ABSTRACT
In yeast, inducible genes such as INO1, PRM1 and HIS4 reposition from the nucleoplasm to nuclear periphery upon activation. This leads to a physical interaction with nuclear pore complex (NPC), interchromosomal clustering, and stronger transcription. Repositioning to the nuclear periphery is controlled by cis-acting transcription factor (TF) binding sites located within the promoters of these genes and the TFs that bind to them. Such elements are both necessary and sufficient to control positioning of genes to the nuclear periphery. We have identified 4 TFs capable of controlling the regulated positioning of genes to the nuclear periphery in budding yeast under different conditions: Put3, Cbf1, Gcn4 and Ste12. In each case, we have defined the molecular basis of regulated relocalization to the nuclear periphery. Put3- and Cbf1-mediated targeting to nuclear periphery is regulated through local recruitment of Rpd3(L) histone deacetylase complex by transcriptional repressors. Rpd3(L), through its histone deacetylase activity, prevents TF-mediated gene positioning by blocking TF binding. Many yeast transcriptional repressors were capable of blocking Put3-mediated recruitment; 11 of these required Rpd3. Thus, it is a general function of transcription repressors to regulate TF-mediated recruitment. However, Ste12 and Gcn4-mediated recruitment is regulated independently of Rpd3(L) and transcriptional repressors. Ste12-mediated recruitment is regulated by phosphorylation of an inhibitor called Dig2, and Gcn4-mediated gene targeting is up-regulated by increasing Gcn4 protein levels. The ability to control spatial position of genes in yeast represents a novel function for TFs and different regulatory strategies provide dynamic control of the yeast genome through different time scales.
Keywords: chromatin; histone deacetylase; interchromosomal clustering; lamina; nuclear periphery; nuclear pore complex; transcription, transcription factor; translational regulation; transcriptional repressor
Introduction
The eukaryotic genome is functionally and spatially organized. During interphase, chromosomes fold into topologically associated domains (TADs) and divide into heterochromatin and euchromatin. Also, chromosomes associate with nuclear structures and occupy distinct territories within the nucleus.1 Within these territories, individual genes are positioned with respect to each other and with respect to stable nuclear structures. For example, in metazoa large transcriptionally repressed Lamina Associate Domains (LADs) position along the nuclear periphery and interact with the nuclear lamina.2 Importantly, the spatial organization of the genome is dynamic and the position of individual genes often changes upon activation or repression.3 For example, during development, LADs can be remodeled to accommodate the repositioning of genes; the β-globin and MyoD genes move away from the nuclear lamina upon transcriptional activation.4,5 Many active genes also interact with nuclear pore proteins (Nups) in diverse organisms including yeast, flies, worms, and mammalian cells.6-10 In both yeast and metazoa these interactions positively correlate with transcription.6,8,11-14 In yeast these interactions occur at the nuclear periphery, presumably in contact with the nuclear pore complex (NPC),13 whereas in higher eukaryotes these interactions often occur in the nucleoplasm with soluble Nups.6,14 Finally, coregulated loci throughout the genome can cluster. Active genes colocalize with RNAP II foci in subnuclear compartments called transcription factories.15,16 In flies, Polycomb-repressed sites cluster together.17,18 Likewise, in yeast, tRNA genes, silenced telomeres and NPC-associated loci each exhibit specific interchromosomal clustering.19-21 These observations support the idea that the spatial organization of the eukaryotic genome compartmentalizes the nucleus into functionally distinct subnuclear environments and that the spatial positioning of a gene both impacts and reflects its transcriptional state.
Transcription factors reposition genes to the nuclear periphery
As a model for these phenomena, we have studied the spatial repositioning of inducible yeast genes from the nucleoplasm to the nuclear periphery. Inducible genes such as INO1, PRM1 and HIS4 are recruited from the nucleoplasm to the nuclear periphery upon activation.7 These genes are inducible under very different conditions (INO1 is activated by inositol starvation, PRM1 is induced by mating pheromone and HIS4 is induced by amino acid starvation) and they are targeted to the periphery only under the conditions that lead to their expression. This recruitment leads to a physical interaction with the nuclear pore complex (NPC) and promotes stronger expression.13,22 Targeting to the periphery is controlled by cis-acting transcription factor (TF) binding sites located within the promoters of these genes.13 These elements and the TFs that bind to them are not only necessary for recruitment, but are also sufficient to target an ectopic locus to the nuclear periphery.13 Furthermore, genes recruited to the NPC by the same TF frequently cluster together.21 Thus, these cis-acting binding sites function as DNA zip codes to control the dynamic, conditional positioning and interchromosomal clustering of genes. This suggests that one of the functions of TFs is to control the spatial organization of the genome.
