ABSTRACT
Chromatin is a highly dynamic structure that imparts structural organization to the genome and regulates the gene expression underneath. The decade long research in deciphering the significance of epigenetics in maintaining cellular integrity has embarked the focus on chromatin remodeling enzymes. These drivers have been categorized as readers, writers and erasers with each having significance of their own. Largely, on the basis of structure, ATP dependent chromatin remodelers have been grouped into 4 families; SWI/SNF, ISWI, IN080 and CHD. It is still unclear to what degree these enzymes are swayed by local DNA sequences when shifting a nucleosome to different positions. The ability of regulating active and repressive transcriptional state via open and close chromatin architecture has been well studied however, the significance of chromatin remodelers in regulating transcription at each step i.e. initiation, elongation and termination require further attention. The authors have highlighted the significance and role of different chromatin remodelers in transcription, DNA repair and histone variant deposition.
KEYWORDS: Chromatin remodelers, histone variants, transcription
Introduction: Chromatin and remodelers
The term chromatin was coined by Walther Flemming for the unique stainable fibrous structures observed in nucleus. Chromatin exists in a highly condensed form and is composed of nucleosomes possessing an octamer of histones H2A, H2B, H3 and H4, wrapped by 146bp of DNA and a linker histone H1. The distinction between condensed heterochromatin and open euchromatin structures were reported by Emil Heitz. Structural alteration in chromatin structure facilitates the downstream gene expression specific to cellular demand and thereby hold significant importance in gene regulatory network.1
Chromatin is highly dynamic structure and its plasticity is provided by (a) remodeling of nucleosomes, (b) chemical modification of histones or incorporation of variants, (c) non-histone DNA binding proteins and (d) non-coding RNAs. The affinity of histones for DNA and DNA-associated proteins is governed by combination of histone variants and post-transcriptional modifications (PTMs) of histones that further regulate the transcriptional activity and the accessibility of DNA for recombination, replication and repair. These alterations in chromatin structure are brought about by distinct class of remodeling enzymes, specified as chromatin remodelers.2
Molecular mechanisms of remodeler actions
Chromatin remodelers are versatile tools that catalyze broad range of chromatin changing reactions including sliding of an octamer across the DNA (nucleosome sliding), changing the conformation of nucleosomal DNA and altering the composition of the octamers (histone variant exchange).3 On the basis of their mode of action, these evolutionary conserved remodeling enzymes have been grouped in 2 categories: (a) mediates histone post-translational modifications on histone and (b) alter histone-DNA contact within the nucleosome through ATP hydrolysis.4
In order to restructure nucleosomes, ATP-dependent chromatin remodelers utilize energy from ATP hydrolysis (∼7.3 kcal/mole) to disrupt the contacts between histones and DNA, and thus regulates the dynamic access to the packaged DNA.5 A conserved core ATPase is shared by all remodelers belonging to the superfamily 2 (SF2) of DEAD/H-box helicases.6 The Snf2 ATPase which is bi-lobed and consists of 2 tandem RecA-like folds (DEXX and HELIC), uses ATP that guides toward translocation down the DNA minor groove.7 On the contrary to usual helicases, remodeler ATPases lack the “pin” motif (wedge domain),8 that is essential for strand separation and therefore only act as DNA translocases.
Classification of chromatin remodelers
ATP-dependent chromatin remodelers are classified into 4 distinct families (Table 1): SWI/SNF (switch/sucrose-non-fermenting), ISWI (imitation switch), CHD (chromodomain-helicase-DNA binding) and INO80 (inositol requiring 80). SNF2-family ATPase domain present in all remodeler families is split in 2 parts; DExx and HELIC. The ATPase domain of the SWI/SNF, ISWI, and CHD remodeler families possesses a short insertion, whereas remodelers of the INO80 family consist of a long insertion. The exclusive domains reside adjacent to the ATPase domain. SWI/SNF remodelers contain bromodomains; ISWI remodelers - SANT-SLIDE modules; CHD remodelers - tandem chromodomains and members of INO80 family possess HAS (helicase SANT) domains (Fig. 1). Each of these domains plays role in remodeler recruitment to chromatin or interacting to specific histone modifications and/or they are involved in the regulation of the ATPase activity of the remodeler.4
Table 1.
Function of chromatin remodelers and associated diseases.
| Chromatin Remodeller | Species | Interacting partner/partners | Function | Disease | ||
|---|---|---|---|---|---|---|
| SWI/SNF (switching defective) Family | Complex | Swi2/Snf2 | Yeast | Transcriptional activation / repression | Coffin-Siris syndrome and Nicolaides-Baraitser syndrome, congenital heart disease, cardiac hypertrophy, malignant rhabdoidtumors,such as choroid plexus carcinoma, medulloblastoma | |
| BAP (Brahma Associated Protein) | Drosophila | |||||
| PBAP (Polybromo-associated BAP) | ||||||
| BAF (BRG1- associated factors) | Human | |||||
| PBAF (Polybromo-associated BAF) | ||||||
| ISWI (imitation switch) family | Complex | NURF | Drosophila | Nucleosome spacing, DNA damage repair, transcriptional repression. | William'S syndrome, Melanotictumour,anencephaly | |
| ACF | ||||||
| CHRAC | ||||||
| ISW1 | Yeast | |||||
| ISW2 | ||||||
| NURF | Human | |||||
| ACF | ||||||
| CHRAC | ||||||
| NoRC | ||||||
| RSF | ||||||
| WICH | ||||||
| CHD (Chromodomain-Helicase-DNA binding) family | Subfamily 1 | SSRP1 protein H3K4me | ATPase activity and relocatesnucleosomes. HDAC activity. | Prostate cancer, Hereditary diffuse gastric cancer (HDGC), Ehlers-Danlos syndrome | ||
| Chd2 | A+T-rich DNA | Helicase activity | Lennox-Gastaut Syndrome, epileptic encephalopathies, autism | |||
| Subfamily2 | Chd3 | H3K36, HDAC1, HDA2, ATR,TRIM27 | HDAC activity | Dermatomyositis, Hodgkin's lymphoma | ||
| Chd4 | HDAC1, HDAC2, TRIM28 | DNA dependent ATPase activity, Epigenetic transcriptional repression | Dermatomyositis | |||
| Chd5 | Unmodified Histone, H3K27me3, | Expressed in neuronal cells, forms nucleosome remodeling and deacetylation complex | Neuroblastoma | |||
| Subfamily3 | Chd6 | RNA Polymerase II, NRF2, NQO1 | Transcriptional activation, Role in redox homeostasis | Pitt- Hopkin syndrome | ||
| Chd7 | Chromatin | Develompment of neural crest cells | CHARGE syndrome | |||
| Chd8 | CTCF, Duplin | Transcriptional repressor, developmental regulation | Autism spectrum disorder (ASD) | |||
| Chd9 | PPAR1α, CBAf1, osteocalcin, myosin | Transcriptional and developmental regulation, Nuclear receptor activation | osteogenic differentiation | |||
| INO80 (inositol requiring 80) | Complex | INO 80 | transcription factor YY1, Rvb1,Rvb2, NFR B, Arp4,Arp5, Arp8 | DNA helicase activity, DNA repair and replication | Aortic hypoplasia, premature atherosclerosis, Immunoglobulin class-switch recombination defects (CSR-D) | |
| Swr 1 | Arp4, Arp 6, Swc2, Rvb1, Rvb2 H2AZ, H2B | |||||
Figure 1.
