ABSTRACT
Samp1 is a transmembrane protein of the inner nuclear membrane (INM), which interacts with the nuclear lamina and the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex in interphase and during mitosis, it localizes to the mitotic spindle. Samp1 was recently found to coprecipitate a protein complex containing Ran, a GTPase with fundamental regulatory functions both in interphase and in mitosis. To investigate the interaction between Samp1 and Ran in further detail, we have designed and expressed recombinant fusion proteins of the Chaetomium thermophilum homolog of Samp1 (Ct.Samp1) and human Ran. Pulldown experiments show that Samp1 binds directly to Ran and that Samp1 binds better to RanGTP compared to RanGDP. Samp1 also preferred RanGTP over RanGDP in living tsBN2 cells. We also show that the Ran binding domain is located between amino acids 75–135 in the nucleoplasmically exposed N-terminal tail of Samp1. This domain is unique for Samp1, without homology in any other proteins in fungi or metazoa. Samp1 is the first known transmembrane protein that binds to Ran and could provide a unique local binding site for RanGTP in the INM. Samp1 overexpression resulted in increased Ran concentrations in the nuclear periphery supporting this idea.
KEYWORDS: EDMD, laminopathies, LINC complex, nucleus, nuclear membrane, Ran
Introduction
In Eukaryotic cell, the nuclear envelope (NE) forms the interface between the nucleus and the cytoplasm.1-3 The nuclear envelope is composed of the inner nuclear membrane (INM), the outer nuclear membrane (ONM), the nuclear lamina and the nuclear pores. The recently discovered Linker of Nucleoskeleton and Cytoskeleton (LINC) complex4,5 forms transcisternal connections spanning the nuclear envelope and connects the cytoskeleton with the nuclear interior. Despite the fact that the INM contains hundreds of unique transmembrane proteins that are variably expressed across different tissues,6-8 only a handful of them have been characterized.9
Ran is a small monomeric GTPase with many regulatory functions in nuclear processes.10-13 Regulators of the Ran nucleotide state are located on opposite sides of the nuclear envelope14 generating a high RanGTP concentration in the nucleus.15-17 In mitosis, a high RanGTP gradient is maintained around the chromosomes9 essential for releasing and activating the spindle assembly factors that controls the mitotic spindle assembly.18 After mitosis, Ran is involved in reformation of NE.19
Lately, it has been realized that in addition to its structural role, the NE, also plays fundamental roles in chromatin organization20 and is involved in a diverse group of diseases collectively called laminopathies.21 One such disease, Emery-Dreifuss Muscular Dystrophy (EDMD) can be caused by point mutations in genes encoding either Lamin A or Emerin, respectively.22 The INM protein Samp123,24 (also known as known as TMEM 201 or Net 5, in rat liver7) has been reported to functionally interact with both Lamin A25 and Emerin.26 More recently, Samp1 was shown to bind directly to Emerin.26 Furthermore, Samp1 depletion phenocopies the centrosomal detachment23 observed in cells from EDMD patients,27 suggesting that Samp1 is involved in a functional network in common with Emerin and Lamin A.
In a recent study, Samp1 was also found to co-precipitate with Ran in live cells using Membrane protein Cross-Link ImmunoPrecipitation (MCLIP).26 Here we investigate Samp1-Ran interaction in detail using the Chaetomium thermophilum homolog of Samp1, and human Ran. We find that the binding is direct and depends on the nucleotide state of Ran. The fact that Samp1 is the only known transmembrane protein that binds to Ran is discussed.
Results
Samp1 binds directly to Ran
Recently, we have reported that the transmembrane INM protein Samp1 co-precipitates with Ran in live U2OS cells using MCLIP.26 To investigate whether the interaction between Samp1 and Ran is direct, we carried out pulldown experiment with recombinantly expressed proteins. Due to the solubility problems with human Samp1 fragments, we took advantage of the homolog of Samp1 in the thermophilic fungus Chaetomium thermophilum (Ct). The N-terminal domain of Ct.Samp1 displays a high homology to human Samp1 (Fig. 1A) with 4 conserved CXXC (C-cysteine, X- any amino acid) motifs (Fig. 1B) and binds the INM protein Emerin,26 as human Samp1 does,24 demonstrating functional conservation. The first hydrophobic domain of Samp1, been shown not to be a transmembrane domain24 and is not conserved in Ct. Recombinant strep tagged N-terminus of Ct.Samp1 (str-Ct.Samp1(1–180)) and His6-Ran, respectively, were subjected to pulldown experiments using strep tactin beads. The results show that the nucleoplasmic N-terminal half of Ct.Samp1 binds directly to human Ran (Fig. 1C).
Figure 1.
