same template, and occasionally the same regulatory proteins.
Access to DNA is largely regulated by the chromatin environment in which DNA is embedded. DNA is wrapped around histone octamers (nucleosomes), which form the basic units of chromatin. The histones can be modified on multiple residues by the addition of chemical groups, such as acetyl or methyl.1 These modifications modulate the electrostatic attraction, and thereby the binding strength between histones and DNA. Further, specific modifications directly recruit regulatory factors to enable specific functions.2
Differential modification of histones along the genome can potentially influence all DNA-related processes. This modification pattern, termed the ‘epigenetic landscape’ is shaped by chromatin modifiers that modify histones at specific positions both in preparation to processes such as gene transcription or DNA replication, or as a consequence of these same processes.3 As such, histone modification provides a means for communication and cross-regulation between different DNA regulated processes (Fig. 1A).
Figure 1.
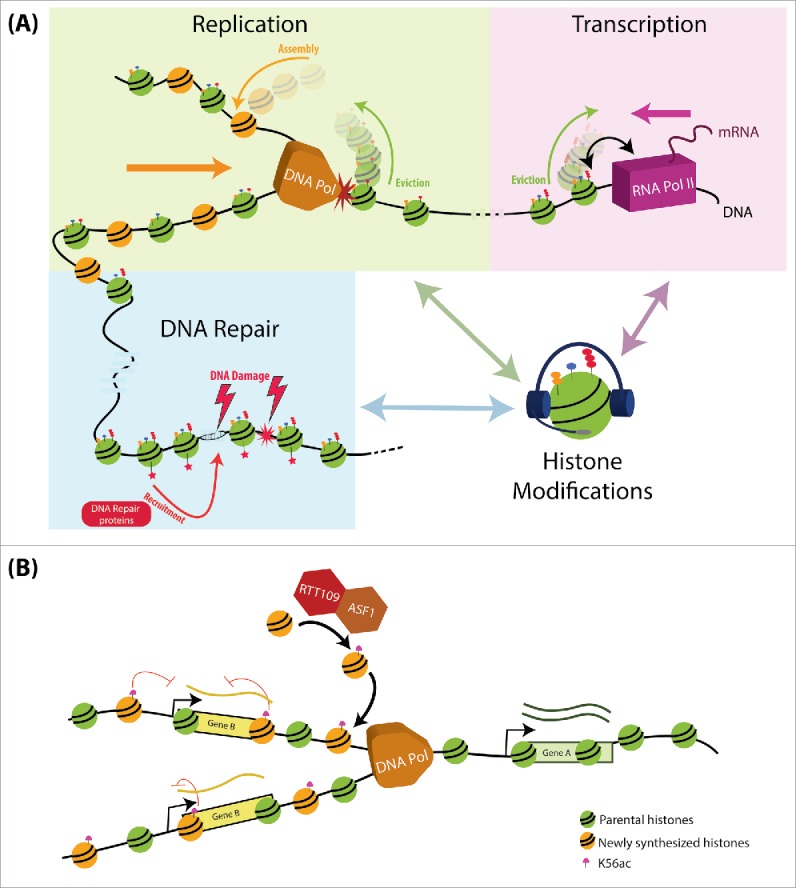
Histone modification as a means for communication and cross-regulation between different DNA regulated processes. (A) Histone modifications can serve as a platform for communication between the different DNA-dependent processes: DNA-related processes are coupled to changes in histone modifications and histone turnover. These include the eviction and assembly of histones during DNA replication and transcription, the modification of histones, in preparation and during these processes, and the recruitment of specific proteins (e.g. DNA repair machinery following DNA damage). We propose that through the platform of histone modifications, one process can pass information needed for the regulation of other DNA-dependent processes. (B) H3K56ac as a signal of communication between DNA replication and transcription: During replication, newly synthesized histones are acetylated by Rtt109 and Asf1 on H3K56. These histones are incorporated in replicated loci, allowing the discrimination between replicated and non-replicated regions. In turn, this information deposited by a replication-associated process, allows tuning expression to buffer changes in DNA dosage.