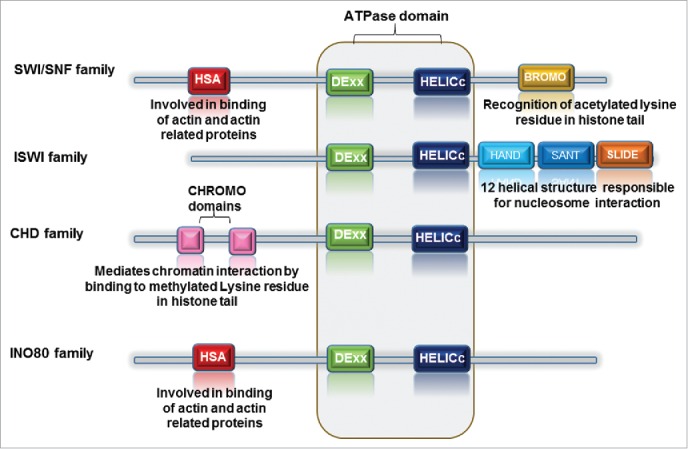
Diagrammatic representation of chromatin remodeler family highlighting the conserved domain with each family member. The DEX and HELICc domains are conserved throughout the family. However, HAND, SANT and SLIDE domains are specific to ISWI family, whereas BROMO domain distinguishes the SWI/SNF family. The presence of CHROMO domains is characteristic of CHD family.
SWI/SNF
The yeast SWI/SNF complex was the first ATP-dependent chromatin remodeler described. Different subunits of SWI/SNF complex were encoded by 2 independent genetic screens for altered gene expression involved in regulating mating type switching (SWI) and sucrose fermentation in yeast (Sucrose Non-Fermenting).9,10 The SWI/SNF subunits are 1-2 MDa in size and consist of 9-12 subunits containing Brg1/Brm and their associated factors (BAFs).11 The complex is targeted to acetylated histone tails through bromodomain subunit within the ATPases.12 An additional feature of SWI/SNF complexes is the presence of actin and/or actin related proteins (Arps). It has been hypothesized that actin and Arps modulate binding of the remodeling complex to chromatin, stimulate the DNA-dependent ATPase activity, facilitates complex assembly and stability, binding of histone, or remodeling and translocation.13-15 Yeast contains 2 SWI/SNF ATPases (Swi2/Snf2 and Sth1) that constitute 2 complexes; ySWI/SNF and RSC, respectively.16 Corresponding to yeast in Drosophila occurs a single protein Swi2/Snf2, called Brahma (BRM), which is present in 2 complexes – BAP and PBAP.17,18 In humans, 2 discrete Swi2/Snf2-like ATPase subunits, named hBRM (human Brahma) and BRG1 (Brahma-Related Gene 1), constitutes the subunits of BAF and PBAF complexes.19,20 In mammals, the presence of unique compositions of BAF complexes in embryonic stem cells and during developmental transitions suggests their potential role in guiding cell fate decisions.21
Diverse functions of SWI/SNF-like complexes have been studied so far. The majority of the studies have implicated SWI/SNF complexes in active gene transcriptional regulation.22 The cooperative association of ySWI/SNF with histone acetytransferase complexes results in activation of gene transcription.23,24 On contrary, literature also suggests role of the SWI/SNF complexes in gene repression.25 In Drosophila, BRM has been reported to be involved in transcription regulation of majority of the genes and is largely associated with transcriptionally active sites on polytene chromosomes. In addition to playing significant role in gene transcription reports also suggests that most of SWI/SNF-family have a direct involvement in other processes such as DNA replication or DNA repair. In yeast, the role of ySWI/SNF in promoting replication initiation 26 and nucleotide excision repair on reconstituted nucleosomal substrates in vitro 27,28 has been reported.
Imaging studies of SWI/SNF complex using electron microscopy directs that these complexes form multilobed C-shaped assemblies that mount the nucleosome in a central cavity with the entry and exit sites of DNA exposed.29,30 Cross-linking and DNA footprinting experiments proposed that the ATPases localize sites at which DNA−histone binding is fragile, such that the torsional strain generated may be endured for loop proparation.31 For the SWI/SNF complex, it has been specifically hypothesized that the ATPase protein binds to a specific site on the nucleosome and subsequently hires 3′ translocase activity to draw DNA from one entry/exit location and extend it to the other in a directional movement.32 Even in the absence of ATP hydrolysis, chromatin remodeler complexes binding to their site of nucleosome attachment generates significant repositioning of DNA relative to the histone octamer, which is believed to assist the formation of a DNA loop necessary for ATP-dependent translocation.33
ISWI
ISWI ATPase encoding gene was first identified in Drosophila homologus to yeast Swi2/Snf2 gene exclusively at the region of the ATPase domain and consequently it was named as imitation switch (ISWI).34 Among eukaryotes ISWI family chromatin remodelers are highly conserved and play a critical role in nucleosome assembly and spacing as well as in the organization of chromatin at a greater level in the cell.35 ISWI complexes, compared to SWI/SNF remodelers are smaller and composed of 2 to 4 subunits; each having the nucleosome-dependent ATPase ISWI. ISWI possess highly conserved SWI2/SNF2 family ATPase domain that provides the motor for chromatin remodeling and a characteristic HANDSANT-SLIDE domains with DNA binding activity.36 By means of DNA-dependent ATPase activity, ISWI remodelers alter nucleosome positioning to promote chromatin assembly, resulting into the transcription suppression. Similarly to primary histone chaperones they are also involved in facilitating the de novo assembly of nucleosomes.
Variability exists within the complex of different species, such as; yeast possesses 2 ISWI ATPases – Isw1 and Isw2, which exist in 4 different complexes. The two different ISWI variants (Isw1 and Isw2) in Saccharomyces cerevisiae in combination with different subunits, form a total of 4 diverse complexes.37 Along with Ioc3 subunit, ISWI (Isw1) forms Isw1a complex and binds to Pol4 II promoters which rejects the basal Pol II transcription machinery, thus inhibiting transcription initiation.38 On the other hand, Isw1 association with Ioc2 and Ioc4 subunits results in the formation of Isw1b complex, which possess regulatory role in Pol II transcription elongation and termination.39 Whereas, Drosophila contains only one ISWI ATPase, which is a constituent of 3 complexes: dNURF (Nucleosome Remodeling Factor), dACF (ATP utilizing chromatin assembly and remodeling factor) and dCHRAC (chromatin accessibility complex).40,41 Six diverse functional ISWI complexes have been identified in Drosophila melanogaster, which include CHRAC, ACF, NURF, RSF, ToRC, and NoRC, each comprising ISWI bound to varied combinations of 9 different subunits.42 In general, the CHRAC and ACF complexes seem to function in assisting nucleosome sliding.43 NURF acts particularly in the epigenetic regulation of stem cells within the testis and RSF, in addition to chromatin remodeling activities plays a crucial role in chromatin assembly through the replacement of histone variants.44
In mammals there exists a huge complex with 2 ISWI ATPases: SNF2H and SNF2L that reside in at least 8 different complexes.45 The characteristic of ISWI complex is the presence of a SANT domain (structurally related to the c-Myb DNA-binding domains) which interacts with the unmodified histone tails, a SLIDE (SANT like ISWI domain) domain that binds with the nucleosomal DNA near the dyad axis, and a HAND domain occupied in both histone and DNA binding/recognition.4 Other specialized subunits constitute supplementary domains to the complexes, that include DNA-binding histone fold motifs (in hCHRAC), plant homeodomain zinc fingers (PHD fingers), bromodomains (hBPTF and hACF1), and extra DNA-binding motifs (HMGI(Y), for dNURF301).4
ISWI association with transcription activation was confirmed experimentally in vitro where dNURF directly facilitated GAL4-mediated transcription from chromatin templates.46 Consequently, the interaction was reported with other sequence-specific transcriptional regulators, including dGAF and dHSF in vivo, thereby facilitating gene expression.47 Further, the transcription repressive role of ISWI remodelers was shown in the transcription of yeast meiotic genes during mitotic growth that was repressed by Isw2. In corroboration, the nuclease digestion analysis revealed that Isw2 complex generates nuclease protected chromatin structure near these genes promoters.48 Interestingly, the Isw2 mutant analysis in nucleosome mapping on a genome wide scale showed the role of Isw2 in prevention of antisense transcription from intergenic region and from cryptic initiation sites.49