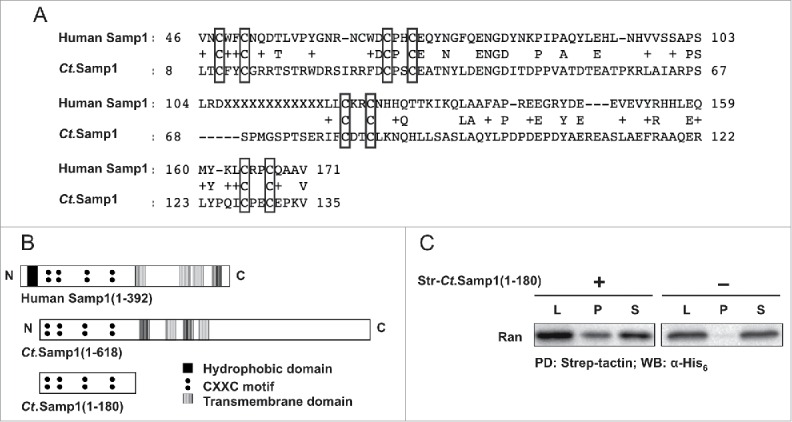
The nucleoplasmically exposed N-terminal half of Samp1 directly binds Ran. (A) Alignment of human Samp1 and Ct.Samp1, performed using the Ct genome resource database (ct.bork.embl.de). (B) Conserved diagrams of human Samp1, Ct.Samp1 and Ct.Samp1 N-terminal domain (1–180). (C) Strep-tactin beads were incubated with His6-Ran in the presence (+) or absence (−) of Str-Ct.Samp1(1–180). Equivalent amounts of Lysate (L), bound (P) and unbound (S) fractions were separated by SDS PAGE and analyzed by Western blotting using anti-His6 antibody as indicated. (PD: pulldown; WB: Western Blot).
Samp1 preferentially binds to RanGTP
Ran is a small monomeric G-protein that mainly exists in two different forms, RanGTP (active) and RanGDP (inactive).28 To investigate whether Samp1 binds to either or both forms, we purified recombinant Ran and specifically loaded it with either GTP or GDP as described in Bischoff et al.29 Pulldown experiments with either RanGTP or RanGDP and bacterial lysates containing str-Ct.Samp1(1–180) were performed and analyzed for Ran binding using Western blotting. The results (Fig. 2A) show that the interaction between Samp1 and RanGTP is significantly stronger than that between Samp1 and RanGDP (Fig. 2B). At a 1:1 molar ratio Samp1 was able to pulldown 40% of RanGTP.
Figure 2.
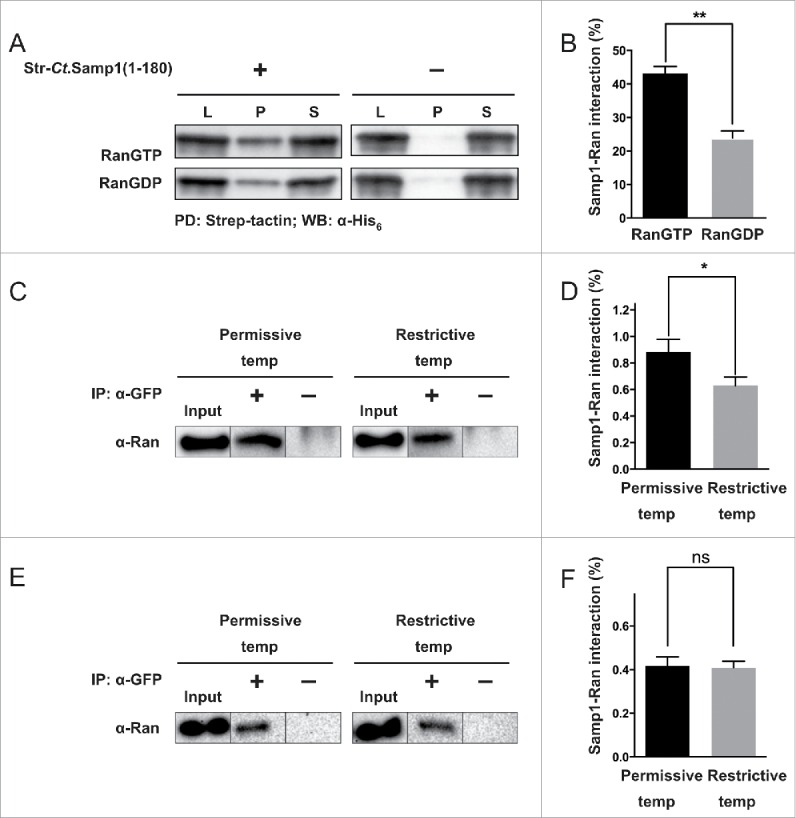
Samp1 preferentially binds to RanGTP. (A-B) In vitro binding. (A) Recombinantly expressed and affinity purified His6-Ran was loaded with either GTP or GDP and subjected to pulldown experiment as described in Fig. 1B. (B) Quantification shows that the binding of Samp1 to RanGTP was 1.8-fold stronger than that to RanGDP (P < 0.005, n = 3). (C-F) In vivo experiments. (C) tsBN2 cells or wt BHK-21 cells (E) transiently expressing Samp1-YFP were grown at 33°C or 37°C. After 24 h, cells were incubated for 4 h at permissive (33°C) or restrictive temperature (39.5°C) before subjection to MCLIP as indicated. Input (cell lysates), the solubilized and diluted protein fractions were subjected to immunoprecipitation (IP) with (+) or without (−) α-GFP antibodies. The proteins were separated by SDS-PAGE and analyzed by Western blotting using antibodies specific for Ran. Note the reduced interaction between Samp1 and Ran at the restrictive temperature. (D) Normalized quantification shows that the interaction between Samp1 and Ran decreases by 1.4-fold on shifting the cells from permissive to restrictive temperature (P < 0.05, n = 3). There was no difference in binding at the two different temperatures when using wt BHK-21 cells (F).
