Abstract
Centronuclear myopathies (CNMs) are genetic diseases whose symptoms are muscle weakness and atrophy (wasting) and centralised nuclei. Recent human genetic studies have isolated several groups of mutations. Among them, many are found in two interacting proteins essential to clathrin-mediated endocytosis, dynamin and the BIN-Amphiphysin-Rvs (BAR) protein BIN1/amphiphysin 2. In this review, by using structural and functional data from the study of endocytosis mainly, we discuss how the CNM mutations could affect the structure and the function of these ubiquitous proteins and cause the muscle-specific phenotype. The literature shows that both proteins are involved in the plasma membrane tubulation required for T-tubule biogenesis. However, this system also requires the regulation of the dynamin-mediated membrane fission, and the formation of a stable protein-scaffold to maintain the T-tubule structure. We discuss how the specific functions, isoforms and partners (myotubularin in particular) of these two proteins can lead to the establishment of muscle-specific features.
Keywords: Centranuclear myopathy, Clathrin-mediated endocytosis, T-tubule, BIN1, Dynamin, BAR domain, Membrane fission, Myotubularin, Muscle, Structure
1. Introduction
Muscles are essential to animal life. Among the different types of muscles, skeletal muscles are involved in coordinated movement, such as walk. Brain neuronal influx orchestrates the contraction and relaxation cycles of the many muscles involved in a specific move. The importance of the correct transduction of such signals within muscle cells is revealed in human diseases where it is impaired. One of these is a group of hereditary diseases with several genetic determinants called centronuclear myopathies (CNMs). The onset, severity and progressiveness vary a lot between different CNM subtypes, but the common symptoms are muscle weakness and the histological phenotype that cell nuclei are abnormally located in the centre of the muscle fibres instead of at the periphery, just below the plasma membrane, giving its name to the disease. This histological phenotype may be a signature of muscle fibres not being fully differentiated. Central nuclei are present in many forms of myopathies, as a consequence of excessive muscle fibre regeneration, due to fibre loss. However, in CNM, no excessive regeneration occurs, and the central position of nuclei is thought to arise from a block in muscle fibre development. However, it could also be that the mutation interferes with the process that locates the nuclei at the periphery, without affecting the muscle functionality.
Almost a decade ago, the first cases of CNM patients with a mutation in the BAR domain (Bin-Amphiphysin-Rvs) protein BIN1 and in dynamin 2 were reported (Bitoun et al., 2005, Nicot et al., 2007). These proteins are ubiquitously expressed and important players of clathrin-mediated endocytosis, where they have a membrane remodelling activity. The aim of this review is to describe how the properties and functions of these proteins discovered in endocytosis can help in understanding the muscle-specific function of these proteins, and how mutations causing CNM would trigger the symptoms.
2. BIN1 in CNM
2.1. BIN1, a crescent-shaped protein able to tubulate membranes
BIN1, a close homologue of the endocytic protein amphiphysin 1 (54% identity) and therefore also called amphiphysin 2, was initially identified as the box-dependent myc-interacting protein 1 (Sakamuro et al., 1996) able to suppress tumours. Quickly after its discovery, it was observed to be located in the proximity of T-tubules in skeletal muscles (Butler et al., 1997). T-tubules are long tubular invaginations from the myocyte plasma membrane into the actin fibre organization. They are parallel to the Z-lines of sarcomeres and allow the propagation of the myocyte plasma membrane depolarisation, triggered by the neuronal action potential. Once in the T-tubules, the depolarization induces the release of calcium from sarcoplasmic membranes adjacent to T-tubules. The calcium further induces myosin activity, and muscle contraction.
BIN1 was first proposed to play a role in muscle cell differentiation based on the results that myoblasts had difficulties to differentiate into myotubes when BIN1 was downregulated (Wechsler-Reya et al., 1998). But it was intriguing to find such similarities between amphiphysin 1 in clathrin-mediated endocytosis (CME) and BIN1 in the T-tubule system. In Drosophila, the single amphiphysin gene accounts for both the endocytic function of human amphiphysin 1 and the muscular function of BIN1, as it is located at both endocytic pits and in the proximity of tubular structures resembling T-tubules in flies (Razzaq et al., 2001). Strikingly, the homozygous null mutant was viable and did not have a significant phenotype in endocytosis of synaptic vesicles, but had a severe phenotype in the muscle contraction and was therefore flightless. Confocal sections revealed that the T-tubule system was present but highly disorganised in the fly muscle, suggesting that the Drosophila amphiphysin/BIN1 homolog participates to the T-tubule biogenesis.
BIN1 belongs to the BAR protein family (BIN1/Amphiphysin/Rvs167; Sakamuro et al., 1996), a protein family which shares the BAR domain. The BAR domain is the lipid membrane binding domain, and forms dimers of the shape of a crescent (Peter et al., 2004). The BAR domain dimer binds to negatively charged lipids via its positively charged concave face (Casal et al., 2006, Peter et al., 2004). Because of its crescent shape, it was shown to bind membranes in a curvature-dependent manner, and is thus considered as a membrane-curvature sensing module (Antonny, 2011). BIN1 further belongs to the subclass of N-BAR proteins as it also contains an N-terminal amphipathic helix. Amphipathic helices are sequences of 15–30 amino acids in which, once folded into an α-helix, the hydrophobic residues are positioned on one face and the hydrophilic residues on the opposite face of the helix (see also Fig.2A; Segrest et al., 1974). They are unfolded in solution and fold into an α-helix while binding to the lipids (velcro model, Antonny, 2011). Point mutations of hydrophobic residues within the amphipathic helices strongly affect liposome binding and tubulation of the two N-BAR proteins endophilin and amphiphysin 1 (Farsad et al., 2001). Thus, these amphipathic helices are often considered as curvature-inducer modules. But amphipathic helices are also proposed to sense curvature and other intra-membrane stresses (Campelo and Kozlov, 2014) as well as to be involved in membrane fission (Boucrot et al., 2012). The structure of BIN1’s N-terminal amphipathic helix in micelles was solved by NMR by Löw et al. (2008) and shows that roughly 20 residues belong to BIN1’s N-terminal amphipathic helix. Thus, the BAR domain of BIN1 has structural features, like other N-BAR domains, with curvature sensing, and curvature-inducing capabilities. As it was shown that the balance between those two activities strongly depends on the BAR domain density on the membrane (Sorre et al., 2012), one can wonder whether the BAR domain of BIN1 is a curvature sensor or a curvature inducer in the physiological context of the T-tubule biogenesis.
Fig. 2.
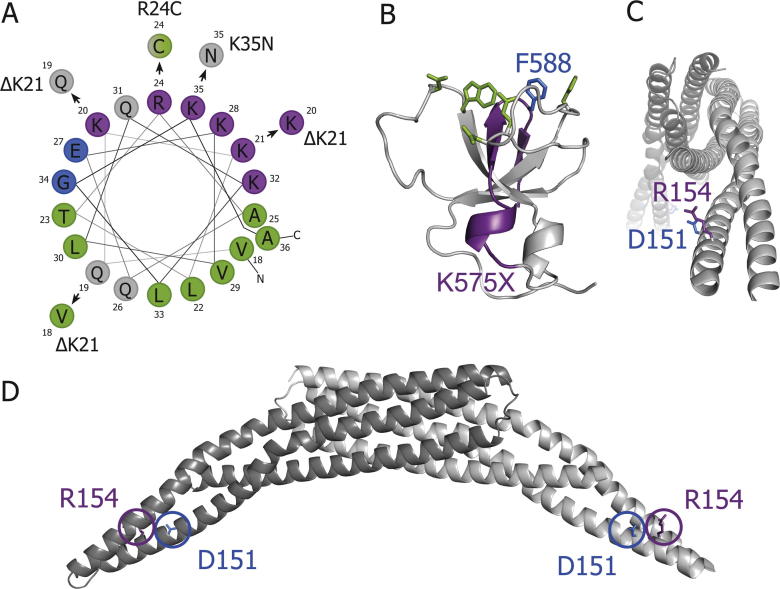
Structural consequences of CNM-linked mutations in BIN1. A. Helical wheel projection of residues 18-36 (amphipathic helix) from human BIN1 and likely new positioning due to ΔK21, R24C and K35N. B. Affected residues of truncation of SH3 domain. Lacking parts in human K575X mutant in violet; human F588 (blue) is part of dynamin binding face (green); crystal structure from rat BIN1 (Owen et al., 1998). Charged residues are marked in violet (positive) and blue (negative), hydrophobic residues in green and polar uncharged residues in grey. C, D. D151 and R154 are on the tip of the BIN1 BAR domain dimer and point sideways. Visualisation of a BAR domain dimer from the front, with each monomer in a different grey shade (C). Side view (D).