Significant implications of ISWI in maintaining higher order chromatin structure came from studies on the polytene chromosomes from 3rd instar Drosophila larvae where the loss of zygotic ISWI in larval salivary glands leads to extensive decondensation of the X chromosome.50 Moreover, human hSNF2h complex was shown to interact with cohesins.51 Human ISWI complexes participate in nucleosome positioning over several kilobases and thereby regulate chromatin folding into loop domains.52 In addition, it helps to promote replication fork progression as it gets enriched at the sites of active replication.53 In human cells, hSNF2h, in concert with ACF1, is essential for facilitating DNA replication through highly condensed heterochromatin.54
INO80
This family of remodeler was first discovered in S. cerevisiae, the yeast Ino80 gene product is responsible for regulation of inositol-responsive gene expression.55 The homologous proteins exist in Drosophila as well as humans. The chromatin remodeling enzymes of the INO80 family are: Ino80 and Swr1 in S. cerevisiae; INO80, and p400 in Drosophila melanogaster and in mammals, as Snf2-related CBP activator protein (SRCAP) and p400. The individual complex is highly conserved and composed of 14 to 15 subunits. The unique proteins for INO80 and SWR1 complexes, are RuvB-like helicases that are functionally related to the bacterial RuvB helicase, involved in DNA repair.56,57
The family has been reported to have the ability to perform unique and specialized functions by binding to the replication forks and Holliday junctions. They also bind to the histone variants of H2A; H2A.X and H2A.Z. In vivo INO80 complex influences nucleosome eviction, however the replacement of a canonical H2A-H2B dimer with an H2AZ-H2B variant dimer is catalyzed by SWR1 complex.58-61 INO80 complexes in Drosophila and mammals contain a YY1 subunit, a zinc finger containing Polycomb group transcription factor associated with growth and development regulatory genes.62
The ATPase subunits of the INO80 family are distinguished from other ATPases in the SNF2 helicases by the presence of a long spacer region that splits the conserved ATPase domain. This region was shown to be bound by RuVB-like subunits and Arps.63 The motor subunits of INO80 protein also contain a HAS domain (Helicase-Sant domain) which is required for Arps and actin components binding.64 The presence of RuvB-like helicases in INO80 complexes suggested an involvement in DNA repair. Indeed, INO80 complex associates with γ-H2AX at sites of DSB and participates in eviction of nucleosomes surrounding DSBs.60,65,66 Conversely, it was suggested that SWR1 complex can exchange γ-H2AX for H2A.Z around DSBs.67 Ultimately, deletion of histone H2A.Z (HTZ1) in yeast, results in changes in chromatin structure at DSBs which consequently leads to reduced association of DNA repair and check point factors.68 The proposal based on above findings suggest antagonistic function of both complexes at chromatin neighboring a DSB site, and that they regulate the incorporation of different histone H2A variants that subsequently can either promote or block cell cycle checkpoint adaptation.67 In addition, both complexes have been shown in genetic screens in yeast to be involved in telomere regulation and proper chromosome segregation.69
CHD
The characteristic feature of CHD family (Chromodomain - Helicase - DNA binding) is the presence of 2 signature sequence motifs: tandem chromodomains are located at the N terminal region, and the SNF2-like ATPase domain positioned in the central region of the protein. Subsequently, several proteins belonging to this highly conserved family have been identified in Drosophila, yeast and other species.70,71 A large group of ATP-dependent chromatin remodelers is constituted by the CHD family, that is further divided into 3 subfamilies based on the presence or absence of additional domains.72,73
The founding member of the CHD family, CHD1, was identified as a murine nuclear protein that interacts immunoglobulin promoter DNA sequences.74 Chd1 (yChd1) is the only CHD family member present in S. cerevisiae, and Hrp1 and Hrp3 in S. pombe. At their C-terminal region Chd1 and Chd2 proteins contain a DNA-binding domain that preferentially binds to AT-rich DNA motifs though, the function of this interaction remains elusive.74,75 The CHD3 and CHD4 (called also Mi-2α and Mi-2β respectively) belong to the second subfamily that lacks the standard DNA binding domain in the C-terminus. Instead, at the N-terminal region they harbour a pair of PHD Zn-finger-like domains. The third subfamily comprises proteins from CHD6 to CHD9. This subfamily is highly variable as it is defined by additional functional motifs in the C-terminal region, like SANT domain or BRK domains.72,73 CHD5 possess both the PHD fingers and a SANT domain, hence there is a discrepancy in protein classification of CHD5.
The CHD family is defined by the presence of tandem chromodomains as the signature motifs (Fig. 2). It was at first characterized in Drosophila HP1 and Polycomb proteins, where it contributes in binding to histone methylated marks, H3K9me3 and H3K27me3, respectively.76,77 The typical chromodomain module was refined to encompass ∼50 amino acids and it folds into 3-stranded anti-parallel β-sheets and one α-helice.78 Human CHD1 chromodomains have been shown to bind H3K4me2/3.79 Additionally, the PHD Zn-finger-like domains are found in various nuclear proteins involved in chromatin-based transcriptional regulation.80,81 The exact function of PHD fingers in CHD remodeler is yet unknown. In CHD3 and CHD4 the PHD fingers interact with histone deacetylase HDAC1 within NuRD.82 Further, studies revealed the association of the second PHD finger of CHD4 with the N-terminus of histone H3 is facilitated by acetylation or methylation of Lys9 (H3K9ac and H3K9me respectively) and inhibited by methylation of Lys4 (H3K4me).83 However, functional correlation between PHD fingers and these interactions needs to be further addressed.
Figure 2.
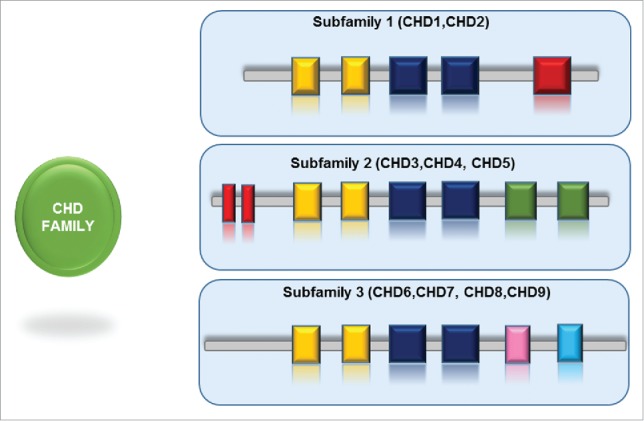
Schematic representation of subfamilies of CHD family of chromatin remodelers. The CHD family is divided into 3 subfamilies, each subfamily is designated with specific set of domains (PHD, SANT, BRK).
Additional domains were mapped in the sequences of CHD remodelers. SANT domains, involved in histone tail binding, were found in several CHD subfamily III members (for example CHD5). The BRK domain (Brahma and Kismet domain) present in several SWI/SNF complexes, was found in Kismet, CHD7, CHD8 and CHD9.72,73,84
Cellular functions of chromatin remodelers: Transcription
The majority of the chromatin remodelers have been studied in relation to transcriptional regulation, both as activators and repressors (Fig. 3). The transcription stage (transcription initiation, elongation and termination) specific involvement of CHD chromatin remodelers has been reported. The only chromatin remodeling complex purified so far is the NuRD complex that couples ATP dependent nucleosome remodeling with histone deacetylation. The transcription repressive ability of the complex has been studied in various genes involved in differentiation and development of C. elegans, D. melanogaster and mammals.85,86 However, not much is known about the mechanism of gene repression by NuRD. In the presence of ATP, purified NuRD disrupts nucleosomes and recombinant Mi-2 slides mononucleosomes in vitro.82,87 On contrary, the disruption in deacteylase activity of NuRD complex does not affect nucleosome remodelling. However, reports do suggest that NuRD along with different gene specific transcription factors get recruited on the target gene promoter and allow histone tails accessibility for deacetylation. Consequently, the recruitment results in generation of more compacted chromatin structure and thus subsequent gene repression.88
Figure 3.