In order to elucidate whether Samp1 has a preference for RanGTP also in live cells, we have used tsBN2 cells, a baby hamster kidney (BHK-21) cell line, which carries a temperature sensitive mutant of RCC1. At the permissive temperature, the tsBN2 cell has a functional RCC1. At the restrictive temperature RCC1, the guanine nucleotide exchange factor for Ran,9 is inactivated30,31 leading to the depletion of RanGTP and accumulation of RanGDP. Interaction studies using MCLIP in tsBN2 cells transiently expressing Samp1-YFP show that Samp1 interacts with Ran in live tsBN2 cells (Fig. 2C). Furthermore, after shifting to the restrictive temperature we observed a significant 1.4-fold decrease in interaction between Samp1 and Ran. The control BHK-21 cells with wild type RCC1 did not show decrease in Samp1 and Ran interaction at restrictive temperature (Fig. 2E and F). The results show that Samp1 interacts stronger with RanGTP compared to RanGDP also in live cells.
Locating the Ran binding domain in Samp1
The nucleoplasmically exposed N-terminal tail of Samp1 is well conserved in evolution and contains four conserved CXXC motifs,23 with potential to form zinc finger(s).24 The N-terminal domain does not share sequence homology with previously characterized Ran binding proteins. Therefore, we decided to elucidate the Ran binding ability in relation to the positions of the CXXC motifs by comparing the Ran binding capacity of shorter fragments i.e., Ct.Samp1(1–180), Ct.Samp1(1–79) and Ct.Samp1(80–180), Ct.Samp1(75–135) and Ct.Samp1(136–180) (Fig. 3A). Ct.Samp1 fragments were expressed in E. coli and the lysates were subjected to pulldown experiments. Both Ct.Samp1(75–135) and Ct.Samp1(80–180) bound Ran as (Fig. 3B) effectively as Ct.Samp1(1–180) (Fig. 1B) whereas Ct.Samp1(1–79) and Ct.Samp1(136–180) did not bind at all (Fig. 3B). This shows that amino acids 75–135 of Samp1 are sufficient to bind to Ran. However, a substitution mutant Ct.Samp1(75–135,C80A) with an alanine instead of the second cysteine of the first CXXC motif, bound equally well as the corresponding wt fragment (Fig. 3B), suggesting that zinc finger formation is not required for Ran binding. In support, the zinc chelator EDTA had no effect on binding between Samp1 and Ran (Fig. 3D). Thus we conclude that it is the amino acid sequence between 75 and 135, rather than potential zinc finger formation that is important for binding to Ran.
Figure 3.
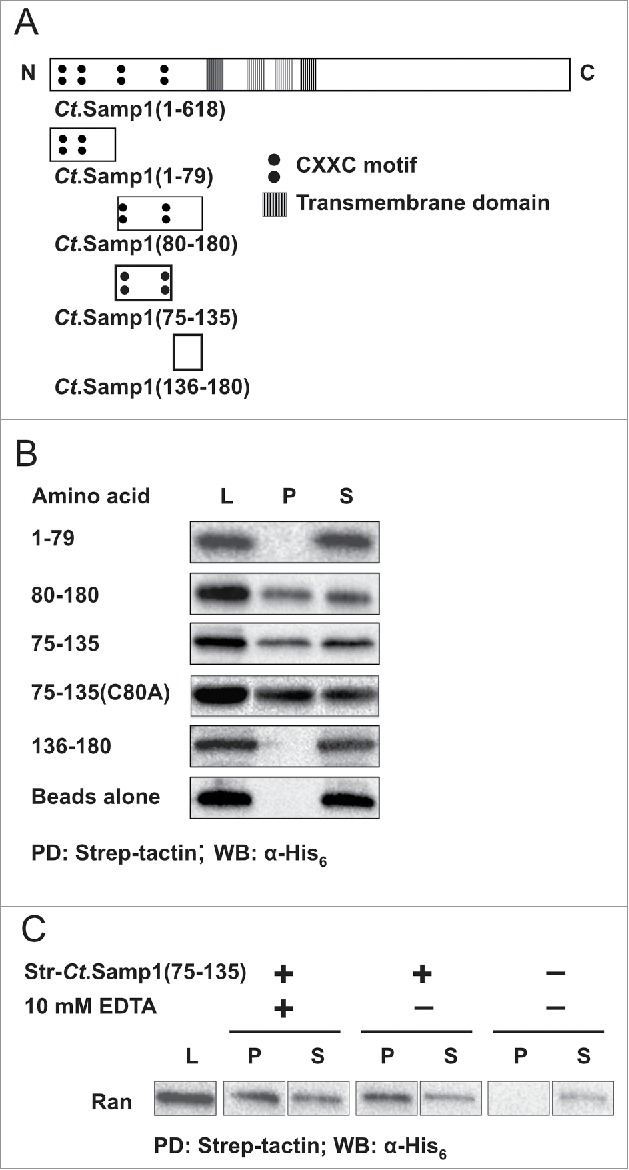