Another domain present in all BIN1 isoforms is an Src homology 3 (SH3) domain which interacts with several Proline-Rich Domains (PRDs), including the one of dynamin (Grabs et al., 1997), another protein mutated in several CNM-patients (see next part). This domain is thus expected to play a major role in the combined action of BIN1 and dynamin in T-tubule biogenesis (see second part).
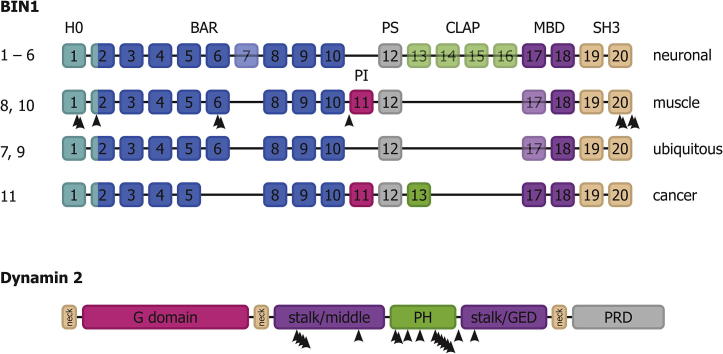
BIN1 has two muscle-specific isoforms (including vs. excluding exon 17), which contain peptide sequences with muscle-specific properties. BIN1 has 20 exons, which are differentially spliced into 11 isoforms (Uniprot ID O00499, UGID 160989; Fig. 1). BIN1 amino acid numbers refer to the position in the canonical isoform 1 in this review. In these muscle-specific isoforms, the clathrin-AP2 (CLAP) binding motif, which targets BIN1 to clathrin-coated pits, is not present, arguing for a role independent of CME in the muscle (Ramjaun and McPherson, 1998). Moreover, only the muscle specific isoforms of BIN1, sometimes referred to as M-Amph2, contain translated exon 11. Exon 11 was found to specifically bind to PI(4,5)P2 and PI(4)P (Lee et al., 2002) and therefore named phosphoinositide (PI) domain. It should be noted that exon 11 was previously named exon 10 (e.g. Lee et al., 2002, Wechsler-Reya et al., 1997) because an additional exon in the BAR domain had been overlooked (current name exon 7 (NCBI), exon a–b from Tsutsui et al., 1997). The overexpression of BIN1 isoform 8, which contains the PI motif, was found to induce membrane tubulation in several cell types (Nicot et al., 2007, Lee et al., 2002), while this was not observed for the neuronal isoform 1 which does not contain the PI motif. This led to the proposition that the PI motif is required for the tubulation activity of BIN1, suggesting a role in the formation of T-tubules. Lee et al. (2002) proposed that PI(4,5)P2, whose concentration increases during myoblast-to-myotube differentiation and is enriched in T-tubules, recruits BIN1 to PI(4,5)P2-rich plasma membrane compartments, leading to membrane tubulation and T-tubule formation. The affinities for the different phosphoinositides PI(3)P, PI(4)P, PI(5)P and PI(4,5)P2 is still a matter of debate (Fugier et al., 2011, Picas et al., 2014), but the PI motif of BIN1 is needed for correct muscle function and localisation of BIN1 in the muscle (Böhm et al., 2013). The role of the PI motif would thus be to locally increase BIN1 protein density onto the membrane in a PIP-dependent manner, promoting curvature induction towards curvature-sensing functions, as shown by Sorre et al. (2012) for amphiphysin 1. In this case, BIN1 would actively participate in the tubulation of the plasma membrane into a T-tubule system. This scenario is consistent with the fact that the T-tubule system is disrupted in patients with mutations within the PI motif (see below).
Fig. 1.
BIN1 and dynamin 2 domain structure. BIN1, upper panel. Dynamin 2, lower panel. Arrowheads indicate positions of CNM-linked mutations. For BIN1, isoform numbers are indicated on the left, the tissues in which the isoforms are expressed on the right. Semi-transparency of exons indicates alternative splicing within the different isoforms in this tissue.
The PI motif was not only found to bind to phosphoinositides but also to the BIN1 SH3 domain (Kojima et al., 2004). Exchanging the PI motif in BIN1 by a different phosphoinositide binding domain without affinity for SH3 domains (PH domain of PLCδ) resulted in abnormal morphology of nascent T-tubules (Kojima et al., 2004). This result could be explained by competitive interactions between PIPs, PI, SH3 and dynamin PRD domains. Wu and Baumgart (2014) determined binding affinities for these different interactions and came to the conclusion that the affinity of the SH3 domain is higher for its own PI motif than for dynamin’s PRD domain (Kd 10 μM and 66 μM, respectively). They also found that the Kd of the PI motif for PI(4,5)P2 was too high to be measured by isothermal titration calorimetry (ITC), but high PRD domain concentrations increased BIN1 membrane binding, suggesting that the SH3 domain is released from the PI motif when the SH3 domain is bound to dynamin’s PRD domain PI motif. Moreover, the Kd values for full-length BIN1 binding to PIPs are very low (all below 2 μM; Fugier et al., 2011), making it the strongest interaction of all three. Thus a possible scenario could be that the phosphoinositide binding of the full-length BIN1 would cause the release of the SH3/PI domain interaction, allowing the SH3 domain to bind to dynamin’s PRD domain, and recruiting dynamin to T-tubules (Lee et al., 2002).
All these domains have mutations causing various forms of CNM. In the following, we review these mutations, and try to pinpoint the specific function disabled in each of these, and discuss the role of BIN1 in the specific context of the muscle.
2.2. BIN1 mutations found in CNM patients
Based on the observation that the Drosophila Amphiphysin/BIN1 null mutant led to a disorganised T-tubule system, Razzaq et al. (2001) proposed that BIN1 mutations might be responsible for myopathies. In 2007, such mutations were indeed reported (Nicot et al., 2007). The authors analysed a group of CNM patients with consanguineous parents, therefore potentially having homozygous mutations. The MTM1 gene, coding for myotubularin, a lipid phosphatase involved in the production of a precursor of PI(4,5)P2 and already shown to cause CNM when mutated, was not mutated in those patients. The authors sequenced BIN1 due to the known link of BIN1 with muscles and PIPs and identified three mutations that were segregating, when homozygous, with the CNM symptoms. Since then, seven further BIN1 mutations were identified (Böhm et al., 2010, Böhm et al., 2013, Böhm et al., 2014, Claeys et al., 2010), out of which four caused a mild version of CNM already when heterozygous, leading to ten currently known CNM-linked BIN1 mutations (see Table 1).
Table 1.
CNM mutations in BIN1.
| Mutation | Exon | Domain | Dominance | Severity | Disease onset | BIN1 phenotype | Reference of discovery |
|---|---|---|---|---|---|---|---|
| ΔK21 | 1 | H0 | Dominant | Mild | Adulthood | Tubulation defect (in vivo) | Böhm et al. (2014) |
| R24C | 1 | H0 | Dominant | Mild | Adulthood | Tubulation defect (in vivo) | Böhm et al. (2014) |
| K35N | 2 | H0 | Recessive | Variable | Neonatal | Tubulation defect (only in vivo) | Nicot et al. (2007) |
| D151N | 6 | BAR | Recessive | Mild | Childhood | Tubulation defect (in vivo/in vitro) | Nicot et al. (2007) |
| R154Q | 6 | BAR | Recessive | Intermediate | Childhood | Tubulation defect (in vivo/in vitro) | Claeys et al. (2010) |
| IVS10-1G > A | 11 | PI | Recessive | Variable | Childhood | Tubulation defect (in vivo/in vitro) | Böhm et al. (2013) |
| Q573X | 20 | SH3 | Recessive | Intermediate | Neonatal | [Not tested] | Böhm et al. (2010) |
| K575X | 20 | SH3 | Recessive | Intermediate | Neonatal or childhood | Dynamin binding defect | Nicot et al. (2007) |
| P593HfsX54 | 20 | SH3 | Dominant | Mild | Adulthood | [Not tested] | Böhm et al. (2014) |
| X594DfsX53 | 20 | SH3 | Dominant | Mild | Adulthood | [Not tested] | Böhm et al. (2014) |
The mutations locate to the BAR domain, its N-terminal amphipathic helix, the PI motif and the C-terminal SH3 domain (see Fig. 1). Mutations in the PI motif are expected to have a muscle-specific phenotype, since only muscle isoforms of BIN1 contain this domain. But it is interesting to note that most of the found CNM-linked mutations affect the N-BAR and SH3 domains that are present in all isoforms of BIN1, leading to the question of how such mutations could lead to a muscle-specific disease.