DNA replication and gene transcription
Particularly notable is the mutual interplay between DNA replication and gene transcription. In eukaryotes, replication initiates from multiple genomic loci called origins of replication, with each origin producing 2 replication forks that progress in opposite directions.4 In most eukaryotic cells, the process of DNA replication occupies a small part of the cell cycle, reaching ~20-30% in rapidly dividing yeast or human cells.5,6 Therefore, the epigenetic landscape is largely defined by processes that are related to gene expression, which occur throughout the cell-cycle. DNA replication, however, largely perturbs this landscape. First, specific modifications are used to define replication origins, or are deposited during the course of DNA polymerase progression.7,8 In addition, histones must be removed from the DNA template to enable replication fork progression and then restored in the wake of the fork.9
The most notable impact of DNA replication on the epigenetic landscape is probably the use of newly synthesized histones for wrapping the newly replicated DNA.10 These new histones lack position-specific modifications, but instead are generally modified on multiple residues. Several regulatory factors participate in this pre-deposition modification, some being dedicated to this task. For example, the histone acetyltransferase Rtt109 acetylates newly synthesized H3 on the internal K56 residue prior to its incorporation onto DNA.10 Other residues that are acetylated prior to incorporation include H4K5 and H4K12.11,12 By contrast, modifications typically associated with active transcription, such as histone tri-methylation,13 are absent from newly synthesized histones. Further, in a recent study we provided evidence that a specific modification, H3K9ac, is deposited ahead of the replication fork, in an apparent preparation for DNA replication.14 This modification appears as a wave that precedes the replication fork by ~5Kb, perhaps enabling smoother fork progression. The replication-dependent deposition is strictly dependent on Rtt109, while Gcn5, the second enzyme catalyzing H3K9 acetylation, fully accounts for expression-dependent H3K9ac.14
The epigenetic landscape is therefore perturbed by DNA replication. Potentially, this could be used for encoding novel regulatory programs that could modulate gene expression. Yet, some modifications should be recovered to relax the genome back to (presumably) expression-optimized state. In some cases, a modification remaining on one copy may be used to define the modification on another copy, either directly or by recruiting appropriate factors, forming an epigenetic memory.15 In other cases, modifications may be relaxed back passively, deposited back progressively through the same regulatory factors or processes that deposited it to begin with, prior to replication.14,16
It is generally difficult to distinguish cases of epigenetic memory from passive retrieval. Still, general trends can be deduced by analyzing the dynamics with which modifications are retrieved following replication. Mass-spectrometry analysis of histones during and post replication in HeLa-cells, for example, reported that most residues, including H3K27, H3K36, and H3K9 monomethylations, recover rapidly, while H3K9 and H3K27 tri-methylation post-replication deposition is slow, and largely delayed, with full deposition requiring several cell cycles.17 We recently studied the dynamics of retrieval by following the genome-wide pattern of multiple histone modifications during and after replication using ChIP-Seq in budding yeast.14 Also here, some modifications appeared on replicated DNA quite immediately (e.g. H4K16ac, H3K4me1), whereas all tri-methylation were deposited with a significant delay that extended for over 20 min. Notably, this delayed deposition was largely correlated with gene transcription strength and gene promoter structure, so that actively transcribing genes were tri-methylated significantly faster than low-expressing ones. Thereby, replication-independent processes, rather than active epigenetic memory account for the recovery of histone tri-methylation pattern following DNA replication.
Maintaining expression homeostasis during DNA replication
In addition to the changes in the epigenetic landscape, DNA replication could potentially perturb gene expression by the mere fact that gene dosage is progressively modified. That is, genes that replicate early in S phase increase in dosage prior to genes that replicate in late S. This gene dosage imbalance could potentially perturb cellular functions. Indeed, while cells are typically insensitive to dosage of individual genes, duplication of some genes can have deleterious effects, as exemplified by disease-associated genes such as SNCA in Parkinson disease and NSD1 in growth retardation.18 Further, whole chromosome duplication (aneuploidy) exerts deleterious effects on growth.19,20 How do cells handle the transient dosage imbalance during DNA replication?
In bacteria, replication-induced dosage imbalance is immediately translated into imbalanced expression.21 In fact, bacteria use this imbalance as means for regulation, capitalizing on this effect for tuning expression with growth rate, inducing specific processes in slow-growing cells, or up-regulating DNA repair genes in response to drugs that slowdown replication.22,23 In sharp contrast, studies in multiple eukaryotic cells suggested the ability to buffer these dosage changes during DNA replication. Classical studies reported that total mRNA synthesis rate does not increase specifically during S phase.24–26 Further, more recently, it has been shown that individual genes do not increase in expression as cells enter S phase using smFISH or live imaging.27,28 Rather, while transcription can now occur from both copies of the DNA, the rate by which transcripts are made is reduced by about 2-folds, largely maintaining the pre-replication transcription rate.