Targeting of INO1 to the nuclear periphery is mediated by 2 DNA elements called GRS I and GRS II.13 The TFs Put3 (Zn+2-binuclear cluster family) and Cbf1 (basic helix-loop-helix family) bind to GRSI and GRSII, respectively and these TFs are necessary for GRS I- and GRS II-mediated targeting to the nuclear periphery.7 The GRS I element is necessary and sufficient to mediate interchromosomal clustering with other GRS I-containing loci.21 Importantly, these TFs are not the factors that control INO1 transcription. INO1 transcription is regulated by the Ino2/Ino4 TFs, which bind to the UASINO elements in the promoter.23 Such a separation between the elements controlling transcription and gene positioning is also seen in other promoters (our unpublished results). Thus, although gene positioning and transcription are coupled, they can be mediated by distinct elements and factors.
In contrast, the positioning of HIS4 and PRM1 is controlled by the same TFs that regulate their expression, Gcn4 and Ste12, respectively. Inserting DNA elements corresponding to the binding sites of Gcn4 (Gcn4BS) and Ste12 (pheromone-response elements, 3xPRE) at the ectopic site URA3, which is normally localized to the nucleoplasm, is sufficient to reposition the locus to the nuclear periphery.7 This also leads to interchromosomal clustering between URA3 and either HIS4 or PRM1. Furthermore, mutant strains lacking Gcn4 or Ste12 fail to target HIS4 or PRM1 to the nuclear periphery, respectively. Finally, artificially tethering Gcn4 or Ste12 to the URA3 locus via a LexA DNA binding domain is sufficient to cause URA3 to reposition to the nuclear periphery. Therefore, these TFs are both necessary and sufficient to control peripheral localization and interchromosomal clustering.
Put3, Cbf1, Gcn4 and Set12 represent 4 different families of transcription factors that mediate spatial repositioning and clustering of these genes. This suggest this ability is a common function of transcription factors. Indeed, in erythroid cells, the transcription factor Klf1 is necessary for clustering of its target genes into specialized transcription factories and in flies, interactions of genes with Nup98 is mediated by the MBD-R2 DNA binding factor.16,24 However, not all transcription factors possess this function. As mentioned above, Ino2/Ino4 binding to the UASINO element within the INO1 promoter is neither necessary nor sufficient to recruit chromatin to the nuclear periphery. Transcription is also separable from gene positioning. The activation domain of Put3 and Gcn4 is dispensable for targeting to the nuclear periphery (our unpublished results). Furthermore, inactivating RNA polymerase II or promoter mutations that block INO1 transcription does not block zip code-dependent recruitment to the nuclear periphery.7,12 Thus, TFs can control gene positioning, separable from their effects on transcription. One important, unaddressed question is how these changes in gene positioning are dynamically regulated. We have explored this question in work described in our recent publication.7
Regulation of transcription factor-mediated gene positioning
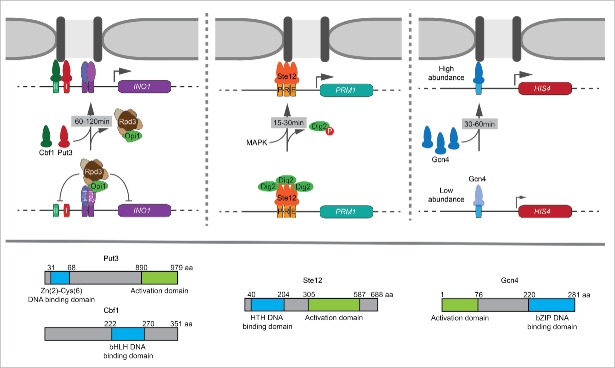
Repositioning of INO1, HIS4 and PRM1 to the nuclear periphery and interchromosomal clustering is conditional and occurs under specific environmental stimuli. This reflects how each zip code is regulated, which is revealed when the zip code is inserted at an ectopic site. GRSI and GRSII are regulated through a context-dependent mechanism: when inserted at an ectopic site, these elements lead to constitutive targeting to the nuclear periphery.13 In other words, they are negatively regulated in the context of the INO1 promoter and only permitted to function when INO1 is induced. Using systematic mutagenesis of cis and trans acting regulators, we find that targeting of INO1 to the nuclear periphery by Put3 and Cbf1 is regulated through local recruitment of Rpd3(L) histone deacetylase complex (Fig. 1).7 Rpd3(L) is recruited to the INO1 promoter under repressing conditions