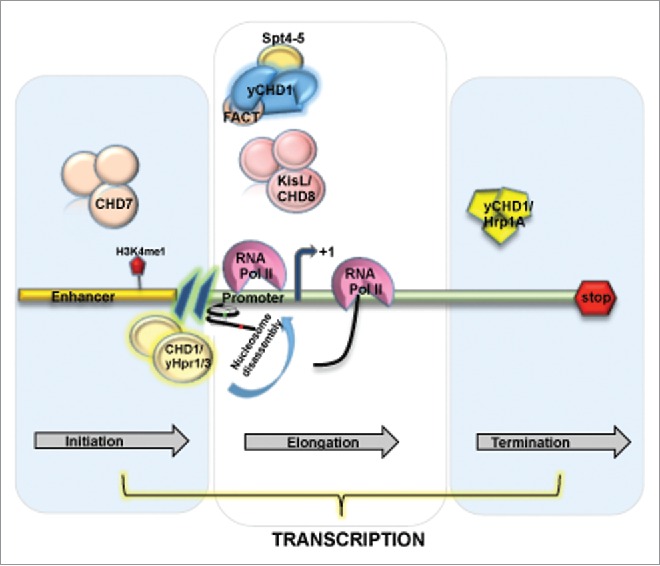
Chromatin remodeler of CHD family and their association with different stages of transcription. CHD7 and CHD8 subfamily in association with other cofactors trigger the transcription initiation and elongation at the enhancer and promoter region of the gene, where as in yCHD1 has been shown to involve in transcription termination.
Transcription initiation
ATP-dependent chromatin remodelers SWI/SNF complexes have been extensively studied in relation to transcription initiation and subsequent assembly of transcription machinery. However, CHD remodelers and their role in transcription initiation are not studied in depth so far. Genome-wide studies on CHD1 homolog of S. pombe, i.e., Hpr1 and Hpr3, suggested their role in nucleosome disassembly at gene promoters.89 Hpr1 and Hpr3 localized at the promoters and to a lesser extent in the ORFs of various S. pombe genes. Hpr1/3 prefer nucleosome dense promoter region. Loss of these remodelers resulted in enhanced H3 density at the promoter regions genome wide, thus indicating their role in nucleosomal disassembly. Further the co purification studies have shown their association with a histone chaperone Nap1, that is linked to nucleosome assembly and disassembly in concert with remodeling complexes.90,91
Numerous studies have implicated CHD remodelers in facilitating transcription activation via binding to enhancers. In particular, human CHD7 genome wide enhancer occupancy has been shown to further overlap with the binding at DNAse I hypersensitive sites enriched in H3K4me1, a hallmark of enhancers. Moreover, CHD7 strongly binds to actively transcribed regions reinforcing its role in gene activation.92 Indeed, another study in Xenopus showed CHD7 mediated direct transcription regulation of core neural crest transcription factors. CHD7 has been shown to interact with BRG1-like complexes, BAF/PBAF and both CHD7 and BRG1 reside in distal regulatory elements of their target genes.93 A weak signal of CHD7 detected at nearby promoters suggests the existence of a looping mechanism.92 Nevertheless, the mechanism of gene activation by CHD7 remains to be elucidated. The enhancer element of an androgen receptor (AR) responsive gene in prostate cancer cells were shown to be occupied by CHD8 in an induction independent manner. CHD8 knockdown studies suggested their plausible involvement in chromatin remodeling at this enhancer and consequently results in gene repression.94 In addition, Mi-2β (CHD4) facilitates enhancer element binding at the CD4 gene during Tcell development. Moreover, Mi-2β was shown to assist recruitment of transcription factor HEB and a histone acetyltransferase, p300, to the CD4 enhancer element, facilitating open chromatin formation and gene activation. Mi-2β remodeler might be implicated in active transcription outside of the NuRD complex since it has been shown to interact with factors in an HDAC independent manner.95
Transcription elongation
Transcription elongation is a highly controlled mechanism and recent studies indicate the significance of chromatin structure in its regulation. The state of transcription elongation to a large extent is defined by the phosphorylation status of the C-terminal domain of the largest subunit of RNAP II (CTD).96 CHD remodeler Kismet (KisL) has been suggested to be involved in an early elongation step. The study have shown the localization of KisL at transcriptionally active sites on polytetene chromosomes. KisL binding pattern highly overalapped with RNAP II phosphorylated at Ser5 and Ser2, Brahma and dCHD1. However, the levels of elongating RNAP II (Ser2) in kisL mutants were drastically reduced, whereas RNAP II phosphorylation at Ser5 remains unaffected. Consequently, chromatin association of the elongation factors Spt6 and dCHD1 was considerably reduced.97 Further studies on KisL mutant showed decrease in association of H3K4 methyltransferases ASH1 and TRX with chromosomes. Additionally, the enhanced H3K27me3 that is essential for Pc function was observed in kisL as well as ash1 and trx mutants.98 In corroboration with above findings, the human homolog of KisL, CHD8, associates with subunits of MLL, a histone H3K4 methyltransferase Ash2L containing complex. CHD8 has been implicated in early transcription elongation or activation of cyclin E2 gene in G1/S cell cycle transition. Interestingly, CHD8 depleted cells were more sensitive toward inhibitors of transcription elongation, like DRB and flavopiridol.99 Though, further studies are required to elucidate the exact functions of CHD8 remodelers in transcription elongation.
CHD1 remodeler also participates in transcription elongation as shown in studies performed on Drosophila where dCHD1 is recruited at the transcription active sites on polytene chromosomes.70 Consequent studies in yeast revealed the interaction of yChd1 with Spt4-Spt5 and Spt16-Pob3 (FACT) and PAF complexes that are exclusively involved in transcription elongation.100-102 In addition to this, dCHD1 participates in deposition of H3.3 variants, nucleosomes spacing and assembly in vivo.91 Collectively, the experiments suggest a role of CHD1 in nucleosome reassembly over elongating RNAP II and in re-establishing of a repressive chromatin structure. However, the function of CHD1 in transcription dependent chromatin assembly demands further investigation.
Transcription termination
The last phase of transcription cycle is transcription termination, which involves the dissociation of the transcription complex from the DNA template and release of the RNA transcript.103 A homolog of yChd1 in S. pombe, identified as Hrp1A is involved in transcription termination by RNAP II. Run on assays on several genes from both hrp1 and chd1 deletion strains showed failure in transcription termination. The chromatin structure analysis indicated alteration in termination region of 800bp that extends ahead of the 3′ end of the gene.104 Similar changes in both induced and uninduced gene states suggest a role of yChd1 in establishment of chromatin structure in the termination regions of yeast ORFs. Furthermore, rDNA (rDNA) genes the transcription termination of RNA Pol I is achieved by yChd1 in combination with Isw1/2. However, in the mutants no effect was observed in the levels of rRNA, thus suggesting that the termination defects were not due to transcription elongation.105
Other cellular function of chromatin remodelers
Chromatin remodelers are also associated with other cellular functions, chromatin compaction/accessibility, replication, histone variant deposition, DNA repair and chromatin maintenance etc.