2.3. CNM-linked mutations in the N-terminal amphipathic helix of BIN1 could decrease the stability of the BIN1 scaffold
In the following, we are going through the different BIN1 mutations in more detail. Three mutations (ΔK21, R24C, K35N; Böhm et al., 2014, Nicot et al., 2007) locate to the amphipathic helix at the N-terminus of the BAR domain of BIN1. In the R24C and K35N mutations, positively charged residues are replaced by hydrophobic or polar uncharged residues. In the ΔK21 mutation, three nucleotides are deleted, leading to a deletion of positively charged lysine 21 without frame shift and a shift in the amino acid sequence on the helical wheel (see Fig.2A). When K21 is absent, the original lysine at this helical wheel position should be unaffected because lysine 20 will likely take over this position. In contrast, the polar uncharged amino acid Q19 should take over the position of the charged K20, meaning that this should be the actually affected helical wheel position of the ΔK21 deletion.
The three residues affected by the above mutations are all in the polar face of the helical wheel and very close to each other. These mutations are thus expected to strongly interfere with the structure of the amphipathic helix (see Fig.2A), and thus with its ability to bind and cause membrane curvature. In the three mutations found in the amphipathic helix of BIN1, positively charged amino acid positions in the wheel projection are replaced by uncharged amino acids, while the hydrophobic face of the helical wheel remains almost unaffected. This underlines the importance of the positively charged residues 20, 24 and 35, which interact with negatively charged lipids of the plasma membrane to trigger proper helix folding into the membrane (Antonny, 2011).
For these three mutations, BIN1’s tubulating activity upon overexpression was decreased in cells (Böhm et al., 2014, Nicot et al., 2007, Wu et al., 2014). No in vitro tubulation data were available for the ΔK21 and R24C mutants but one study tested the K35N mutant’s tubulating activity in vitro and did not observe significant differences to the WT (Wu et al., 2014). However, they observed decreased membrane binding of the K35N BIN1 N-BAR compared to WT. One might speculate that the K35N mutant has a defect in membrane tubulation because of reduced membrane binding, but in vitro, where high protein concentrations are used, the membrane is probably saturated with protein regardless of its binding efficiency. Consistently, ΔK21 and R24C cause CNM already in the heterozygous state, while K35N only when homozygous. The reduced tubulation activity of K35N-BIN1 could be compensated by co-overexpressing wild-type BIN1, while this was not the case for the ΔK21 and R24C mutants (Böhm et al., 2014), confirming the dominant effect of the latter two mutations.
But the deleterious effect of these mutants may come from another reason. Mim et al. (2012) found that the N-terminal amphipathic helix of endophilin, another N-BAR protein, forms homo-interactions between helix rungs, and that these interactions are crucial for the formation of the N-BAR lattice on a membrane tube. A similar observation was recently made for the BIN1 lattice (Adam et al., 2015). Thus, the deleterious effect on T-tubule biogenesis of CNM-linked BIN1 mutations in the amphipathic helix might be linked to its role in the assembly of the BIN1 scaffold.
The possible configurations of interacting amphipathic helices in the helical scaffold had two energetically favourable positions (Mim et al., 2012). In one case, the two helices interacted over the whole length while in the other they were slightly shifted by about a third of their length. In this shifted position, the C-terminal residues of the amphipathic helix should then be less crucial for the polymerization of BIN1 as they would not participate in the rung interactions. This could explain why ΔK21 and R24C cause CNM already in the heterozygous state while adults carrying a heterozygous K35N mutation had no symptoms, as K35 is one of the last residues of the amphipathic helix (Löw et al., 2008). It should be noted that the few adults with known heterozygous K35N mutation (Nicot et al., 2007) were parents of young children and therefore one cannot exclude that they also develop(ed) symptoms of heterozygous CNM later in their life, after publication of the study.
2.4. CNM-linked mutations in the BAR domain of BIN1 decrease its tubulation property
Two recessive mutations causing CNM were found in the BAR domain itself (Claeys et al., 2010, Nicot et al., 2007). In both mutations (D151N, R154Q), single charged amino acids are replaced by polar uncharged ones. From their position in the concave face of the BAR dimer (Fig.2C and D), it is expected that the mutated residues participate in membrane binding. Indeed, the position of the two mutations is at tips of the BIN1 dimer, and a cryo EM structure of BIN1 bound to membrane tubes shows that the tip regions of the BIN1 BAR are deeply inserted into the membrane (Adam et al., 2015). This would fit with the sideways/upwards facing residues affected in these mutants. Also, heterodimers of WT and D151N (or R154Q) could form, explaining the recessive character of these mutations.
Surprisingly, both mutations affect exon 6, which is present in all BIN1 isoforms. All BIN1 isoforms should therefore be affected, and not only the muscle-specific isoform 8. Thus, the observed tissue-specificity of the mutant phenotypes of D151N and R154Q must arise from a tissue-specific function or a tissue-specific set of partners. In contrast to the K35N (N-terminal amphipathic helix) mutant, the decrease in membrane tubulation in cells was also observed in vitro (Picas et al., 2014, Wu et al., 2014). Membrane binding of the D151N and R154Q mutants was decreased (Wu et al., 2014) and their localisation was different from WT BIN1 in the muscle (Böhm et al., 2013, Toussaint et al., 2011). The onset of CNM in patients with D151N or R154Q BIN1 is later than in those having the K35N mutation (childhood vs. embryonic onset). Furthermore, K35N-CNM is more severe (Claeys et al., 2010, Nicot et al., 2007), suggesting that the amphipathic helix function might be more important for BIN1’s muscular function than the one changed in the D151N and R154Q BIN1 mutants. The fact that K35N-CNM is more severe may account for another function of the amphipathic helix than simply the proposed tubulation property (observed in cells), as the D151N and R154Q BIN1 mutants, for which the tubulation decrease was even confirmed in vitro, have a stronger tubulation phenotype but milder CNM symptoms and onset. As proposed above, this “second” function of the amphipathic helix could be the formation of interactions between adjacent turns of the helix, helping polymerization of BIN1.
It has been proposed that the crescent shape of BAR domains and the amphipathic helices have two distinct roles: while the crescent shape of BAR domains would sense membrane curvature, the amphipathic helices would actually induce it (Boucrot et al., 2012, Farsad et al., 2001). The CNM-related BIN1 mutants in these regions were not going along this proposition. While a mutation in the amphipathic helix should have resulted in a membrane tubulation defect, no significant tubulation defect was observed in vitro (K35N, Wu et al., 2014). The D151N and R154Q mutants should have shown a membrane curvature sensing defect instead of showing reduced membrane tubulation, whereas the opposite was observed (Wu et al., 2014).
2.5. CNM-linked BIN1 mutations in the muscle-specific PI motif decrease BIN1’s membrane tubulation properties
Only one known CNM-linked mutation affects the PI motif, the domain which specifically interacts with phosphoinositides (Lee et al., 2002). The progression of this type of CNM was very rapid upon the onset of the disease, two out of three patients died 2–4 years after the onset. This mutation is a splice acceptor site mutation (Böhm et al., 2013). The splice donor site at the end of exon 10 mistakenly binds to the splice acceptor site of exon 12, as the splice acceptor site of exon 11 is mutated (IVS10-1G > A; intervening sequence 10, position −1, G-to-A conversion). Only the muscle-specific isoforms 8 and 10 (Fig. 1) are affected by the mutation because the splice acceptor site of exon 11 is anyway not used in the other isoforms. The tissue-specificity of the IVS-1G > A mutation is thus far more easy to explain than for the other BIN1 mutations causing CNM. With this mutation, isoforms 8 and 10 are reverted to the ubiquitous form, therefore patients do not express muscle-specific isoforms. Thus, it is predicted that in these patients, the muscle-specific properties of BIN1, in particular the specific, phosphoinositide dependent, recruitment to the plasma membrane to induce T-tubule formation, should be lost. Indeed, the localisation of BIN1 in the muscle was observed to be altered in patients with this mutation (Böhm et al., 2013). Also, the tubulation was decreased in cultured myotubes and in vitro experiments (Böhm et al., 2013, Wu and Baumgart, 2014) when comparing BIN1 with and without the PI motif. The CNM-phenotype of the IVS10-1G > A mutation drastically illustrates either or both of the following hypotheses. The first hypothesis would be that the specific recruitment of BIN1 to phosphoinositides is indeed crucial for the proper muscle-specific function of BIN1. The second hypothesis is based on the observation (see next sub-chapter for more detail) that the PI motif can also bind to the SH3 domain (Kojima et al., 2004, Wu and Baumgart, 2014). This interaction could be inhibitory, and participate in the regulation of dynamin recruitment by BIN1 bound to phosphoinositides (see discussion below). This would explain also that mutants lacking the PI motif have similar effects in patients than hyperactive dynamin 2 mutants (see discussion below).