by the transcriptional repressors Opi1 and Ume6.25,26 Rpd3(L) regulates zip code activity by blocking transcription factor binding through its histone deacetylase activity.7 This regulation was abolished by either perturbing the recruitment of Rpd3 or inactivating its catalytic activity (rpd3 H188A), resulting in constitutive targeting of INO1 to the nuclear periphery. Furthermore, Rpd3(L) regulation could be recapitulated at the ectopic URA3 locus by artificially tethering either Opi1 or Ume6 near GRS I or GRS II, blocking targeting to the nuclear periphery and interchromosomal clustering (Fig. 1, left panel).7 To test if this was general function of repressors in yeast, 21 different repressors were tethered beside the GRS I. A majority of repressors could block GRS I function; 16 of the 21 repressors tested blocked Put3-mediated recruitment to the nuclear periphery and 11 of these required Rpd3. Thus, 5 repressors block Put3 binding by either recruiting a different histone deacetylase or through alternative mechanisms. These results suggest that Put3s ability to reposition genes can be blocked through several different mechanisms. Because Put3 controls the positioning of genes that are not perfectly co-regulated,13 cells could use these different strategies to independently regulate Put3-mediated targeting in different contexts, depending on the environmental stimulus.
Figure 1.
Multiple strategies in regulating transcription factor (TF)-mediated recruitment to the nuclear periphery over different time scales. Top: (Left) Put3 and Cbf1 bind to GRSI and GRSII respectively in the promoter of INO1 and mediate its recruitment to the nuclear periphery upon inositol starvation. These TFs are regulated by the local recruitment of Rpd3(L) histone deacetylase by transcriptional repressors Opi1 and Ume6. Repression is relieved between 60–120 min leading to peripheral localization and interchromosomal clustering of INO1. (Middle) Ste12 mediates the recruitment of PRM1 to the nuclear periphery upon mating pheromone stimulation. Ste12 is regulated downstream of DNA binding by MAPK phosphorylation of the inhibitor Dig2. Phosphorylation of Dig2 and the recruitment of PRM1 occurs rapidly between 15–30 min. (Right) Gcn4-mediated recruitment of HIS4 to the nuclear periphery is controlled by Gcn4 abundance. Gcn4 is translationally regulated. Through increased protein levels of Gcn4, maximal peripheral targeting occurs between 30–60 min. Bottom: Graphical depiction of TF domains. Put3 is a Zn+2 – binuclear cluster TF. Cbf1 is a basic helix-loop-helix TF. Ste12 is a helix-turn-helix TF. Gcn4 is a basic leucine zipper TF.
Cells use different strategies to regulate Gcn4- and Ste12-mediated gene positioning. Unlike Put3, regulation of Ste12- and Gcn4-mediated repositioning is context-independent; inserting the Gcn4BS or the 3xPRE at URA3 led to repositioning to the nuclear periphery upon histidine starvation or pheromone treatment, respectively.7 Furthermore, loss of Rpd3 had no effect on recruitment of PRM1 or HIS4 to the nuclear periphery and the 3xPRE was completely resistant to tethering of Opi1 or Ume6.7 Instead, Ste12-mediated gene positioning is regulated downstream of DNA binding. At PRM1, Ste12 is constitutively bound and Ste12-dependent transcription is inhibited by 2 repressors, Dig1 and Dig2.27 Upon mating pheromone stimulation, Dig1 and Dig2 are phosphorylated by the MAPK Fus3, causing them to dissociate from Ste12.28 Dissociation of both Dig1 and Dig2 is required for PRM1 transcriptional activation, but loss of Dig2 alone led to constitutive Ste12-mediated peripheral localization and interchromosomal clustering.7,29 Furthermore, mutation of serine 34 to alanine in Dig2 - blocking phosphorylation - also blocked targeting to the nuclear periphery. Likewise, mutation of serine 34 to aspartate - mimicking phosphorylation - led to constitutive targeting to the nuclear periphery. Thus, Ste12-mediated gene positioning is regulated through post-translational modification of an inhibitor (Fig. 1, middle panel).