Localization of Ran binding domain of Samp1. (A) Schematic diagrams of Ct.Samp1 and Ct.Samp1 fragments. (B) Ct.Samp1 fragments (Str-Ct.Samp1(1–79) or Str-Ct.Samp1(80–180) or Str-Ct. Samp1(75–135) or Str-Ct.Samp1(75–135,C80A) or Str-Ct.Samp1(136–180)) were subjected to pulldown experiment with His6-Ran using strep-tactin beads. Equivalent amounts of the Lysate (L), bound (P) and unbound (S) fractions were separated by SDS PAGE and analyzed by Western blotting using anti-His6 antibody as indicated. (PD-pulldown, WB-Western Blot). (C) Pull-down experiment with Str-Ct.Samp1(75–135) in the absence (−) and presence (+) of 10 mM EDTA, respectively.
Effect of Samp1 on Ran distribution
In order to get an estimate of the physiological significance of the interaction between Samp1 and Ran, we investigated if Samp1 is able to recruit Ran to the nuclear periphery. For this we overexpressed human Samp1-YFP in HeLa cells and analyzed the distribution of Ran in the nuclear rim using pre-extraction and immunostaining. Indeed, Samp1-YFP overexpressing cells showed an increased Ran staining of the nuclear rim (Fig. 4) demonstrating that Samp1 levels affect the Ran concentration in the nuclear periphery.
Figure 4.
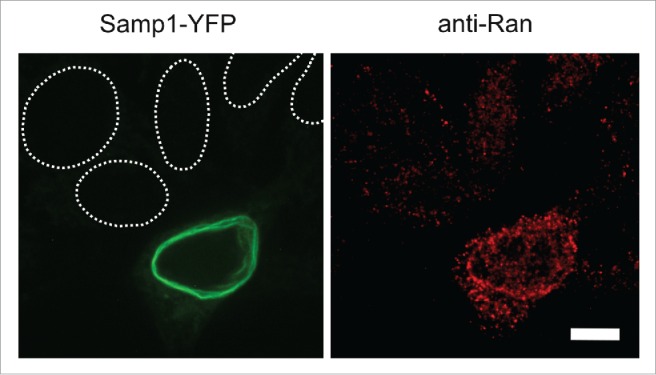
Samp1 redistributes Ran to the nuclear periphery. Immunofluorescence micrographs of pre-extracted HeLa cell expressing Samp1-YFP. Staining of Ran shows an increased signal in Samp1-YFP positive cells compared to untransfected cells (nuclei denoted by dashed white lines). Bar, 10 µm.
Discussion
In the present study, we have shown that the INM protein Samp1 is able to bind Ran directly. We have also shown both in vitro and in live cells that Samp1 binds better to RanGTP compared to RanGDP. The difference might be due to the different conformations of RanGTP and RanGDP, perhaps involving the loop region, which is more accessible in RanGTP.32 In vitro, at 1:1 molar ratio, Samp1 was able to pulldown 40% of RanGTP. Even in live cells with many competing Ran binding partners as much as 1% of total RanGTP was bound to Samp1, suggesting Samp1-Ran interaction plays a significant role in the NE.
Our data also show that the Ran binding domain of Ct.Samp1 is located between amino acids 75–135 in the nucleoplasmically exposed N-terminal part of Samp1. This part of Samp1 does not share sequence homology with other previously characterized Ran binding proteins, including RanBP1, RanBP2, RanBP3, RanBP9, RanBP10, Importin β, NTF2 and Nup153.33-39 The Ran binding domains of RanBP2 and NUP153 share a similar C2C2 Zinc finger motif.36,39 However, despite the conserved CXXC motifs in Samp1 the binding to Ran was not dependent on formation of a potential zinc finger. Thus, the amino acids between residues 75–135 of Samp1 define a novel Ran binding domain.