The variability of the CNM onset may account for different functions of BIN1 during development and maintenance of muscles. Strikingly, the skeletal muscle phenotype of bin1 morphant zebrafish embryos, notably the disorganization of triads, could be rescued by both BIN1 with the PI motif (isoform 8) and BIN1 without the PI motif (isoform 9) (Smith et al., 2014). This supports a specific role of the PI motif during muscle maturation and maintenance rather than during early development (Smith et al., 2014).
2.6. CNM-linked BIN1 mutations in the SH3 domain decrease the interaction with dynamin
The SH3 domain of BIN1 is important for its interaction with PRD-domain-containing proteins like dynamin and is present in all 11 isoforms reported till today (Uniprot), therefore mutations in the SH3 domain also affect the neuronal and ubiquitous isoforms. In this case, explaining the tissue-specificity of the induced muscle phenotype revealed complex. Four CNM mutations were found in this domain so far (Böhm et al., 2010, Böhm et al., 2014, Nicot et al., 2007). Two of these, Q573X and K575X, lead to a 20%-truncation of the SH3 domain amino acid sequence due to premature stop codons (Böhm et al., 2014, Nicot et al., 2007) while the other two, P593HfsX54 and X594DfsX53, cause a frame shift and add 52 additional amino acids to the SH3 domain (Böhm et al., 2014). No functional studies for these last two mutations have been reported yet, making it difficult to draw conclusions on the disease mechanism. Both are dominant mutations and cause mild and late-onset CNM already in the heterozygous case, in contrast to the SH3-domain-truncating mutations Q573X and K575X whose heterozygous carriers are healthy but which cause a more severe CNM with early onset when homozygous.
There are several functional studies about the K575X mutant, which could also account for the effect of the Q573X mutation due to their very similar SH3-domain truncations. In both mutants, the last α-helix and the last two β-sheets of the SH3 domain are missing (Fig.2B and Owen et al., 1998). The first consequence is likely a destabilisation and potential misfolding of the whole SH3 domain, because the two deleted β-sheets are normally part of a β-barrel which forms the inner scaffold of many SH3 domains (Owen et al., 1998). The second consequence is that the dynamin binding site is lacking an important hydrophobic residue (F588) in the K575X mutant, required for dynamin binding in vitro (Owen et al., 1998). The decreased dynamin-binding capacity of the truncated K575X mutant was indeed confirmed in vitro (Nicot et al., 2007). The K575X mutation did not affect the membrane tubulation ability of overexpressed BIN1 in COS-1 cells (Nicot et al., 2007). In contrast, and as expected, dynamin was not recruited to these tubules (Nicot et al., 2007). A recent study tested whether the recruitment of dynamin to BIN1 positive structures was only mediated via direct dynamin 2-BIN1 interactions (Picas et al., 2014) or via dynamin’s known curvature sensing properties (Roux et al., 2010). The results suggested that dynamin 2 recruitment to BIN1 positive membrane tubules might also be due to BIN1’s ability to cluster PIPs, which in turn recruit dynamin 2. Additionally, the SH3 domain of BIN1 could of course still recruit dynamin to these regions via a direct interaction with dynamin’s PRD domain. This would speak for the fact that BIN1 can still partially recruit dynamin, even with a truncated SH3 domain in the case of the K575X mutant (Nicot et al., 2007).
A difficulty in unfolding the disease mechanism of the K575X BIN1 mutant lies in the ability of the PI motif to interact with the WT SH3 domain but not with the K575X SH3 mutant (Wu and Baumgart, 2014). Since this mutant shows an increase in membrane tubulation in vitro (Wu and Baumgart, 2014), it has been proposed that the SH3-PI interaction may play as an intra-molecular inhibition, which could be released by BIN1 binding to either the membrane or dynamin. The increased curvature inducing properties of the K575X-mutated protein would therefore result from a decreased autoinhibition.
But the primary role of BIN1’s SH3 domain is to bind to dynamin. Dynamin is a GTPase that tubulates lipid membranes by forming helical polymers around membrane tubes. It also constricts the membrane, leading to fission upon GTP hydrolysis. In the following, we describe the effect of the mutations found in dynamin 2 in some CNM-related mutations, and discuss the interplay between dynamin and BIN1 in the specific case of the T-tubule biogenesis.
3. Dynamin 2 in CNM
Dynamin is a large GTPase that mediates membrane fission at the final step of clathrin-mediated endocytosis. Dynamin polymerizes around the neck of clathrin-coated buds and severs the membrane upon GTP hydrolysis, in order to release the vesicle in the cytoplasm. Other less known functions comprise roles in clathrin independent endocytosis and actin dynamics (Ferguson and De Camilli, 2012).
In mammalian cells there are three dynamin genes that share a similar sequence and the same domain structure. Dynamin 1 is expressed in the brain, dynamin 2 is ubiquitous and dynamin 3 is expressed in lungs, testis and brain (Cao et al., 1998). Dynamins are constituted of five domains, starting from the N-terminus with the G-domain, in charge of GTP hydrolysis. It is connected to the rest of the protein through a flexible hinge called the Bundle Signaling Element (BSE or neck). At the other side of the BSE, the “stalk” region comprises two distinct sequence parts, the middle domain and the GTPase Enhancing Domain (GED). The stalk region mediates dimer formation and further interactions that allow self-assembly. The Pleckstrin Homology (PH) domain binds the plasma membrane through specific phosphoinositides (especially PI(4,5)P2). As discussed above, the PRD domain is deputed to the binding to and recruitment of other proteins, in particular BIN1. It defines the specific functions of the three dynamins as its sequence is the most variable between variants, allowing for differential binding of dynamin partners (Faelber et al., 2011, Raimondi et al., 2011). Dynamin function in membrane fission involves its GTPase activity that is up regulated by polymerization and lipid binding (Smirnova et al., 1999, Warnock et al., 1996), allowing the action of dynamin to be maximal only when polymerized at the membrane neck. Structural data showed that in absence of nucleotide, the PH domain is flipped-back onto the stalk, having specific interactions (Faelber et al., 2011) with both the stalk and the GTPase domain, that were broken upon polymerization (Reubold et al., 2015). Thus, it was proposed that the PH domain, when flipped and bound to the stalk and the GTPase domain, would be an inhibitory “closed” state of dynamin which would be release when the PH domain would bind the membrane, activating the GTPase and the polymerization of dynamin.
Specific heterozygous mutations in the ubiquitous dynamin 2 (Uniprot ID P50570-1, UGID 164319) lead to autosomal dominant centronuclear myopathy (ADCNM) (Bitoun et al., 2005). The clinical aspects of ADCNM caused by mutations of dynamin 2 are variable depending on the specific mutation involved (Bitoun et al., 2009b), and occasionally even for the same mutation. ADCNM can have mild, intermediate or severe forms. The age of onset is also very variable and ranges from early (neonatal and childhood) and intermediate (adolescence) to late onset (adulthood) (Böhm et al., 2012).
The only tissue that is affected by CNM-related dynamin 2 mutations is the muscle, but the mutations causing CNM involve the ubiquitous dynamin 2 whose functions are necessary in the whole organism. How a tissue-specific disease can be caused by mutations in a ubiquitous protein is still unclear, but analysing the effect of these mutations at the biochemical and functional levels supports a muscle-specific structural role of dynamin 2 independent of its function in membrane fission.