Gcn4-mediated targeting to the nuclear periphery is regulated by a third mechanism; the occupancy of Gcn4 binding to its target genes, controlled by its abundance.7 Unlike the other mechanisms discussed above, which behave in a switch-like manner, Gcn4-mediated targeting to the nuclear periphery and interchomosomal clustering occurs at a lower level under uninducing conditions and at a maximal level under inducing conditions.7 Thus, Gcn4-mediated gene positioning is quantitatively regulated. It is well-established that Gcn4-mediated transcription is regulated through the abundance of Gcn4. In the presence of amino acids, Gcn4 is poorly translated due to several short, upstream open reading frames (uORFs) in the 5′ end of the GCN4 mRNA.30 When cells are starved for amino acids, translational initiation is slowed, permitting the ribosome to skip the uORFs and increase the translation of Gcn4.30 Increasing Gcn4 protein production by mutating the uORFs resulted in constitutive recruitment and clustering of HIS4.7 Thus, under uninducing conditions, low levels of Gcn4 lead to modest targeting to the nuclear periphery and measurable interchromosomal clustering. However, under inducing conditions, higher Gcn4 protein levels increase the occupancy of the Gcn4BS and this leads to maximal peripheral targeting and interchromosomal clustering (Fig. 1, right panel).
Using a small number of genes as models, a number of factors have been implicated in the targeting of genes to the NPC, including mRNA export factors, the SAGA histone acetyltransferase and components of the NPC itself.11,13,31,32 It is unclear if all of these are universally required. We find that the SAGA complex, for example, is required for targeting of INO1 and HIS4 to the nuclear periphery, but is dispensable for targeting of PRM1 to the nuclear periphery.7,13 If SAGA functions to bridge the interaction between chromatin and the NPC, as has been suggested, this result indicates that transcription factors use more than one mechanism to promote repositioning to the nuclear periphery. Alternatively, this dependence on SAGA for gene recruitment may reflect an upstream role. The SAGA complex is a multi-subunit histone modifying enzyme that has broad impacts on gene expression.33 Because the regulation of Put3, Cbf1 and Gcn4 occurs at the level of TF occupancy whereas Ste12 is regulated downstream of binding, it is possible that SAGA may either facilitate TF DNA binding or alter the abundance of the TFs themselves.
These results highlight the critical role of TFs in controlling gene positioning and interchromosomal interactions. TF-mediated gene positioning can be regulated through at least 4 different mechanisms: regulation of TF binding by the Rpd3(L) HDAC, regulation of TF binding (or function) by repressors independent of Rpd3(L), regulation of TF occupancy through changes in TF abundance and regulation of TF function through post-translational modification of an inhibitor. These different strategies operate over different time scales to alter the positioning of individual genes and the arrangement of chromosomes with respect to each other: MAPK signaling leads to rapid repositioning and clustering of Ste12 targets within ∼15–30 min, while changes in Gcn4 protein levels leads to a moderate rate of repositioning and clustering within ∼30–60 min and Rpd3(L) regulated gene positioning is derepressed over ∼60–120 min (Fig. 1).7 Thus, reflecting the regulation of TF function, cells employ different strategies to regulate dynamic TF-mediated gene positioning over very different temporal regimes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors acknowledge members of the Brickner lab, past and present, for contributing to the work described and for helpful discussions.
Funding
The work discussed in this Extra View was supported by NIH R01 GM080484. JHB is the Soretta and Henry Shapiro Research Professor in Molecular Biology.