Histone variant deposition
Different variants of histone proteins exist, having specialized cellular functions along with their fundamental role in DNA structural organization. H3.3 variant of H3 has been largely associated with transcriptional activation and CHD1 might be associated with its deposition and nucleosome assembly during transcription elongation. However, deposition of H3.3 variant outside transcription has been reported in early Drosophila development accomplished by dCHD1 within the paternal pronucleus.106 Additionally, in mammalian cell lines CHD1 has been associated with centromere function. Moreover, knockdown studies on CHD1 reported marked decrease in CENP-A binding to the centromers, which suggest the plausible role of CHD1 in centromeric histone variant deposition.107 However, this function of CHD1 is not necessarily conserved, as in Drosophila this remodeler is not required for CENP-A deposition.108
DNA repair
INO80 complexes in the context of DNA break repair in yeast has been well studied, however its role in ATP-dependent chromatin remodeling at damaged sites within the human genome remains yet to be deciphered. CHD4 association with ataxia telangiectasia mutated serine protein (ATR) kinase and also as a target of the ATM/ATR pathway in a proteomic screen suggested its potential role in DNA repair.109,110 Despite few ambiguities in the results of various studies, the outcome ascertained that CHD4 depleted cells displayed sensitivity toward ionizing radiation, lacking in double strand break (DSB) repair ability and they exhibit extended persistence of the phosphorylated H2AX (γH2AX).111,112 In addition, CHD4 knockdown studies have reported delay in cell cycle progression and apoptosis activation via the p53/p21 pathway. This, suggests the possible role of CHD4 in regulating deacetylation of p53, thereby controlling G1/S cell cycle transition.113 Interestingly, other studies in CHD4 depleted cells have shown decreased accumulation of ubiquitination at the DSB sites. Notably, CHD4 knockdown leads to a significant decrease in histone ubiquitin ligases, RNF168 and BRCA1 accumulation at the DSB sites.111,112 Collectively, these results signify multifaceted and probably mutually dependent roles of CHD4 in DNA damage response and cell cycle progression. Consequently, a more widespread role of CHD4 in chromatin maintenance has been suggested.
Chromatin remodelers association in disease
Chromatin remodeling is an ill-defined multistep process that requires communication, cooperation between the ATPase and auxiliary domains; and their interaction with chromatin. Histone (PTM) and chromatin remodeling in concert play an essential role in embryonic development and disease by altering gene expression profile. Therefore, mutations or alterations affecting the function and targeting of chromatin-remodeling complexes cause several types of cancers, other syndromes and multi-system developmental disorders.113 Studies have shown temporal and tissue specific function of chromatin remodelers during heart development and thereby any mutation leads to the advancement of many cardiovascular tissues.
SWI/SNF complexes recently have gained more attention because of their unusual frequency of mutation in a varied range of human cancers as well as neurodevelopmental disorders.114 SWI/SNF gene mutations occur in a broad spectrum of tumors ranging from early stage stem cell-like cancer to later stage adult cancers such as lung cancers. Mutations in SWI/SNF component are reported in huge spectrum of cancers at a frequency of around 19%. Malignant rhabdoid tumor (MRT) are a highly aggressive group of tumors that usually occur in early childhood in various locations, including the kidney, lung, soft tissue and brain.115,116 They are mostly found in children is characterized by biallelic loss of the SWI/SNF core component.117 The SNF5 gene is found to have undergone bi-allelic loss in the majority of human malignant rhabdoid tumors (MRTs), and some ‘proximal-type’ epithelioid sarcomas.. Mammalian SWI/SNF complexes play siginificant role in cell cycle progression and facilitate double strand break (DSB) repair via H2A.X phosphorylation.117 SWI/SNF complexes contribute to the development of T cells,117 hepatocytes,117 oligodendrocytes,117 and embryonic stem cell self-renewal and pluripotency.117
Also, mutations in other complexes like CHD1, CHD4, CHD5, CHD7, ATRX leads to a variety of disorders and genomic instability. There are many human developmental disorders that are a result of mutation in remodeling genes. Loss of CHD5 leads to microcephaly and features as seen in 1p36 Syndrome. Williams Beuren Syndrome is also the result of deletion of WSTF along with several critical genes required for proper development. CHD7 mutation leads to CHARGE Syndrome. Mutation in ATRX gene leads to α thalassemia X-linked mental retardation syndrome ATRX Syndrome.
The disorders related to dysregulation of chromatin remodeling complexes are as:
ATRX syndrome
The cytogenetic location of ATR gene is Xq21.1. It encodes for a protein that plays role in normal development. Studies have shown ATRX regulating the expression of HBA1 and HBA2 genes that are crucial for hemoglobin production in RBC and this leads to a blood disorder called α thalassemia. It is the only gene found to be mutated in ATRX Syndrome. Patients with this syndrome are reported with intellectual disability, somatic abnormalities, hypotonia and abnormal hemoglobin synthesis.118
CHARGE syndrome
CHARGE syndrome is a multiple organ disorder that stands for Coloboma of eye, heart defects, atresia choanae, retarded growth, genital abnormality and ear abnormality.119 It occurs in approximately 1 in 8500 or 10,000 individuals. Mutations (frameshift and deletion) in CHD7 gene (located on chromosome 8), leads to the production of nonfunctional protein that interrupts chromatin remodeling of other genes involved in normal development and thus leads to the disorder. The identification of 7 heterozygous CHD7 stop-codon mutations and 2 single-copy 8q12 deletions of CHD7 gene designate that haploinsufficiency of this gene could account for most cases of CHARGE syndrome. CHARGE syndrome might also have a genetically heterogeneous etiology, as different genomic abnormalities have been identified in affected individuals.
Neuroblastoma
Neuroblastoma is a tumor of the sympathetic nervous system. It is the most common childhood extracranial solid tumor, accounting for 8%–10% of childhood cancers and 15% of childhood cancer deaths.120 Neuroblastomas demonstrate clinical heterogeneity, from spontaneous regression to relentless progression. Chromo domain helicase DNA binding protein 5 (CHD5) is the strongest candidate tumor suppressor gene that is deleted from 1p36.31 in neuroblastomas, and inactivation of the second allele may occur by an epigenetic mechanism. It is assumed that it functions by forming a nucleosome remodeling and deacetylation (NuRD) complex which regulates transcription of particular genes. CHD5 is mainly expressed in the nervous system and testis. On the basis of its position, pattern of expression, and function in neuroblastoma cells and xenografts, CHD5 was identified as a tumor suppressor gene (TSG). The chromatin remodeler CHD5 is expressed in neural tissue and is frequently deleted in aggressive neuroblastoma. Very few studies have been carried out about the function of CHD5 in the nervous system or its mechanism of action. CHD5 may also play an important role in the development of many other tissues besides the nervous system and testis. CHD5 also plays a major role as a tumor suppressor gene in gliomas and a variety of other tumor types, including breast, colon, lung, ovary, and prostate cancers.117
Coffin-Siris syndrome
It is an autosomal dominant disorder known to be caused by mutation in one of the 5 genes (ARID1A, ARID1B, SMARCA4, SMARCB1, and SMARCE1), but mostly de novo mutations are the main reason. Each of these genes codes for one of the subunit of multi-protein complex SWI/SNF. The mutation in one of the genes thus result in abnormal or deregulated chromatin remodeling followed by expression of several genes that are responsible for different signs and symptoms associated to the disease, which includes hypoplasia of the nail of the fifth digit (fifth digit syndrome), mild to severe intellectual and developmental disability (sitting and walking), distinctive facial features typically with wide nose and a flat nasal bridge.121
COFS and COCKAYNE syndrome type II