How general is the buffering of gene dosage during S phase? While lack of correlation between overall mRNA synthesis and DNA replication may suggest genome-wide buffering, it still allows the alternative interpretation that some process, other than DNA is limiting for gene transcription. For example, if RNA-Pol II levels are limiting, total mRNA synthesis would not increase during DNA replication, yet the distribution of polymerases between the genes would change, biasing transcription toward early replicating genes. We therefore examined whether buffering occurs as a global phenomenon by following genome-wide transcription of cells synchronously progressing through S phase.29 Comparing transcription of early replicating genes to that of late replicating ones supported buffering: mRNA levels of early replicating genes remained largely constant (increasing by only 10-15%) during S phase, despite a significant increase in their DNA content (reaching ~60-70%). Therefore, the increase in gene dosage during S phase is largely suppressed to maintain expression homeostasis.
Expression homeostasis during DNA replication depends on replication-associated chromatin modification
How gene-dosage imbalance accompanying DNA replication is buffered at the molecular level? Key to buffering is the ability to distinguish replicated from non-replicated DNA, suggesting that chromatin modification may be involved. Searching for buffering-related factors would optimally involve assaying expression of all chromatin associated mutants as they progress synchronously through S phase. As a surrogate for this effort, we analyzed a dataset published by the lab of F. Holstege, reporting the transcription profile of 165 chromatin-associated mutants.30 Despite using asynchronous cultures, in which only ~25% are actively replicating, the resolution of the data enabled detecting mutants in which expression of early replicating genes increased relative to that of late replicating ones, indicating loss of buffering.
Two related factors were identified as critical for buffering the gene dosage imbalance during S phase: the acetyltrasferase Rtt109 and its associated chaperone Asf1.31,32 This requirement, detected in asynchronous cells, was validated using time course experiments. In fact, in cells deleted of Rtt109, mRNA synthesis of early replicating genes increased to the same extent as the increase in their DNA content. Both factors are periodically expressed in G1/S-phase and acetylate histone H3 on the internal K56 residue and the tail K9 residue. To define the specific modification mediating buffering, we examined histone mutants incapable of undergoing this modification.33 Unmodifiable H3K9 had no effect on buffering, while H3K56 was required. In fact, buffering was fully lost in the 2 mutants changing H3K56 to mimic either constantly non-acetylated (H3K56A) or to constantly acetylated (H3K56Q) form.
We therefore conclude that Rtt109-dependent H3K56ac mediates the buffering of gene dosage during DNA replication (Fig. 1B). This modification is indeed optimally suited for this task: it is added to H3 prior to its incorporation onto DNA, and marks the newly replicated DNA.10,34 This modification is removed at the end of S phase by the 2 deacetylases HST3 and HST4,35 thereby retrieving wild-type transcription and allowing cells to prepare for the next replication cycle.
The role of Rtt109 as modulator of gene expression
The loss of expression homeostasis during S phase in cells deleted of Rtt109, or in cells that cannot acetylate H3K56ac, suggests that this modification suppresses transcription. Yet, previous studies which analyzed the role of Rtt109 in gene transcription suggested that it in fact acts to enhance, rather than repress gene expression.36–38 While these studies focus on individual genes, and could potentially be explained as specific effects (e.g., Snf5, implicated in activation of histone genes through H3K56ac,36 was included in the Lenstra et al. data set but did not show loss of buffering), experiments which analyzed the genome-wide pattern of H3K56ac found no correlation between the abundance of this modification and gene expression.39,40 What could be the cause of these differences?
The role of Rtt109/H3K56ac in expression homeostasis is specific to S phase, the time in which this modification is most apparent on the genome. By contrast, the potential role of this modification in gene activation did not distinguish this phase. Notably, replication-specific effects of H3K56ac on chromatin structure were described: while H3K56ac promotes nucleosome disassembly during transcription activation,37 it in fact promotes nucleosome assembly during DNA replication (as well as during DNA damage repair).10,41 This role in nucleosome assembly, observed during DNA replication, may shift the balance toward a more closed chromatin structure, consistent with the suppression of gene expression. Alternatively, H3K56ac may also recruit regulatory factors specifically during S phase.42,43 For example, during DNA replication, H3K56ac shows increased affinity to the assembly factors Rtt106 and Caf1.10,43 Supporting this replication-specific effect, H3K56ac abundance in wild-type cells correlates with changes in gene expression during replication in Δrtt109 but not in WT cells.29
Conclusion
Distinguishing replicated from non-replicated regions is critical for buffering the transient gene dosage imbalance during DNA replication. This information is communicated from the replication process to affect transcription through the replication-associated modification H3K56ac. We hypothesize that this role of chromatin as a communication platform between different DNA related processes may extend further. For example, a histone modification added during DNA damage may limit gene expression from genes in the damaged region. Similarly, modifications that are specific to highly expressing genes may signal to influence replication fork progression to limit possible collisions between RNA and DNA polymerase. It would be interesting to explore these, or similar avenues of communication, which could help understand how the genome is co-optimized for multiple, parallel processes.