Samp1 is the first transmembrane protein reported to bind to RanGTP, raising questions concerning the significance of local enrichment of RanGTP close to membranes in which Samp1 distributes. In various processes Ran acts by a common mechanism. RanGTP releases assembly factors (from inhibitory complexes with nuclear transport receptors e.g., Transportin or Importin β) and thereby activates them to initiate assembly processes. For example, on the surface of mitotic chromosomes high RanGTP concentration assures precisely localized assembly of microtubules at kinetochores and not elsewhere. In interphase, it is unclear how NE protein interactions are controlled by RanGTP, which (unlike many other monomeric G-proteins) lacks lipid anchor or amphipathic α-helix for membrane interaction. Because Samp1 is Ran's only known transmembrane binding partner, one could speculate that Samp1 provides a local binding site for RanGTP, which functions as a homing signal for RanGTP triggered processes at precise positions in the INM. Here we report redistribution of Ran to the nuclear periphery as a result of Samp1-YFP overexpression. This observation implies that INM associated Ran can vary due to differential expression of Samp1 and thus might influence many vital cell functions carried out by the NE protein network, including those that become disturbed in EDMD. Future experiments will be needed to explore these interesting possibilities.
Materials and methods
Cell culture and transfection
tsBN2,40 Baby hamster kidney 21 cells (BHK-21) or HeLa were grown and maintained in 1× Dulbecco's modified Eagle's (DMEM) F12 medium (ThermoFischer Scientific, #11330–032) or DMEM glutamax (ThermoFischer Scientific, #21885–025) supplemented with 10% fetal bovine serum (FBS, v/v) (ThermoFischer Scientific, #10500064) and 1% penicillin-streptomycin (v/v) in a humidified atmosphere containing 5% CO2. The BHK-21 and HeLa cells were maintained at 37°C, whereas tsBN2 cells were cultured at 33°C. For interaction studies, the cells were treated for 4 h at 33°C (permissive temperature) or 39.5°C (restrictive temperature) before crosslinking. For transfection of cells with plasmids encoding Samp1-YFP, X-treme gene HP DNA transfection reagent (Sigma, #000000006366236001) was used and cells were analyzed 24 h post-transfection.
MCLIP
MCLIP (Membrane protein Cross-Link ImmunoPrecipitation) was performed as previously described in Jafferali et al.26 tsBN2 or BHK-21 cells that are transiently expressing Samp1-YFP were subjected to crosslinking using 1 mM DSP (dithiobis-succinimidyl-propionate) (ThermoFischer Scientific, #22585) and solubilized with 7 M Urea (Sigma, #33247) and 1% Triton X-100 (Sigma, #93443). The solubilized and diluted lysates were subjected to immunoprecipitation (IP) with or without α-GFP antibodies (Chromotek, #GTA-20). Input (whole cell lysate) and pellets from IP step of MCLIP were loaded, separated on 10% SDS PAGE precast gels (Bio-Rad, #456–1094) and analyzed by Western blot using goat polyclonal anti-Ran (c-20) (1:200) (Santa Cruz Biotechnology, #Sc1156) antibody.
Synthesis of strep-tag pET 33B+ vector
The His6-tag was exercised from pET33B+ vector (Novagen, #69054–3) using NcoI and BamHI according to manufacturer´s (ThermoFisher Scientific) protocol. The strep-tag encoding oligos (forward, 5′-CATGGGCTGGAGCCATCCGCAGTTTGAAAAGGGG-3′ and reverse 5′-GATCCCCCTTTTCAAACTGCGGATGGCTCCAGCC-3′) were annealed by heating to 98°C for 2 min and left to cool. The resulting oligo pair was ligated to the complementary open vector using T4 DNA ligase (ThermoFischer Scientific, #EL0014) according to manufacturer´s (ThermoFischer Scientific) instructions.