3.1. Biochemical, structural and cellular aspects of CNM-related dynamin 2 mutations
ADCNM shares the major cellular and sub-cellular phenotypes with other forms of CNM such as centralized nuclei, predominance of type I fibres (slowly contracting and less power-generating than type II fibres), muscle weakness and atrophy. But it also implicates an abnormal radial distribution of sarcoplasmic strands not found in CNM caused by the mutations of other genes (Böhm et al., 2012).
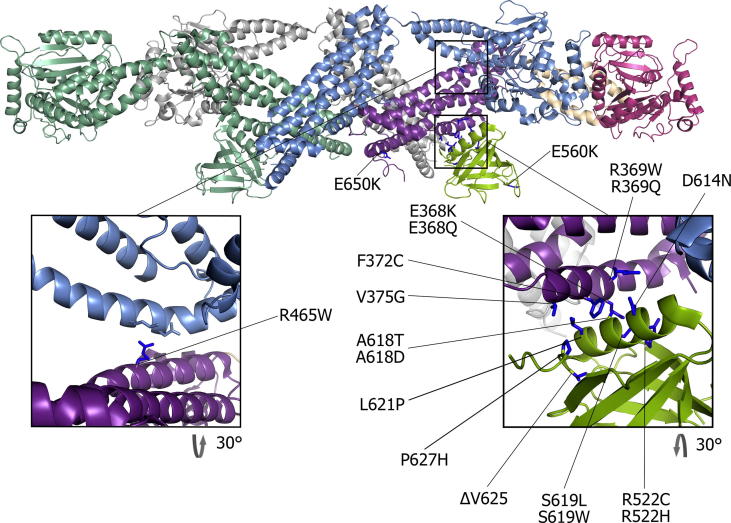
Among the mutations of dynamin 2 that cause CNM, the majority are missense mutations located in the stalk (middle and GED domains) and in the PH domain. All the mutations that have been biochemically characterized (R465W, S619L, A618T, E368K, ΔV625 and R369W) show an increase in basal GTPase activity and less GTP-induced disassembly as compared to wild type dynamin 2 (Chin et al., 2015, James et al., 2014, Kenniston and Lemmon, 2010, Wang et al., 2010). R369W, S619L and ΔV625 have also been shown to have a higher ability to self-assemble, which may account for their reduced disassembly rate. Their basal GTPase activity is higher than the wild type, but does not increase upon assembly. Thus, biochemical data available for these mutations show a gain of function phenotype, both for assembly and for GTPase activity, suggesting that the auto-inhibitory mechanism discussed above for the PH domain is not functional in these mutants. Indeed, among the CNM-related dynamin 2 mutations, the majority is positioned at the interface (or in close proximity) between the stalk and the PH domain in the dimer structure in absence of nucleotide (see Table 2 and Fig. 3). Moreover, the majority of CNM-related mutations in this interface locally change the charge or the hydrophobicity of the residues and could be accounted for a major disruption of the interface. Although no data is available for other CNM-related mutations in this interface (E368Q, R369Q, F372C, V375G, D614N, A618D, L621P, P627R and P627H) (Bitoun et al., 2005, Böhm et al., 2012, Catteruccia et al., 2013, Echaniz-Laguna et al., 2007, Jungbluth et al., 2010, Kierdaszuk et al., 2013, Neto et al., 2015, Susman et al., 2010), one could predict that they would have the same effect on the auto-inhibitory conformation of dynamin.
Table 2.
CNM mutations in Dynamin 2.
| Mutation | Exon | Domain | Dominance | Severity | Disease onset | Hyperactivitya of dynamin | Reference of discovery |
|---|---|---|---|---|---|---|---|
| E368 K | 8 | Stalk (middle) | Dominant | Severe | Neonatal or childhood | Yes | Bitoun et al. (2005) |
| E368Q | 8 | Stalk (middle) | Dominant | Variable | Variable | [Not tested] | Echaniz-Laguna et al. (2007) |
| R369W | 8 | Stalk (middle) | Dominant | Mild | Adolescence or adulthood | Yes | Bitoun et al. (2005) |
| R369Q | 8 | Stalk (middle) | Dominant | Mild | Variable | [Not tested] | Bitoun et al. (2005) |
| F372C | 8 | Stalk (middle) | Dominant | Severe | Childhood | [Not tested] | Neto et al. (2015) |
| V375G | 8 | Stalk (middle) | Dominant | Severe | Neonatal | [Not tested] | Catteruccia et al. (2013) |
| R465W | 11 | Stalk (middle) | Dominant | Mild | Variable | Yes | Bitoun et al. (2005) |
| R522C | 15 | PH | Dominant | Mild | Variable | [Not tested] | Böhm et al. (2012) |
| R522H | 15 | PH | Dominant | Mild | Variable | [Not tested] | Susman et al. (2010) |
| R523G | 15 | PH | Dominant | Mild | Adulthood | [Not tested] | Böhm et al. (2012) |
| E540K | 15 | PH | Dominant | Severe | Adulthood | [Not tested] | Catteruccia et al. (2013) |
| E560K | 16 | PH | Dominant | Severe | Childhood | Yes | Bitoun et al., 2009a, Bitoun et al., 2009b |
| D614N | 17 | PH | Dominant | Mild | Adulthood | [Not tested] | Kierdaszuk et al. (2013) |
| A618D | 17 | PH | Dominant | Mild | Neonatal | [Not tested] | Melberg et al. (2010) |
| A618T | 17 | PH | Dominant | Severe | Neonatal | Yes | Bitoun et al. (2007) |
| S619L | 17 | PH | Dominant | Severe | Neonatal | Yes | Bitoun et al. (2007) |
| S619W | 17 | PH | Dominant | Severe | Neonatal | Yes | Bitoun et al. (2007) |
| L621P | 17 | PH | Dominant | Severe | Neonatal | [Not tested] | Bitoun et al. (2007) |
| ΔV625 | 17 | PH | Dominant | Severe | Neonatal | Yes | Bitoun et al. (2007) |
| P627R | 17 | PH | Dominant | Intermediate | Neonatal or childhood | [Not tested] | Böhm et al. (2012) |
| P627H | 17 | PH | Dominant | Severe | Childhood | [Not tested] | Susman et al. (2010) |
| E650K | 18 | Stalk (GED) | Dominant | Mild | Childhood | Yes | Bitoun et al., 2009a, Bitoun et al., 2009b |
| Splice site mut. | 17-18 | PH-stalk (GED) | Dominant | Mild | Variable | [Not tested] | Böhm et al. (2012) |
Basal activity and/or lipid-coupled activity.
Fig. 3.
Dynamin tetramer structure. Structure of the dynamin tetramer (Reubold et al., 2015) with represented dynamin 2 CNM-related mutations. The inlets show the interface between the stalk and the PH domain (right panel) and mutation R465 W (left panel).
The most common dynamin 2 mutation R456W that causes CNM (Böhm et al., 2012) is also positioned in the stalk domain, but targets another intermolecular interface of the protein. It localises to the outer monomer, where it interfaces the BSE of the inner monomer facing an asparagine and a lysine residue (Reubold et al., 2015) (Fig. 3). The mutation of a polar and positively charged residue (arginine) to a hydrophobic residue (tryptophan) could cause a disruption of this interface, causing the same release of the auto-inhibitory conformation than for mutations in the PH-stalk interface. Consistently, the R456W mutation shares many aspects of the phenotype of the stalk-PH mutations, being hyperactive and more stable in the presence of GTP, suggesting a lack of the auto-inhibitory regulation.
The remaining known CNM-related dynamin 2 mutations (R522C, R522H, R523G, E540K, E560K, E650K and donor splice site deletion between exon 17 and 18; Bitoun et al., 2009a, Bitoun et al., 2009b, Böhm et al., 2012, Catteruccia et al., 2013, Jeub et al., 2008, Susman et al., 2010) have not yet been biochemically or biologically characterized and they all reside in the PH domain (except E650K, located in the stalk domain) in regions probably necessary for the interactions with lipids or the regulation of self-assembly upon lipid interaction.