Mutations in yeast homolog Rad26, ERCC6 (excision repair cross complementing rodent repair deficiency, complementing group 6) that codes for a SWI/SNF related ATPase causes both COFS (cerebro-oculo-facio-skeletal) and Cockayne syndrome. It is inherited in an autosomal recessive manner. ERCC6 protein promotes transcriptional elongation by RNA Polymerase I,II,III and functions in transcription coupled DNA repair. COFS is a rare genetic progressive neurodegenerative disorder that affects mainly brain and spinal cord. It is said that COFS syndrome is prenatal extreme case of Cockayne Syndrome. The symptoms include postnatal growth failure, microcephaly, congenital cataracts, severe mental retardation, dysmorphic features, progressive skeletal defects, retinopathy. The most common symptom between the 2 syndromes is increased sensitivity to UV induced DNA damage and oxidative stress.4
Williams Syndrome
An autosomal dominant disorder caused by deletion of specific genetic segment from chromosome 7 (7q11.2). The segment contains more than 25 genes and some of the genes that are deleted in this disorder includes CLIP2, ELN, GTF2I, GTF2IRD1, and LIMK1. It has been observed that deletion or loss of function of ELN gene leads to cardiovascular disabilities and connective tissues abnormalities. Studies report that gene deletion of CLIP2, GTF2I, GTF2IRD1, LIMK1 explains the characteristic visual-spatial difficulties, dysmorphism, aberrant vitamin D metabolism, hypercalcemia, behavioral imbalance and other cognitive disabilities in patients of Williams Syndrome. An indirect correlation exists between BRG1 and BRM associated chromatin remodeling complexes and in the pathogenesis of this syndrome.122
Schimke immuno osseus dysplasia
An autosomal recessive multisystem disorder, caused by mutation in the SMARCAL1 gene (SWI/SNF-related matrix associated, actin-dependent regulator of chromatin, subfamily A-like protein 1) whose specific function is still unknown Being a pleiotropic disorder, it leads to a spectrum of symptoms that includes T-cell immunodeficiency, renal defects, spondyloepiphyseal dysplasia (SED) resulting in short stature, nephropathy, hyperpigmentation, lordosis (exaggerated curvature of the lower back) facial dysmorphism. It is also known to regulate several genes required for cellular proliferation. More is yet to be discovered about this gene and disorder.123
Conclusion
In summary, chromatin remodelers have significant role in molecular and cellular function viz nucleosome sliding, eviction, histone exchange, histone post-translational modification, chromatin compaction, accessibility, transcription, replication, DNA repair etc. We can summarize that defect in chromatin remodelling complexes lead to a diverse spectrum of human disorders like cancer, developmental disorders and birth defects. These disease phenotypes depict the role of these complexes in DNA replication, regulation of gene expression and DNA repair. Chromatin remodelers have implications in gene regulation that associates with gene reprogramming and disease conditions like in cancer progression. Further study of chromatin remodelers is required to understand their mode of action and to help us in designing novel strategies for prevention and cure of cancer and other chromatin related diseases. Structural aspects of remodeler will be essential for defining precisely the molecular pathophysiology and scope of chromatin remodeling and epigenetics in human diseases. The field is an open platform to decipher the possible therapeutic potential of these chromatin modulation proteins in future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
M.T. and N.I was supported by fellowship from grant SB/FT/LS-429/2012 from Department of Science and Technology (DST), India K.V. was supported by research fellowships from the University Grants Commission (UGC) India. A.K.P. was supported by a Ramalingaswami fellowship from DBT and financial support from IIT Delhi and Kusuma Trust, UK.
References
- [1].van Holde K, Zlatanova J. Chromatin fiber structure: Where is the problem now? Semin Cell Dev Biol 2007; 18:651-8; PMID:17905614; http://dx.doi.org/ 10.1016/j.semcdb.2007.08.005 [DOI] [PubMed] [Google Scholar]
- [2].Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41-5; PMID:10638745; http://dx.doi.org/ 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- [3].Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci U S A 2007; 104:15635-40; PMID:17893337; http://dx.doi.org/ 10.1073/pnas.0702430104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Ann Rev Biochem 2009; 78:273-304; PMID:19355820; http://dx.doi.org/ 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- [5].Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell 2004; 116:259-72; PMID:14744436; http://dx.doi.org/ 10.1016/S0092-8674(04)00044-3 [DOI] [PubMed] [Google Scholar]
- [6].Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 1994; 265:53-60; PMID:8016655; http://dx.doi.org/ 10.1126/science.8016655 [DOI] [PubMed] [Google Scholar]
- [7].Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem 2007; 76:23-50; PMID:17506634; http://dx.doi.org/ 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- [8].Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 2005; 121:363-73; PMID:15882619; http://dx.doi.org/ 10.1016/j.cell.2005.03.026 [DOI] [PubMed] [Google Scholar]
- [9].Carlson M, Osmond BC, Neigeborn L, Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 1984; 107:19-32; PMID:6373495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol 1984; 178:853-68; PMID:6436497; http://dx.doi.org/ 10.1016/0022-2836(84)90315-2 [DOI] [PubMed] [Google Scholar]
- [11].Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 1993; 12:4279-90; PMID:8223438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marmorstein R, Berger SL. Structure and function of bromodomains in chromatin-regulating complexes. Gene 2001; 272:1-9; PMID:11470504; http://dx.doi.org/ 10.1016/S0378-1119(01)00519-4 [DOI] [PubMed] [Google Scholar]
- [13].Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Ann Rev Biochem 2002; 71:755-81; PMID:12045110; http://dx.doi.org/ 10.1146/annurev.biochem.71.110601.135507 [DOI] [PubMed] [Google Scholar]
- [14].Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A 2002; 99:2824-9; PMID:11880634; http://dx.doi.org/ 10.1073/pnas.032662899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J 2003; 22:3175-87; PMID:12805231; http://dx.doi.org/ 10.1093/emboj/cdg296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell 1996; 87:1249-60; PMID:8980231; http://dx.doi.org/ 10.1016/S0092-8674(00)81820-6 [DOI] [PubMed] [Google Scholar]
- [17].Zraly CB, Marenda DR, Nanchal R, Cavalli G, Muchardt C, Dingwall AK. SNR1 is an essential subunit in a subset of Drosophila brm complexes, targeting specific functions during development. Dev Biol 2003; 253:291-308; PMID:12645932; http://dx.doi.org/ 10.1016/S0012-1606(02)00011-8 [DOI] [PubMed] [Google Scholar]
- [18].Crosby MA, Miller C, Alon T, Watson KL, Verrijzer CP, Goldman-Levi R, Zak NB. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol 1999; 19:1159-70; PMID:9891050; http://dx.doi.org/ 10.1128/MCB.19.2.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 1994; 370:477-81; PMID:8047169; http://dx.doi.org/ 10.1038/370477a0 [DOI] [PubMed] [Google Scholar]
- [20].Wang D, Marsh JL, Ayala FJ. Evolutionary changes in the expression pattern of a developmentally essential gene in three Drosophila species. Proc Natl Acad Sci U S A 1996; 93:7103-7; PMID:8692952; http://dx.doi.org/ 10.1073/pnas.93.14.7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ho L, Crabtree GR. Chromatin remodelling during development. Nature 2010; 463:474-84; PMID:20110991; http://dx.doi.org/ 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Armstrong JA, Emerson BM. Transcription of chromatin: these are complex times. Curr Opin Genet Dev 1998; 8:165-72; PMID:9610406; http://dx.doi.org/ 10.1016/S0959-437X(98)80137-8 [DOI] [PubMed] [Google Scholar]