PCR amplification
His6-Ct.Samp1(1–180) plasmid26 or pKH3-Ran plasmid38 were used as cDNA template for amplifying Ct.Samp1 or Ran PCR products. PCR reactions were performed as previously described in Jafferali et al.26 and PCR products were cloned into strep-tag pET 33B+ vector or pET 28B+ vector (Novagen, #69865–3). The following primers were used: Ct.Samp1(1–180), forward, 5′-GCAGGATCCAATGCCCCTCCGTAC-3′and reverse, 5′-CAGCTCGAGTTACCTCTTTCCCAGCGC-3′; Ct.Samp1(1–79), forward, 5′-GCAGGATCCAATGCCCCTCCGTAC-3′ and reverse,
5′-CAGCTCGAGTTAAAAGATGCGTTCGCTC-3′; Ct.Samp1(80–180), forward, 5′-GCAGGATCCAATGTGCGACACCTG-3′ and reverse,
5′-CAGCTCGAG TTACCTCTTTCCCAG-3′; Ct.Samp1(75–135), forward,
5′-GGGGGATCCAATGAGCGAACGCATCTTT-3′and reverse,
5′-CAGCTCGAGTTACTTCGGCTCGCACTC-3′; Ct.Samp1(136–180), forward,
5′-GGGGGATCCAATGGTACGCGCTCGGC-3′ and reverse,
5′-CAGCTCGAGTTACCTCTTTCCCAG-3′; Ran, forward,
5′-GCAGGATCCATGGCTGCGCAG-3′ and reverse,
5′-CAGAAGCTTTTACAGGTCATCATCCTCATC-3′. Bacterial expression was performed as previously described in Jafferali et al.26
His-trap purification of Ran
One milliliter of cell lysis buffer (25 mM Hepes, pH 7.4, 100 mM NaCl, 1% Triton X-100 and protease inhibitor) (All chemicals were purchased from Sigma) was added to 5 × 109 bacterial cells expressing His6-Ran and incubated on ice for 45 min. The bacterial cells were sheared by brief sonication and soluble proteins were recovered in the supernatant following centrifugation at 18,000 × g for 30 min at 4°C. His-Trap Chelating HP column (GE Healthcare, #17-5248-02) was equilibrated with 10 column volumes of buffer A (50 mM Hepes, pH 7.4, 500 mM NaCl, 20 mM Imidazole) (All chemicals were purchased from Sigma). After loading the sample, the column was washed with 10 volumes of buffer B (50 mM Hepes, pH 7.4, 400 mM NaCl, 30 mM Imidazole) (All chemicals were purchased from Sigma). The protein was eluted from the column with 200 mM of Imidazole in buffer B. An overnight dialysis was performed to remove the excess Imidazole using dialysis buffer (25 mM Hepes, pH 7.4, 100 mM NaCl) (All chemicals were purchased from Sigma) and concentrated with a 10-kDa molecular weight cut-off Amicon Ultra-4 centrifugal filter devices (Millipore, #UFC901096).
Pulldown experiment
Bacterial cells (5 × 109) expressing different Samp1 fragments were treated with 1 ml of lysis buffer (100 mM Hepes, 200 mM NaCl, 1mg/ml lysozyme, 0.1% Triton X-100, 10% glycerol and protease inhibitor, pH 7.4) (All chemicals were purchased from Sigma) on ice for 45 min. The bacterial cells were sheared by brief sonication and soluble proteins were recovered in the supernatant following centrifugation at 15,500 × g for 30 min at 4°C. Fifty μl of settled strep-tactin agarose beads (iba, #2-1201-002) were blocked with 5% BSA in PBS. To the blocked beads, 25 μmol of bait protein or bacterial cell lysate containing 200 μg bait protein was added and the total volume was brought to 500 μl by addition of lysis buffer and incubated at 4°C for 1 h with end-over-end rotation. The unbound bacterial cell lysate was separated by centrifugation at 800 × g for 3 min. To the beads, 25 μmol of prey protein or bacterial cell lysate containing 200 μg prey protein was added along with lysis buffer as before and incubated at 4°C for 1 h with end-over-end rotation. The beads were washed 3x with wash buffer (100 mM Hepes, 500 mM NaCl, 0.1% Triton X-100, pH 7.4) (All chemicals were purchased from Sigma). The unbound lysates containing both bait and prey proteins were TCA precipitated. Equivalent amounts of bound, unbound and lysate fractions were loaded onto 10% SDS PAGE precast gels (Bio-Rad, #456–1094) and analyzed by Western Blotting using mouse monoclonal anti-His (1:2500) (ThermoFischer Scientific, # MA121315) antibody.
Western blot analysis
SDS PAGE separated proteins were transferred onto PVDF (Bio-Rad, #10600021) membranes and blocked with 5% milk in PBS-T (blocking solution) for 1 h at RT. The membranes were incubated with primary antibodies in the blocking solution for 1 h or overnight at 4°C for in vivo studies. After 3 × 10 min washes in PBS-T, the membranes were incubated with secondary antibody horseradish-peroxidase-coupled rabbit anti-goat IgG (Abcam, #ab6741) or horseradish-peroxidase-coupled donkey anti-mouse IgG (GE health care, #NA931) in the blocking solution for 1 h. After 4 × 10 min washes in PBS-T, the membranes were subjected to ECL detection (SuperSignal West Dura, ThermoFisher Scientific, #34075). The emitted chemiluminescent signal was imaged by ChemiDoc XRS+imaging system (Bio-Rad). The binding percentage was calculated using the following equation: (IP/IL) - (IP/IL)background; IP:band intensity of bound fraction; IL:band intensity of Lysate (input). The % binding was analyzed using the Prism 6 software. Three replicates were performed and statistically analyzed using Student's t-test.