But what could be the consequence of this gain of function onto T-tubule biogenesis, and why would it be muscle-specific, since these mutations appear on the ubiquitous form of dynamin 2? Important clues came out from the in vivo analysis of these mutants, both in endocytic and T-tubule biogenesis function of dynamin 2: while overexpression of these mutations dramatically impairs endocytosis in cultured cells, heterozygous patients' cells express the mutant and the wild-type at similar levels, and show no defect in CME (Bitoun et al., 2009b, Koutsopoulos et al., 2011, Sidiropoulos et al., 2012). Furthermore, in Mouse Embryonic Fibroblasts (MEFs) from mice bearing the heterozygous R465W mutation, CME functions normally, while it is slightly impaired in MEFs homozygous for these mutations (homozygous mice die after birth) (Durieux et al., 2010). These findings show that the disease is probably not related to the function of dynamin in CME, whose alteration causes embryonic lethality, and that these mutations seem to have very little effect on CME. Moreover, a gain of function of dynamin 2 in CME is likely to have no physiological impact, as CME would just go faster. However, a recent study has shown a slight impairment in clathrin-independent EGF, cholera toxin uptake and EGFR downregulation as well as a defect in trafficking of p75 receptor from the Golgi to the plasma membrane in stable cell lines expressing physiological levels of the hyperactive dynamin 2 mutations R465W and E368K (Liu et al., 2011). This may cause signalling deregulation in these patients, which may account for the severity of some forms.
As a conclusion, the biochemical, biological and clinical analysis of CNM-related dynamin 2 mutations has shown that they have a global gain of function with no dramatic impact on the ubiquitous functions of dynamin 2 in CME. The pathology must therefore arise from the alteration of another function of dynamin 2 delimited to a muscle-specific role.
3.2. Gain of function dynamin 2 mutations alter a tissue-specific balance
The fact that none of the ubiquitous functions of dynamin 2 are defective in a heterozygous background of CNM-related dynamin 2 mutations leads to consider tissue-specific functions of dynamin 2 in the muscle. In muscle fibres, dynamin 2 has been found along the Z-line, co-localizing with α-actinin and desmin. Dynamin 2 is located in close proximity to dihydropyridine receptor (DHPR), a marker of T-tubules (Cowling et al., 2011, Durieux et al., 2010), which suggests that dynamin 2, like BIN1, could be implicated in T-tubule biogenesis. The presence of dynamin 2 on T-tubules still has to be shown. The analysis of CNM-related dynamin 2 mutations in animal models and in cells strongly implicates that the hyperactivation of dynamin 2 is deleterious to the maintenance and stabilization of T-tubules and triads, suggesting a tissue-specific role of dynamin in muscles related to T-tubule and triad shaping. The expression of human dynamin 2 carrying the mutation R465W in adult mice results in a decrease in muscle mass due to the appearance of atrophic fibres. Histological analysis showed misalignment of Z-lines and triad defects such as abnormally shaped T-tubules and a swollen sarcoplasmic reticulum (Cowling et al., 2011). In heterozygous mice for the same mutation, a slight disorganization of T-tubules, sarcoplasmic reticulum and an increase in cytosolic Ca2+ are also observed, although defects in the triad structure are milder (Durieux et al., 2010).
Similar results have been obtained in non-mammalian organisms, supporting a conserved role of dynamin 2 in T-tubule development through evolution. In larvae of transgenic Drosophila ectopically expressing one of the human dynamin 2 mutations R465W, A618T or S619L in the muscle displayed fragmented T-tubules even when expressed later in development when T-tubules are already formed (Chin et al., 2015). In zebrafish, where extensive disorganization of triad structures and T-tubule fragmentation can be observed when the human dynamin 2 S619L mutant is expressed during development, these defects led to impaired excitation-contraction coupling (Gibbs et al., 2014).
Altogether, these results suggest a tissue-specific function of dynamin 2 in the establishment of the T-tubule structures in muscle fibres. T-tubules are a result of plasma membrane tubulation carried out most probably by caveolin-3 and BIN1. Dynamin 2 could either serve as a scaffolding protein as it is able to form a more rigid scaffold then N-BAR proteins (Frost et al., 2008), which could help the maintenance and the resistance of the T-tubule system needed for muscle contraction. An alternative hypothesis is that dynamin 2 could be involved in T-tubule length regulation through membrane fission. Another hint of a possible function of dynamin in T-tubule shaping comes from the BIN1 mutant K575X: its impairment of the SH3 domain, thus loss of interaction with dynamin 2, could account for its pathological outcome in patients (Nicot et al., 2007).
However, the fission activity of dynamin 2 must be blocked or tightly regulated to play this role, otherwise dynamin would excessively cut these T-tubules, leading to a fragmented T-tubule system. In a wild type background, dynamin 2 fission activity should be somehow inhibited or regulated at the T-tubule. This also explains why gain-of function mutations would have a dramatic effect specifically at the T-tubule, as enhancing the fission activity of dynamin 2 would cause disruption of the T-tubule system. But how dynamin 2 fission activity would be specifically inhibited at the T-tubule? As described in the following, this may arise from its specific interaction with BIN1.
3.3. Interplay between dynamin, BIN1 and MTM1 functions in T-tubule biogenesis
BIN1 may specifically recruit dynamin 2 to the T-tubule system (Lee et al., 2002) and inhibit its fission activity. Supporting this assertion, other N-BAR proteins such as endophilin and amphiphysin regulate dynamin’s fission activity, but the nature of the regulation (activation versus inhibition) seemed to vary for different N-BAR proteins and in different conditions (Farsad et al., 2001, Meinecke et al., 2013, Neumann and Schmid, 2013, Takei et al., 1999). In some experiments (Boucrot et al., 2012, Farsad et al., 2001), N-BAR proteins associated with dynamin have displayed an inhibitory action on dynamin fission activity. Thus, one could postulate that BIN1 and dynamin 2 form a heteropolymer that would block dynamin 2’s ability to sever the membrane. However, this effect has not been reproducibly established, and contradictory results have been obtained (Meinecke et al., 2013, Neumann and Schmid, 2013, Takei et al., 1999, Yoshida et al., 2004).
In muscle fibres expressing CNM-related dynamin 2 mutations, the abnormal hyperactivity of these mutants could overcome the inhibition of dynamin-fission activity, shifting the equilibrium between dynamin 2 scaffolding with a controlled membrane fission function towards unregulated membrane fission. This would result in the fragmented and abnormally shaped T-tubules, as observed in the animal models expressing hyperactive dynamin 2 mutations (Chin et al., 2015, Gibbs et al., 2014).
Evidences supporting this model come from experiments in cells. Upon overexpression of BIN1 (or other N-BAR proteins) in cultured cells, an extensive membrane tubulation is observed. This effect can be counteracted by overexpressing dynamin (Itoh et al., 2005) or by expressing a CNM-related hyperactive mutation (Chin et al., 2015, Gibbs et al., 2014). In these examples, the plasma membrane tubulation is the result of an imbalance between the tubulation activity of BIN1 (higher) and the fission activity of dynamin (lower): the balance can be restored by either increasing the amount of dynamin, or by expressing a dynamin mutant with increased activity.
Also, as described above, mutations in the SH3 domain of BIN1 could impair its interaction with dynamin 2, releasing dynamin’s inhibition. In these mutants, dynamin 2 would not be inhibited enough to allow for proper T-tubule maintenance. However, as BIN1 also participates in the recruitment of dynamin 2 to the T-tubule system, together with phosphoinositides, one may question if the reduced recruitment of dynamin 2 in the SH3 BIN1 mutants may counteract the reduced inhibition of dynamin fission activity. In this context, the role of the phosphoinositides may be essential to understand the mechanism of the disease. Myotubularin, the third protein causing CNM when mutated, may be critical as it is a 3-phosphatase that can interact with BIN1. In Royer et al. (2013), the authors found that myotubularin (MTM1) binds BIN1 in vitro and in skeletal muscle. Similar to the effect of binding of dynamin’s PRD domain to BIN1, the authors propose that MTM1 may help dynamin and BIN1 tubulating activity. It would also require MTM1’s phosphatase activity. MTM1 is a 3-phosphatase and its substrates are PI(3)P and PI(3,5)P2. As BIN1 was shown to bind to PI(5)P as well as PI(4,5)P2 in contrast to PI(3,5)P2 (Fugier et al., 2011, Lee et al., 2002, Picas et al., 2014), one can hypothesize that MTM1 might promote the formation of 5-phosphate phosphoinositides, such as PI(5)P and PI(4,5)P2, by increasing the level of their precursors (PI and PI(5)P). These 5-phosphate phosphoinositides will then recruit BIN1 and dynamin to the plasma or T-tubule membrane.