- [23].Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 1997; 147:451-65; PMID:9335585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev 1999; 13:1412-21; PMID:10364158; http://dx.doi.org/ 10.1101/gad.13.11.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A 1997; 94:11268-73; PMID:9326598; http://dx.doi.org/ 10.1073/pnas.94.21.11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Flanagan JF, Peterson CL. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res 1999; 27:2022-8; PMID:10198436; http://dx.doi.org/ 10.1093/nar/27.9.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hara R, Sancar A. Effect of damage type on stimulation of human excision nuclease by SWI/SNF chromatin remodeling factor. Mol Cell Biol 2003; 23:4121-5; PMID:12773556; http://dx.doi.org/ 10.1128/MCB.23.12.4121-4125.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gaillard H, Fitzgerald DJ, Smith CL, Peterson CL, Richmond TJ, Thoma F. Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J Biol Chem 2003; 278:17655-63; PMID:12637512; http://dx.doi.org/ 10.1074/jbc.M300770200 [DOI] [PubMed] [Google Scholar]
- [29].Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol 2005; 12:747-55; PMID:16086025; http://dx.doi.org/ 10.1038/nsmb973 [DOI] [PubMed] [Google Scholar]
- [30].Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem 2004; 279:39933-41; PMID:15262970; http://dx.doi.org/ 10.1074/jbc.M406060200 [DOI] [PubMed] [Google Scholar]
- [31].Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 2006; 13:339-46; PMID:16518397; http://dx.doi.org/ 10.1038/nsmb1071 [DOI] [PubMed] [Google Scholar]
- [32].Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol 2008; 28:6010-21; PMID:18644858; http://dx.doi.org/ 10.1128/MCB.00693-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A 2010; 107:3458-62; PMID:20142505; http://dx.doi.org/ 10.1073/pnas.1000398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol 1994; 14:2225-34; PMID:7908117; http://dx.doi.org/ 10.1128/MCB.14.4.2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Erdel F, Rippe K. Chromatin remodelling in mammalian cells by ISWI-type complexes–where, when and why? FEBS J 2011; 278:3608-18; PMID:21810179; http://dx.doi.org/ 10.1111/j.1742-4658.2011.08282.x [DOI] [PubMed] [Google Scholar]
- [36].Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet 2013; 29:621-9; PMID:23870137; http://dx.doi.org/ 10.1016/j.tig.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Toto M, D'Angelo G, Corona DF. Regulation of ISWI chromatin remodelling activity. Chromosoma 2014; 123:91-102; PMID:24414837; http://dx.doi.org/ 10.1007/s00412-013-0447-4 [DOI] [PubMed] [Google Scholar]
- [38].Vary JC Jr., Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol 2003; 23:80-91; PMID:12482963; http://dx.doi.org/ 10.1128/MCB.23.1.80-91.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mellor J, Morillon A. ISWI complexes in Saccharomyces cerevisiae. Biochim Biophys Acta 2004; 1677:100-12; PMID:15020051; http://dx.doi.org/ 10.1016/j.bbaexp.2003.10.014 [DOI] [PubMed] [Google Scholar]
- [40].Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 1995; 83:1021-6; PMID:8521502; http://dx.doi.org/ 10.1016/0092-8674(95)90217-1 [DOI] [PubMed] [Google Scholar]
- [41].Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 1997; 388:598-602; PMID:9252192; http://dx.doi.org/ 10.1038/41587 [DOI] [PubMed] [Google Scholar]
- [42].Bouazoune K, Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res 2006; 14:433-49; PMID:16821138; http://dx.doi.org/ 10.1007/s10577-006-1067-0 [DOI] [PubMed] [Google Scholar]
- [43].Chioda M, Vengadasalam S, Kremmer E, Eberharter A, Becker PB. Developmental role for ACF1-containing nucleosome remodellers in chromatin organisation. Development 2010; 137:3513-22; PMID:20843858; http://dx.doi.org/ 10.1242/dev.048405 [DOI] [PubMed] [Google Scholar]
- [44].Cherry CM, Matunis EL. Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell 2010; 6:557-67; PMID:20569693; http://dx.doi.org/ 10.1016/j.stem.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yadon AN, Tsukiyama T. SnapShot: Chromatin remodeling: ISWI. Cell 2011; 144:453- e1; PMID:21295704; http://dx.doi.org/ 10.1016/j.cell.2011.01.019 [DOI] [PubMed] [Google Scholar]
- [46].Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell 1997; 1:141-50; PMID:9659911; http://dx.doi.org/ 10.1016/S1097-2765(00)80015-5 [DOI] [PubMed] [Google Scholar]
- [47].Badenhorst P, Finch JT, Travers AA. Tramtrack co-operates to prevent inappropriate neural development in Drosophila. Mech Dev 2002; 117:87-101; PMID:12204250; http://dx.doi.org/ 10.1016/S0925-4773(02)00183-1 [DOI] [PubMed] [Google Scholar]
- [48].Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 2000; 103:423-33; PMID:11081629; http://dx.doi.org/ 10.1016/S0092-8674(00)00134-3 [DOI] [PubMed] [Google Scholar]
- [49].Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 2007; 450:1031-5; PMID:18075583; http://dx.doi.org/ 10.1038/nature06391 [DOI] [PubMed] [Google Scholar]
- [50].Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al.. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 2000; 5:355-65; PMID:10882076; http://dx.doi.org/ 10.1016/S1097-2765(00)80430-X [DOI] [PubMed] [Google Scholar]
- [51].Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A 2002; 99:7420-5; PMID:12032298; http://dx.doi.org/ 10.1073/pnas.112008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 2002; 419:641-5; PMID:12374985; http://dx.doi.org/ 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- [53].Vincent M, Lauriault P, Dubois MF, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res 1996; 24:4649-52; PMID:8972849; http://dx.doi.org/ 10.1093/nar/24.23.4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Marenda DR, Zraly CB, Dingwall AK. The Drosophila Brahma (SWI/SNF) chromatin remodeling complex exhibits cell-type specific activation and repression functions. Dev Biol 2004; 267:279-93; PMID:15013794; http://dx.doi.org/ 10.1016/j.ydbio.2003.10.040 [DOI] [PubMed] [Google Scholar]
- [55].Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Molecular microbiology 1999; 32:741-51; PMID:10361278; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01390.x [DOI] [PubMed] [Google Scholar]
- [56].Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem 1999; 274:22437-44; PMID:10428817; http://dx.doi.org/ 10.1074/jbc.274.32.22437 [DOI] [PubMed] [Google Scholar]
- [57].Qiu J, Guan MX, Bailis AM, Shen B. Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic Acids Res 1998; 26:3077-83; PMID:9628902; http://dx.doi.org/ 10.1093/nar/26.13.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al.. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 2003; 12:1565-76; PMID:14690608; http://dx.doi.org/ 10.1016/S1097-2765(03)00497-0 [DOI] [PubMed] [Google Scholar]
- [59].Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004; 303:343-8; PMID:14645854; http://dx.doi.org/ 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- [60].Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 2005; 438:379-83; PMID:16292314; http://dx.doi.org/ 10.1038/nature04148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 2007; 26:4113-25; PMID:17762868; http://dx.doi.org/ 10.1038/sj.emboj.7601835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 2006; 20:1110-22; PMID:16618800; http://dx.doi.org/ 10.1101/gad.377406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell 2004; 16:465-77; PMID:15525518; http://dx.doi.org/ 10.1016/j.molcel.2004.09.033 [DOI] [PubMed] [Google Scholar]
- [64].Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 2008; 15:469-76; PMID:18408732; http://dx.doi.org/ 10.1038/nsmb.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004; 119:767-75; PMID:15607974; http://dx.doi.org/ 10.1016/j.cell.2004.11.037 [DOI] [PubMed] [Google Scholar]
- [66].van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004; 119:777-88; PMID:15607975; http://dx.doi.org/ 10.1016/j.cell.2004.11.033 [DOI] [PubMed] [Google Scholar]
- [67].Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev 2006; 20:2437-49; PMID:16951256; http://dx.doi.org/ 10.1101/gad.1440206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 2009; 33:335-43; PMID:19217407; http://dx.doi.org/ 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- [69].Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al.. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A 2004; 101:13513-8; PMID:15353583; http://dx.doi.org/ 10.1073/pnas.0405753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci U S A 1996; 93:7137-42; PMID:8692958; http://dx.doi.org/ 10.1073/pnas.93.14.7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A 1997; 94:11472-7; PMID:9326634; http://dx.doi.org/ 10.1073/pnas.94.21.11472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol 2007; 85:463-76; PMID:17713581; http://dx.doi.org/ 10.1139/O07-063 [DOI] [PubMed] [Google Scholar]