Immunofluorescence
HeLa cells expressing Samp1-YFP were cultured on coverslips. Prior to fixation cells were pre-extracted in 37°C cytoskeleton buffer (CB: 60 mM PIPES, 27 mM HEPES, 10 mM EGTA, 4 mM MgSO4, pH 7.0) supplemented with 5 mM ATP, 1 mM GTP and 300 ng/ml saponin (All chemicals were purchased from Sigma) for 2 min. Cells were fixed using 3% paraformaldehyde in CB for 20 min at 37°C and permeabilized in 0.5% Triton X-100 in CB at RT. Followed by washing and blocking for 1 h in blocking solution (2% BSA, 0.05% Triton X-100 and Na Azide in CB) (All chemicals were purchased from Sigma). Samples were incubated with primary antibody for 1 h washed 6 × 2 min in blocking solution and incubated with secondary antibody and counterstained for 1 h followed by 6 × 2 min washes in CB prior to mounting in Fluoromount (Southern Biotech, #0100–01). For Ran anti-Ran antibody C-20 (1:500, Santa Cruz, #sc-1156) and secondary antibody anti-Goat Alexa568 antibody (1:2500, ThermoFischer Scientific, #A-11057) was used.
Samples were detected on a custom made spinning disk confocal, Zeiss Axiovert 200 body, Yokogawa CSU22 head, Hamamatsu Flash 4.0 Orca sCMOS camera, Chrysta Laser Nordic combiner with 488 nm, 568 nm and 632 nm solid state laser light source and Prior ProScan 2 filter wheel with Chroma dichroic filter sets. Powered by micro manager41 and customized arduino controllers.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Marry Dasso (NIHCD, Bethesda, USA) for sharing tsBN2 cells, Dr. Dan Sjostrand (DBB, Stockholm University, Stockholm, Sweden) for synthesis of strep-tag pET33B+ vector and Dr. Ian Macara (Vanderbilt University School of Medicine, Nashville, USA) for providing pKH3-Ran plasmid.
Funding
This work was made possible by grants from the Swedish Research Council #621-2010-448, Cancerfonden #110590 and the foundation Olle Engkvists minne.
References
- [1].Gerace L, Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol 1988; 4:335-74; PMID:2461721; http://dx.doi.org/ 10.1146/annurev.cb.04.110188.002003 [DOI] [PubMed] [Google Scholar]
- [2].Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; PMID:20300205; http://dx.doi.org/ 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci 2010; 123:1973-8; PMID:20519579; http://dx.doi.org/ 10.1242/jcs.019042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 2015; 208:11-22; PMID:25559183; http://dx.doi.org/ 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006; 172:41-53; PMID:16380439; http://dx.doi.org/ 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Korfali N, Wilkie GS, Swanson SK, Srsen V, de Las Heras J, Batrakou DG, Malik P, Zuleger N, Kerr ARW, Florens L, et al.. The nuclear envelope proteome differs notably between tissues. Nucl Austin Tex 2012; 3:552-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schirmer EC, Florens L, Guan T, Yates JR 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003; 301:1380-2; PMID:12958361; http://dx.doi.org/ 10.1126/science.1088176 [DOI] [PubMed] [Google Scholar]
- [8].Wilkie GS, Korfali N, Swanson SK, Malik P, Srsen V, Batrakou DG, de las Heras J, Zuleger N, Kerr ARW, Florens L, et al.. Several Novel Nuclear Envelope Transmembrane Proteins Identified in Skeletal Muscle Have Cytoskeletal Associations. Mol Cell Proteomics 2011; 1-16; PMID:20876400; http://dx.doi.org/ 10.1074/mcp.M110.003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell 2004; 16:319-30; PMID:15525506 [DOI] [PubMed] [Google Scholar]
- [10].Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci 1991; 88:10830-10834; PMID:1961752; http://dx.doi.org/ 10.1073/pnas.88.23.10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 2008; 9:464-77; PMID:18478030; http://dx.doi.org/ 10.1038/nrm2410 [DOI] [PubMed] [Google Scholar]
- [12].Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 1993; 365:661-3; PMID:8413630; http://dx.doi.org/ 10.1038/365661a0 [DOI] [PubMed] [Google Scholar]
- [13].Sazer S, Dasso M. The ran decathlon: multiple roles of Ran. J Cell Sci 2000; 113 (Pt 7):1111-8; PMID:10704362 [DOI] [PubMed] [Google Scholar]
- [14].Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 1999; 15:607-60; PMID:10611974; http://dx.doi.org/ 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- [15].Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol 1990; 111:309-321; PMID:2116418; http://dx.doi.org/ 10.1083/jcb.111.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 1996; 135:1457-1470; PMID:8978815; http://dx.doi.org/ 10.1083/jcb.135.6.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol 1993; 123:1649-1659; PMID:8276887; http://dx.doi.org/ 10.1083/jcb.123.6.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Forbes DJ, Travesa A, Nord MS, Bernis C. Reprint of “Nuclear transport factors: global regulation of mitosis.” Curr Opin Cell Biol 2015; 34:122-34; PMID:26196321; http://dx.doi.org/ 10.1016/j.ceb.2015.07.005 [DOI] [PubMed] [Google Scholar]