References
- [1].Hubner MR, Eckersley-Maslin MA, Spector DL. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev 2013; 23:89-95; PMID:23270812; http://dx.doi.org/ 10.1016/j.gde.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948-51; PMID:18463634; http://dx.doi.org/ 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- [3].Egecioglu D, Brickner JH. Gene positioning and expression. Curr Opin Cell Biol 2011; 23:338-45; PMID:21292462; http://dx.doi.org/ 10.1016/j.ceb.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lee H, Quinn JC, Prasanth KV, Swiss VA, Economides KD, Camacho MM, Spector DL, Abate-Shen C. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev 2006; 20:784-94; PMID:16600910; http://dx.doi.org/ 10.1101/gad.1392006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 2006; 20:1447-57; PMID:16705039; http://dx.doi.org/ 10.1101/gad.1419506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010; 140:360-71; PMID:20144760; http://dx.doi.org/ 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- [7].Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Garvey Brickner D, Ahmed S, Watchmaker L, Brickner JH. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biol 2016; 212:633-46; PMID:26953353; http://dx.doi.org/ 10.1083/jcb.201508068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2004; 2:e342; PMID:15455074; http://dx.doi.org/ 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117:427-39; PMID:15137937; http://dx.doi.org/ 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- [10].Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol 2013; 200:589-604; PMID:23460676; http://dx.doi.org/ 10.1083/jcb.201207024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006; 441:774-8; PMID:16760983; http://dx.doi.org/ 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- [12].Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 2007; 5:e81; PMID:17373856; http://dx.doi.org/ 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 2010; 12:111-8; PMID:20098417; http://dx.doi.org/ 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372-83; PMID:20144761; http://dx.doi.org/ 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 2006; 172:177-87; PMID:16418531; http://dx.doi.org/ 10.1083/jcb.200507073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al.. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 2010; 42:53-61; PMID:20010836; http://dx.doi.org/ 10.1038/ng.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet 2011; 7:e1001343; PMID:21455484; http://dx.doi.org/ 10.1371/journal.pgen.1001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011; 144:214-26; PMID:21241892; http://dx.doi.org/ 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- [19].Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science 2003; 302:1399-401; PMID:14631041; http://dx.doi.org/ 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 1991; 66:1279-87; PMID:1913809; http://dx.doi.org/ 10.1016/0092-8674(91)90049-5 [DOI] [PubMed] [Google Scholar]
- [21].Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 2012; 22:1234-46; PMID:22579222; http://dx.doi.org/ 10.1016/j.devcel.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ahmed S, Brickner JH. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet 2007; 23:396-402; PMID:17566592; http://dx.doi.org/ 10.1016/j.tig.2007.05.009 [DOI] [PubMed] [Google Scholar]
- [23].Graves JA, Henry SA. Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics 2000; 154:1485-95; PMID:10747047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pascual-Garcia P, Jeong J, Capelson M. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep 2014; 9:433-42; PMID:25310983; http://dx.doi.org/ 10.1016/j.celrep.2014.09.002 [DOI] [PubMed] [Google Scholar]
- [25].Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 1998; 392:831-5; PMID:9572144; http://dx.doi.org/ 10.1038/33952 [DOI] [PubMed] [Google Scholar]
- [26].Heyken WT, Repenning A, Kumme J, Schuller HJ. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol Microbiol 2005; 56:696-707; PMID:15819625; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04499.x [DOI] [PubMed] [Google Scholar]
- [27].Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature 2010; 464:1187-91; PMID:20237471; http://dx.doi.org/ 10.1038/nature08934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol 1997; 7:228-38; PMID:9094309; http://dx.doi.org/ 10.1016/S0960-9822(06)00118-7 [DOI] [PubMed] [Google Scholar]
- [29].Olson KA, Nelson C, Tai G, Hung W, Yong C, Astell C, Sadowski I. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol Cell Biol 2000; 20:4199-209; PMID:10825185; http://dx.doi.org/ 10.1128/MCB.20.12.4199-4209.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 2005; 59:407-50; PMID:16153175; http://dx.doi.org/ 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- [31].Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 2007; 282:3042-9; PMID:17158105; http://dx.doi.org/ 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- [32].Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet 2007; 8:507-17; PMID:17549064; http://dx.doi.org/ 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- [33].Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 2007; 26:5329-40; PMID:17694076; http://dx.doi.org/ 10.1038/sj.onc.1210603 [DOI] [PMC free article] [PubMed] [Google Scholar]