Finally, it is worthwhile noting that the involvement of Rtt109 in the homeostasis mechanism described was first observed in a published dataset that was reported for different purposes. This emphasizes the great potential and possibilities that are now presented with the accumulation of high quality genome-wide data that is open access to all.44
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of our laboratory for productive discussions and comments on this review.
Funding
This work was supported by the European Research Council and the Israel Science Foundation.
References
- [1].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381-95; PMID:21321607; http://dx.doi.org/ 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics 2012; 190:351-87; PMID:22345607; http://dx.doi.org/ 10.1534/genetics.111.132266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 2013; 20:259-66; PMID:23463310; http://dx.doi.org/ 10.1038/nsmb.2470 [DOI] [PubMed] [Google Scholar]
- [4].Hand R. Eucaryotic DNA: organization of the genome for replication. Cell 1978; 15:317-25; PMID:719745; http://dx.doi.org/ 10.1016/0092-8674(78)90001-6 [DOI] [PubMed] [Google Scholar]
- [5].Rew DA, Wilson GD. Cell production rates in human tissues and tumours and their significance. Part II: clinical data. Eur J Surg Oncol 2000; 26:405-17; PMID:10873364; http://dx.doi.org/ 10.1053/ejso.1999.0907 [DOI] [PubMed] [Google Scholar]
- [6].Koren A, Soifer I, Barkai N. MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Res 2010; 20:781-90; PMID:20219942; http://dx.doi.org/ 10.1101/gr.102764.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 2005; 436:294-8; PMID:16015338; http://dx.doi.org/ 10.1038/nature03714 [DOI] [PubMed] [Google Scholar]
- [8].Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 2010; 17:430-7; PMID:20228802; http://dx.doi.org/ 10.1038/nsmb.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 2005; 280:12065-8; PMID:15664979; http://dx.doi.org/ 10.1074/jbc.R400039200 [DOI] [PubMed] [Google Scholar]
- [10].Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008; 134:244-55; PMID:18662540; http://dx.doi.org/ 10.1016/j.cell.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A 1995; 92:1237-41; PMID:7862667; http://dx.doi.org/ 10.1073/pnas.92.4.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem 2006; 281:9287-96; PMID:16464854; http://dx.doi.org/ 10.1074/jbc.M512956200 [DOI] [PubMed] [Google Scholar]
- [13].Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al.. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005; 122:517-27; PMID:16122420; http://dx.doi.org/ 10.1016/j.cell.2005.06.026 [DOI] [PubMed] [Google Scholar]
- [14].Bar-Ziv R, Voichek Y, Barkai N. Chromatin dynamics during DNA replication. Genome Res 2016; PMID:27225843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Almouzni G, Cedar H. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 2016; 8:a019372; PMID:27141050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Radman-Livaja M, Liu CL, Friedman N, Schreiber SL, Rando OJ. Replication and active demethylation represent partially overlapping mechanisms for erasure of H3K4me3 in budding yeast. PLoS Genet 2010; 6:e1000837; PMID:20140185; http://dx.doi.org/ 10.1371/journal.pgen.1000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Alabert C, Barth TK, Reverón-Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 2015; 29:585-90; PMID:25792596; http://dx.doi.org/ 10.1101/gad.256354.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet 2007; 8:17-35; PMID:17386002; http://dx.doi.org/ 10.1146/annurev.genom.8.021307.110233 [DOI] [PubMed] [Google Scholar]
- [19].Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A 2012; 109:14746-53; PMID:22908297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007; 317:916-24; PMID:17702937; http://dx.doi.org/ 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- [21].Chandler MG, Pritchard RH. The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol Gen Genet 1975; 138:127-41; PMID:1105148; http://dx.doi.org/ 10.1007/BF02428117 [DOI] [PubMed] [Google Scholar]
- [22].Slager J, Kjos M, Attaiech L, Veening J-W. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 2014; 157:395-406; PMID:24725406; http://dx.doi.org/ 10.1016/j.cell.2014.01.068 [DOI] [PubMed] [Google Scholar]
- [23].Narula J, Kuchina A, Lee D-YD, Fujita M, Süel GM, Igoshin OA. Chromosomal Arrangement of Phosphorelay Genes Couples Sporulation and DNA Replication. Cell 2015; 162:328-37; PMID:26165942; http://dx.doi.org/ 10.1016/j.cell.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barnes A, Nurse P, Fraser RS. Analysis of the significance of a periodic, cell size-controlled doubling in rates of macromolecular synthesis for the control of balanced exponential growth of fission yeast cells. J Cell Sci 1979; 35:41-51; PMID:422679 [DOI] [PubMed] [Google Scholar]