Furthermore, Royer et al. (2013) observed that both Q573X and K575X BIN1 mutants have an increased interaction with MTM1 in vitro. A plausible mechanism would thus be that the same SH3 mutants of BIN1 have a decreased interaction with dynamin and an increased interaction with MTM1. The defective recruitment of dynamin by the BIN1 SH3 mutant could thus be compensated by the increase of 5-phosphate phosphoinositides resulting from a higher recruitment of MTM1 to BIN1 structures. However in this case, dynamin’s fission activity would not be inhibited, since the interaction with BIN1 is lost, and would thus lead to a disrupted T-tubule system.
The BIN1-MTM1 interaction is also interesting when compared with other N-BAR/phosphatase pairs: for example, endophilin physically interacts with and recruits the lipid phosphatase synaptojanin to the membrane of clathrin-coated pits (Perera et al., 2006). In the synaptojanin-endophilin case, the synaptojanin recruitment is proposed to result in a decrease of the PI(4,5)P2 concentration and to allow for fission and uncoating of clathrin-coated pits (Perera et al., 2006). The striking difference to the synaptojanin-endophilin pair is that MTM1 recruitment by BIN1 should, via increasing the concentration of the PI(4,5)P2 precursor, lead to an increase of PI(4,5)P2, which in turn would recruit more BIN1 and dynamin. This opposite effect correlates nicely with the idea that the scaffold which stabilises T-tubules would have a positive feedback loop where the scaffold proteins increase their respective recruitment, whereas the scaffold and coat implicated in the formation of a clathrin-coated vesicle is temporary.
4. General conclusions
We have reviewed in this article how different groups of mutations disturb the structure and function of BIN1, leading to CNM. BIN1 mutations in the amphipathic helix could decrease BIN1’s scaffolding properties on membrane tubes. Mutations in the tip region of the BAR domain dimer may decrease BIN1’s tubulation properties. Also in the case of splice-site mutations resulting in a loss of the PI motif, BIN1 tubulates less and the T-tubule formation and/or maintenance is impaired. In contrast, mutations in the SH3 domain do not impair BIN1’s curvature inducing features but might affect both dynamin and MTM1 levels at the T-tubules.
CNM-related mutations in dynamin 2 are hyperactive and thus correspond to a gain of function. As long as they are heterozygous, they cannot impair clathrin-mediated endocytosis because they maintain the ability to mediate membrane fission and their action is regulated by redundant factors and regulators. In T-tubule biogenesis and maintenance, it is primarily the membrane scaffolding and tubulation activities or a moderate fission activity of dynamin that are at work. T-tubule biogenesis thus requires a down regulation of dynamin 2’s intrinsic fission activity. Thus, dynamin 2 mutations that have increased fission activity which are not harmful for CME, probably cause massive problems in the T-tubule system because of their inability to be down regulated. Thus, the balance between membrane tubulating and fission activities of dynamin is crucial for the T-tubule biogenesis.
Given that CNM-associated dynamin mutants are hyperactive and BIN1 mutants have a decrease-of-function, CNMs are one example where an N-BAR protein seems to negatively regulate dynamin’s fission activity. To test this hypothesis, one could overexpress dynamin 2 GTPase defective mutants in cells with CNM-associated BIN1 mutations. If this rescues the low-function BIN1 phenotype (fragmented T-tubules), this would clearly speak for a role of BIN1 in preventing dynamin from cutting T-tubules. Fitting with this theory, a patent has been published for the idea to treat CNM patients with dynamin inhibitors (Laporte and Cowling, European Patent EP2862928A1).
Another interesting matter of the CNMs is its tissue-specificity to muscles, while other cellular functions seemed unaffected by the CNM mutations, even though most of the mutations affect ubiquitously expressed exons of BIN1 and dynamin 2. We hypothesize that the tissue-specificity of the pathology is due to the fact that T-tubules are one of few cases where membrane tubules are formed in cells and are required to be incredibly stable over time, while they represent ideal targets for dynamin’s fission activity. Thus a tight regulation of the levels of BIN1 and dynamin, as well as of the balance between their specific activities is required in the T-tubule system, with very little redundancy as compared to the CME system. Most probably, the triple complex between dynamin 2, BIN1 and MTM1 is at the heart of this balance between membrane tubulation and fission activity, as many of the mutations found in these three proteins affect the protein-protein interaction domains. Overall, if membrane deformation properties are too low, the T-tubule formation is impaired. If fission activity is too high, T-tubule are snapped too early after formation.
Acknowledgements
We thank Andreja Vujičić-Žagar for help with rendering of the protein figures; Marko Kaksonen and Jean Gruenberg for helpful discussion. AR acknowledges funding support from: Human Frontier Science Program (HFSP), Young Investigator Grant #RGY0076-2008: the European Research Council (ERC), starting (consolidator) grant #311536-MEMFIS: the Swiss National Fund for Research, grants #131003A_130520 and #131003A_149975.
References
- Adam J., Basnet N., Mizuno N. Structural insights into the cooperative remodeling of membranes by amphiphysin/BIN1. Sci. Rep. 2015;5:15452. doi: 10.1038/srep15452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Bitoun M., Maugenre S., Jeannet P.-Y., Lacène E., Ferrer X., Laforêt P., Martin J.-J., Laporte J., Lochmüller H., Beggs A.H., Fardeau M., Eymard B., Romero N.B., Guicheney P. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- Bitoun M., Bevilacqua J.A., Prudhon B., Maugenre S., Taratuto A.L., Monges S., Lubieniecki F., Cances C., Uro-Coste E., Mayer M., Fardeau M., Romero N.B., Guicheney P. Dynamin 2 mutations cause sporadic centronuclear myopathy with neonatal onset. Ann. Neurol. 2007;62 doi: 10.1002/ana.21235. [DOI] [PubMed] [Google Scholar]
- Bitoun M., Bevilacqua J.A., Eymard B., Prudhon B., Fardeau M., Guicheney P., Romero N.B. A new centronuclear myopathy phenotype due to a novel dynamin 2 mutation. Neurology. 2009;72:93–95. doi: 10.1212/01.wnl.0000338624.25852.12. [DOI] [PubMed] [Google Scholar]
- Bitoun M., Durieux A.-C., Prudhon B., Bevilacqua J.A., Herledan A., Sakanyan V., Urtizberea A., Cartier L., Romero N.B., Guicheney P. Dynamin 2 mutations associated with human diseases impair clathrin-mediated receptor endocytosis. Hum. Mutat. 2009;30:1419–1427. doi: 10.1002/humu.21086. [DOI] [PubMed] [Google Scholar]
- Böhm J., Yiş U., Ortaç R., Cakmakçı H., Kurul S.H., Dirik E., Laporte J. Case report of intrafamilial variability in autosomal recessive centronuclear myopathy associated to a novel BIN1 stop mutation. Orphanet J. Rare Dis. 2010;5:35. doi: 10.1186/1750-1172-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J., Biancalana V., DeChene E.T., Bitoun M., Pierson C.R., Schaefer E., Karasoy H., Dempsey M.A., Klein F., Dondaine N., Kretz C., Haumesser N., Poirson C., Toussaint A., Greenleaf R.S., Barger M.A., Mahoney L.J., Kang P.B., Zanoteli E., Vissing J., Witting N., Echaniz-Laguna A., Wallgren-Pettersson C., Dowling J., Merlini L., Oldfors A., Ousager L.B., Melki J., Krause A., Jern C., Oliveira A.S.B., Petit F., Jacquette A., Chaussenot A., Mowat D., Leheup B., Cristofano M., Aldea J.J.P., Michel F., Furby A., Llona J.E.B., Van Coster R., Bertini E., Urtizberea J.A., Drouin-Garraud V., Roud C.B., Prudhon B., Bedford M., Mathews K., Erby L.A.H., Smith S.A., Roggenbuck J., Crowe C.A., Spitale A.B., Johal S.C., Amato A.A., Demmer L.A., Jonas J., Darras B.T., Bird T.D., Laurino M., Welt S.I., Trotter C., Guicheney P., Das S., Mandel J.L., Beggs A.H., Laporte J. Mutation spectrum in the large gtpase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum. Mutat. 2012;33:949–959. doi: 10.1002/humu.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J., Vasli N., Maurer M., Cowling B.S., Cowling B., Shelton G.D., Kress W., Toussaint A., Prokic I., Schara U., Anderson T.J., Weis J., Tiret L., Laporte J. Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 2013;9:e1003430. doi: 10.1371/journal.pgen.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J., Biancalana V., Malfatti E., Dondaine N., Koch C., Vasli N., Kress W., Strittmatter M., Taratuto A.L., Gonorazky H., Laforet P., Maisonobe T., Olive M., Gonzalez-Mera L., Fardeau M., Carriere N., Clavelou P., Eymard B., Bitoun M., Rendu J., Faure J., Weis J., Mandel J.-L., Romero N.B., Laporte J. Adult-onset autosomal dominant centronuclear myopathy due to BIN1 mutations. Brain. 2014;137:3160–3170. doi: 10.1093/brain/awu272. [DOI] [PubMed] [Google Scholar]