- [73].Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res 2007; 618:30-40; PMID:17350655; http://dx.doi.org/ 10.1016/j.mrfmmm.2006.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A 1993; 90:2414-8; PMID:8460153; http://dx.doi.org/ 10.1073/pnas.90.6.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Stokes DG, Perry RP. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 1995; 15:2745-53; PMID:7739555; http://dx.doi.org/ 10.1128/MCB.15.5.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A 1991; 88:263-7; PMID:1898775; http://dx.doi.org/ 10.1073/pnas.88.1.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. BioEssays 2004; 26:133-40; PMID:14745831; http://dx.doi.org/ 10.1002/bies.10392 [DOI] [PubMed] [Google Scholar]
- [78].Ball LJ, Murzina NV, Broadhurst RW, Raine AR, Archer SJ, Stott FJ, Murzin AG, Singh PB, Domaille PJ, Laue ED. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J 1997; 16:2473-81; PMID:9171360; http://dx.doi.org/ 10.1093/emboj/16.9.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 2005; 438:1181-5; PMID:16372014; http://dx.doi.org/ 10.1038/nature04290 [DOI] [PubMed] [Google Scholar]
- [80].Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet 2006; 22:320-9; PMID:16631276; http://dx.doi.org/ 10.1016/j.tig.2006.03.008 [DOI] [PubMed] [Google Scholar]
- [81].Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, et al.. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 2010; 38:165-78; PMID:20346720; http://dx.doi.org/ 10.1016/j.molcel.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 1998; 2:851-61; PMID:9885572; http://dx.doi.org/ 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- [83].Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem 2011; 286:11779-91; PMID:21278251; http://dx.doi.org/ 10.1074/jbc.M110.208207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 1999; 126:1175-87; PMID:10021337 [DOI] [PubMed] [Google Scholar]
- [85].Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 2000; 16:351-6; PMID:10904264; http://dx.doi.org/ 10.1016/S0168-9525(00)02066-7 [DOI] [PubMed] [Google Scholar]
- [86].Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 2009; 4:532-6; PMID:19923891; http://dx.doi.org/ 10.4161/epi.4.8.10108 [DOI] [PubMed] [Google Scholar]
- [87].Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Müller J, Becker PB. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J 2000; 19:4332-41; PMID:10944116; http://dx.doi.org/ 10.1093/emboj/19.16.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 1999; 13:1924-35; PMID:10444591; http://dx.doi.org/ 10.1101/gad.13.15.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 2007; 26:2868-79; PMID:17510629; http://dx.doi.org/ 10.1038/sj.emboj.7601728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A 2006; 103:3090-3; PMID:16492771; http://dx.doi.org/ 10.1073/pnas.0511050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 2005; 12:160-6; PMID:15643425; http://dx.doi.org/ 10.1038/nsmb884 [DOI] [PubMed] [Google Scholar]
- [92].Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, et al.. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res 2009; 19:590-601; PMID:19251738; http://dx.doi.org/ 10.1101/gr.086983.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010; 463:958-62; PMID:20130577; http://dx.doi.org/ 10.1038/nature08733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Menon T, Yates JA, Bochar DA. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol Endocrinol 2010; 24:1165-74; PMID:20308527; http://dx.doi.org/ 10.1210/me.2009-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity 2007; 27:723-34; PMID:17980631; http://dx.doi.org/ 10.1016/j.immuni.2007.09.008 [DOI] [PubMed] [Google Scholar]
- [96].Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 2006; 20:2922-36; PMID:17079683; http://dx.doi.org/ 10.1101/gad.1477006 [DOI] [PubMed] [Google Scholar]
- [97].Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 2005; 132:1623-35; PMID:15728673; http://dx.doi.org/ 10.1242/dev.01713 [DOI] [PubMed] [Google Scholar]
- [98].Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS genetics 2008; 4:e1000217; PMID:18846226; http://dx.doi.org/ 10.1371/journal.pgen.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rodriguez-Paredes M, Ceballos-Chavez M, Esteller M, Garcia-Dominguez M, Reyes JC. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res 2009; 37:2449-60; PMID:19255092; http://dx.doi.org/ 10.1093/nar/gkp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 2002; 277:10753-5; PMID:11805083; http://dx.doi.org/ 10.1074/jbc.C200023200 [DOI] [PubMed] [Google Scholar]
- [101].Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 2003; 22:1846-56; PMID:12682017; http://dx.doi.org/ 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 2007; 27:6103-15; PMID:17576814; http://dx.doi.org/ 10.1128/MCB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Current opinion in cell biology 2005; 17:257-61; PMID:15901494; http://dx.doi.org/ 10.1016/j.ceb.2005.04.003 [DOI] [PubMed] [Google Scholar]
- [104].Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 2002; 10:1441-52; PMID:12504018; http://dx.doi.org/ 10.1016/S1097-2765(02)00778-5 [DOI] [PubMed] [Google Scholar]
- [105].Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol 2007; 14:123-30; PMID:17259992; http://dx.doi.org/ 10.1038/nsmb1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al.. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 2007; 317:1087-90; PMID:17717186; http://dx.doi.org/ 10.1126/science.1145339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Molecular biology of the cell 2009; 20:3986-95; PMID:19625449; http://dx.doi.org/ 10.1091/mbc.E09-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Podhraski V, Campo-Fernandez B, Worle H, Piatti P, Niederegger H, Bock G, Fyodorov DV, Lusser A. CenH3/CID incorporation is not dependent on the chromatin assembly factor CHD1 in Drosophila. PloS One 2010; 5:e10120; PMID:20396651; http://dx.doi.org/ 10.1371/journal.pone.0010120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Schmidt DR, Schreiber SL. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry 1999; 38:14711-7; PMID:10545197; http://dx.doi.org/ 10.1021/bi991614n [DOI] [PubMed] [Google Scholar]
- [110].Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; http://dx.doi.org/ 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- [111].Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al.. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 2010; 190:731-40; PMID:20805324; http://dx.doi.org/ 10.1083/jcb.200912135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 2010; 190:741-9; PMID:20805320; http://dx.doi.org/ 10.1083/jcb.201001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 2010; 29:3130-9; PMID:20693977; http://dx.doi.org/ 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lopez AJ, Wood MA. Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Frontiers in behavioral neuroscience 2015; 9:100; PMID:25954173; http://dx.doi.org/ 10.3389/fnbeh.2015.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Roberts CW, Orkin SH. The SWI/SNF complex–chromatin and cancer. Nat Rev Cancer 2004; 4:133-42; PMID:14964309; http://dx.doi.org/ 10.1038/nrc1273 [DOI] [PubMed] [Google Scholar]
- [116].Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998; 394:203-6; PMID:9671307; http://dx.doi.org/ 10.1038/28212 [DOI] [PubMed] [Google Scholar]
- [117].Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends in molecular medicine 2007;13:37–80. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. BioEssays 2003; 25:1192-200; PMID:14635254; http://dx.doi.org/ 10.1002/bies.10359 [DOI] [PubMed] [Google Scholar]
- [119].Johnson CA. Chromatin modification and disease. J Med Genet 2000; 37:905-15; PMID:11106353; http://dx.doi.org/ 10.1136/jmg.37.12.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Martin DM. Chromatin remodeling in development and disease: focus on CHD7. PLoS Genet 2010; 6:e1001010; PMID:20657659; http://dx.doi.org/ 10.1371/journal.pgen.1001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. Role of CHD5 in human cancers: 10 years later. Cancer Res 2014; 74:652-8; PMID:24419087; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, et al.. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF). Proc Natl Acad Sci U S A 2009; 106:9280-5; PMID:19470456; http://dx.doi.org/ 10.1073/pnas.0901184106 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [123].Elizondo LI, Huang C, Northrop JL, Deguchi K, Clewing JM, Armstrong DL, Boerkoel CF. Schimke immuno-osseous dysplasia: a cell autonomous disorder? American journal of medical genetics Part A 2006; 140:340-8; PMID:16419127; http://dx.doi.org/ 10.1002/ajmg.a.31089 [DOI] [PubMed] [Google Scholar]