- [19].Joseph J. Ran at a glance. J Cell Sci 2006; 119:3481-4; PMID:16931595; http://dx.doi.org/ 10.1242/jcs.03071 [DOI] [PubMed] [Google Scholar]
- [20].Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22:832-53; PMID:18381888; http://dx.doi.org/ 10.1101/gad.1652708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Worman HJ. Nuclear lamins and laminopathies. J Pathol 2012; 226:316-25; PMID:21953297; http://dx.doi.org/ 10.1002/path.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Helbling-Leclerc A, Bonne G, Schwartz K. Emery-Dreifuss muscular dystrophy. Eur J Hum Genet EJHG 2002; 10:157-61; PMID:11973618; http://dx.doi.org/ 10.1038/sj.ejhg.5200744 [DOI] [PubMed] [Google Scholar]
- [23].Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci 2009; 122:2100-7; PMID:19494128; http://dx.doi.org/ 10.1242/jcs.047373 [DOI] [PubMed] [Google Scholar]
- [24].Gudise S, Figueroa RA, Lindberg R, Larsson V, Hallberg E. Samp1 is functionally associated with the LINC complex and A-type lamina networks. J Cell Sci 2011; 124:2077-85; PMID:21610090; http://dx.doi.org/ 10.1242/jcs.078923 [DOI] [PubMed] [Google Scholar]
- [25].Borrego-Pinto J, Jegou T, Osorio DS, Auradé F, Gorjánácz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci 2012; 125:1099-105; PMID:22349700; http://dx.doi.org/ 10.1242/jcs.087049 [DOI] [PubMed] [Google Scholar]
- [26].Jafferali MH, Vijayaraghavan B, Figueroa RA, Crafoord E, Gudise S, Larsson VJ, Hallberg E. MCLIP, an effective method to detect interactions of transmembrane proteins of the nuclear envelope in live cells. Biochim Biophys Acta BBA - Biomembr 2014; 1838:2399-403; http://dx.doi.org/ 10.1016/j.bbamem.2014.06.008 [DOI] [PubMed] [Google Scholar]
- [27].Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol 2007; 178:897-904; PMID:17785515; http://dx.doi.org/ 10.1083/jcb.200702026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 1996; 15:5584-94; PMID:8896452 [PMC free article] [PubMed] [Google Scholar]
- [29].Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol 1995; 257:135-44; PMID:8583915; http://dx.doi.org/ 10.1016/S0076-6879(95)57019-5 [DOI] [PubMed] [Google Scholar]
- [30].Khuperkar D, Helen M, Magre I, Joseph J. Inter-cellular transport of ran GTPase. PloS One 2015; 10:e0125506; PMID:25894517; http://dx.doi.org/ 10.1371/journal.pone.0125506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nishitani H, Ohtsubo M, Yamashita K, Iida H, Pines J, Yasudo H, Shibata Y, Hunter T, Nishimoto T. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J 1991; 10:1555-64; PMID:1851087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 1999; 399:230-7; PMID:10353245; http://dx.doi.org/ 10.1038/20375 [DOI] [PubMed] [Google Scholar]
- [33].Hayashi N, Yokoyama N, Seki T, Azuma Y, Ohba T, Nishimoto T. RanBP1, a Ras-like nuclear G protein binding to Ran/TC4, inhibits RCC1 via Ran/TC4. Mol Gen Genet MGG 1995; 247:661-9; PMID:7616957; http://dx.doi.org/ 10.1007/BF00290397 [DOI] [PubMed] [Google Scholar]
- [34].Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 2001; 153:1391-402; PMID:11425870; http://dx.doi.org/ 10.1083/jcb.153.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nakamura M, Masuda H, Horii J, Kuma K i, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J Cell Biol 1998; 143:1041-52; PMID:9817760; http://dx.doi.org/ 10.1083/jcb.143.4.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 1999; 18:1982-95; PMID:10202161; http://dx.doi.org/ 10.1093/emboj/18.7.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Percipalle P, Clarkson WD, Kent HM, Rhodes D, Stewart M. Molecular interactions between the importin alpha/beta heterodimer and proteins involved in vertebrate nuclear protein import. J Mol Biol 1997; 266:722-32; PMID:9102465; http://dx.doi.org/ 10.1006/jmbi.1996.0801 [DOI] [PubMed] [Google Scholar]
- [38].Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol CB 1998; 8:1403-6; PMID:9889103; http://dx.doi.org/ 10.1016/S0960-9822(98)00023-2 [DOI] [PubMed] [Google Scholar]
- [39].Yaseen NR, Blobel G. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin nup358. J Biol Chem 1999; 274:26493-502; PMID:10473610; http://dx.doi.org/ 10.1074/jbc.274.37.26493 [DOI] [PubMed] [Google Scholar]
- [40].Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 2005; 7:626-32; PMID:15908946; http://dx.doi.org/ 10.1038/ncb1263 [DOI] [PubMed] [Google Scholar]
- [41].Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods 2014; 1; e10; 1; PMID:25606571; http://dx.doi.org/ 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]