- [25].Elliott SG, McLaughlin CS. Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1978; 75:4384-8; PMID:360219; http://dx.doi.org/ 10.1073/pnas.75.9.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Skog S, Tribukait B. Discontinuous RNA and protein synthesis and accumulation during cell cycle of Ehrlich ascites tumour cells. Exp Cell Res 1985; 159:510-8; PMID:4029278; http://dx.doi.org/ 10.1016/S0014-4827(85)80024-0 [DOI] [PubMed] [Google Scholar]
- [27].Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, Wu AR, Churchman LS, Singh A, Raj A. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell 2015; 58:339-52; PMID:25866248; http://dx.doi.org/ 10.1016/j.molcel.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods 2010; 7:631-3; PMID:20639867; http://dx.doi.org/ 10.1038/nmeth.1482 [DOI] [PubMed] [Google Scholar]
- [29].Voichek Y, Bar-Ziv R, Barkai N. Expression homeostasis during DNA replication. Science 2016; 351:1087-90; PMID:26941319; http://dx.doi.org/ 10.1126/science.aad-1162 [DOI] [PubMed] [Google Scholar]
- [30].Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NACH, Margaritis T, van de Pasch LAL, van Heesch SAAC, Brok MO, Groot Koerkamp MJA, et al.. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 2011; 42:536-49; PMID:21596317; http://dx.doi.org/ 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Han J, Zhou H, Horazdovsky B, Zhang K, Xu R-M, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 2007; 315:653-5; PMID:17272723; http://dx.doi.org/ 10.1126/science.113-3234 [DOI] [PubMed] [Google Scholar]
- [32].Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 2007; 315:649-52; PMID:17272722; http://dx.doi.org/ 10.1126/science.1135862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 2008; 134:1066-78; PMID:18805098; http://dx.doi.org/ 10.1016/j.cell.2008.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Han J, Zhou H, Li Z, Xu R-M, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem 2007; 282:14158-64; PMID:17369253; http://dx.doi.org/ 10.1074/jbc.M70-0611200 [DOI] [PubMed] [Google Scholar]
- [35].Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The Sirtuins Hst3 and Hst4p Preserve Genome Integrity by Controlling Histone H3 Lysine 56 Deacetylation. Curr Biol 2006; 16:1280-9; PMID:16815704; http://dx.doi.org/ 10.1016/j.cub.2006.06.023 [DOI] [PubMed] [Google Scholar]
- [36].Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005; 121:375-85; PMID:15882620; http://dx.doi.org/ 10.1016/j.cell.2005.03.011 [DOI] [PubMed] [Google Scholar]
- [37].Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A 2008; 105:9000-5; PMID:18577595; http://dx.doi.org/ 10.1073/pnas.0800057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lin L, Schultz MC. Promoter regulation by distinct mechanisms of functional interplay between lysine acetylase Rtt109 and histone chaperone Asf1. Proc Natl Acad Sci U S A 2011; 108:19599-604; PMID:22106264; http://dx.doi.org/ 10.1073/pnas.1111501108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Weiner A, Hsieh T-HSS, Appleboim A, Chen HV V, Rahat A, Amit I, Rando OJJ, Friedman N. High-Resolution Chromatin Dynamics during a Yeast Stress Res-ponse. Mol Cell 2015; 58:371-86; PMID:25801168; http://dx.doi.org/ 10.1016/j.molcel.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kuang Z, Cai L, Zhang X, Ji H, Tu BP, Boeke JD. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat Struct Mol Biol 2014; 21:854-63; PMID:25173176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated Lysine 56 on Histone H3 Drives Chromatin Assembly after Repair and Signals for the Completion of Repair. Cell 2008; 134:231-43; PMID:18662539; http://dx.doi.org/ 10.1016/j.cell.2008.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal K, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999; 399:491-6; PMID:10365964; http://dx.doi.org/ 10.1038/20974 [DOI] [PubMed] [Google Scholar]
- [43].Su D, Hu Q, Li Q, Thompson JR, Cui G, Fazly A, Davies BA, Botuyan MV, Zhang Z, Mer G. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature 2012; 483:104-7; PMID:22307274; http://dx.doi.org/ 10.1038/nature10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Longo DL, Drazen JM. Data Sharing. N Engl J Med 2016; 374:276-7; PMID:26789876; http://dx.doi.org/ 10.1056/NEJMe1516564 [DOI] [PubMed] [Google Scholar]