- Boucrot E., Pick A., Çamdere G., Liska N., Evergren E., McMahon H.T., Kozlov M.M. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.H., David C., Ochoa G.C., Freyberg Z., Daniell L., Grabs D., Cremona O., De Camilli P. Amphiphysin II (SH3p9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F., Kozlov M.M. Sensing membrane stresses by protein insertions. PLoS Comput. Biol. 2014;10:e1003556. doi: 10.1371/journal.pcbi.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Garcia F., McNiven M.A. Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal E., Federici L., Zhang W., Fernandez-Recio J., Priego E.M., Miguel R.N., DuHadaway J.B., Prendergast G.C., Luisi B.F., Laue E.D. The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition. Biochemistry. 2006;45:12917–12928. doi: 10.1021/bi060717k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteruccia M., Fattori F., Codemo V., Ruggiero L., Maggi L., Tasca G., Fiorillo C., Pane M., Berardinelli A., Verardo M., Bragato C., Mora M., Morandi L., Bruno C., Santoro L., Pegoraro E., Mercuri E., Bertini E., D’Amico A. Centronuclear myopathy related to dynamin 2 mutations: clinical, morphological, muscle imaging and genetic features of an Italian cohort. Neuromuscul. Disord. 2013;23:229–238. doi: 10.1016/j.nmd.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y.-H., Lee A., Kan H.-W., Laiman J., Chuang M.-C., Hsieh S.-T., Liu Y.-W. Dynamin-2 mutations associated with centronuclear myopathy are hypermorphic and lead to T-tubule fragmentation. Hum. Mol. Genet. 2015;24:5542–5554. doi: 10.1093/hmg/ddv285. [DOI] [PubMed] [Google Scholar]
- Claeys K.G., Maisonobe T., Böhm J., Laporte J., Hezode M., Romero N.B., Brochier G., Bitoun M., Carlier R.Y., Stojkovic T. Phenotype of a patient with recessive centronuclear myopathy and a novel BIN1 mutation. Neurology. 2010;74:519–521. doi: 10.1212/WNL.0b013e3181cef7f9. [DOI] [PubMed] [Google Scholar]
- Cowling B.S., Toussaint A., Amoasii L., Koebel P., Ferry A., Davignon L., Nishino I., Mandel J.L., Laporte J. Increased expression of wild-type or a centronuclear myopathy mutant of dynamin 2 in skeletal muscle of adult mice leads to structural defects and muscle weakness. Am. J. Pathol. 2011;178:2224–2235. doi: 10.1016/j.ajpath.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux A.C., Vignaud A., Prudhon B., Viou M.T., Beuvin M., Vassilopoulos S., Fraysse B., Ferry A., Lainé J., Romero N.B., Guicheney P., Bitoun M. A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum. Mol. Genet. 2010;19:4820–4836. doi: 10.1093/hmg/ddq413. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A., Nicot A.-S., Carré S., Franques J., Tranchant C., Dondaine N., Biancalana V., Mandel J.-L., Laporte J. Subtle central and peripheral nervous system abnormalities in a family with centronuclear myopathy and a novel dynamin 2 gene mutation. Neuromuscul. Disord. 2007;17:955–959. doi: 10.1016/j.nmd.2007.06.467. [DOI] [PubMed] [Google Scholar]
- Faelber K., Posor Y., Gao S., Held M., Roske Y., Schulze D., Haucke V., Noé F., Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011 doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Farsad K., Ringstad N., Takei K., Floyd S.R., Rose K., De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.M., De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 2012:13. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E.H., De Camilli P., Unger V.M. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C., Klein A.F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N., Kokunai Y., Tsuburaya R., de la Grange P., Dembele D., Francois V., Precigout G., Boulade-Ladame C., Hummel M.-C., de Munain A.L., Sergeant N., Laquerrière A., Thibault C., Deryckere F., Auboeuf D., Garcia L., Zimmermann P., Udd B., Schoser B., Takahashi M.P., Nishino I., Bassez G., Laporte J., Furling D., Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Gibbs E.M., Davidson A.E., Telfer W.R., Feldman E.L., Dowling J.J. The myopathy-causing mutation DNM2-S619L leads to defective tubulation in vitro and in developing zebrafish. Dis. Model. Mech. 2014;7:157–161. doi: 10.1242/dmm.012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabs D., Slepnev V.I., Songyang Z., David C., Lynch M., Cantley L.C., De Camilli P. The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. J. Biol. Chem. 1997;272:13419–13425. doi: 10.1074/jbc.272.20.13419. [DOI] [PubMed] [Google Scholar]
- Itoh T., Erdmann K.S., Roux A., Habermann B., Werner H., De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- James N.G., Digman M.A., Ross J.A., Barylko B., Wang L., Li J., Chen Y., Mueller J.D., Gratton E., Albanesi J.P., Jameson D.M. A mutation associated with centronuclear myopathy enhances the size and stability of dynamin 2 complexes in cells. Biochim. Biophys. Acta. 2014;1840:315–321. doi: 10.1016/j.bbagen.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeub M., Bitoun M., Guicheney P., Kappes-Horn K., Strach K., Druschky K.F., Weis J., Fischer D. Dynamin 2-related centronuclear myopathy: clinical, histological and genetic aspects of further patients and review of the literature. Clin. Neuropathol. 2008;27:430–438. doi: 10.5414/npp27430. [DOI] [PubMed] [Google Scholar]
- Jungbluth H., Cullup T., Lillis S., Zhou H., Abbs S., Sewry C., Muntoni F. Centronuclear myopathy with cataracts due to a novel dynamin 2 (DNM2) mutation. Neuromuscul. Disord. 2010;20:49–52. doi: 10.1016/j.nmd.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Kenniston J.A., Lemmon M.A. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdaszuk B., Berdynski M., Karolczak J., Redowicz M.J., Zekanowski C., Kaminska A.M. A novel mutation in the DNM2 gene impairs dynamin 2 localization in skeletal muscle of a patient with late onset centronuclear myopathy. Neuromuscul. Disord. 2013;23:219–228. doi: 10.1016/j.nmd.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Kojima C., Hashimoto A., Yabuta I., Hirose M., Hashimoto S., Kanaho Y., Sumimoto H., Ikegami T., Sabe H. Regulation of Bin1 SH3 domain binding by phosphoinositides. EMBO J. 2004;23:4413–4422. doi: 10.1038/sj.emboj.7600442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos O.S., Koch C., Tosch V., Böhm J., North K.N., Laporte J. Mild functional differences of dynamin 2 mutations associated to centronuclear myopathy and Charcot-Marie Tooth peripheral neuropathy. PLoS One. 2011;6:e27498. doi: 10.1371/journal.pone.0027498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte, J., Cowling, B., 2015. Dynamin 2 inhibitor for the treatment of centronuclear myopathies. European Patent EP2862928A1.
- Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O.A., Ochoa G.-C., Farsad K., Wenk M.R., De Camilli P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- Liu Y.-W., Lukiyanchuk V., Schmid S.L. Common membrane trafficking defects of disease-associated dynamin 2 mutations. Traffic. 2011;12:1620–1633. doi: 10.1111/j.1600-0854.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löw C., Weininger U., Lee H., Schweimer K., Neundorf I., Beck-Sickinger A.G., Pastor R.W., Balbach J. Structure and dynamics of helix-0 of the N-BAR domain in lipid micelles and bilayers. Biophys. J. 2008;95:4315–4323. doi: 10.1529/biophysj.108.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M., Boucrot E., Camdere G., Hon W.C., Mittal R., McMahon H.T. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J. Biol. Chem. 2013;288:6651–6661. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melberg A., Kretz C., Kalimo H., Wallgren-Pettersson C., Toussaint A., Böhm J., Stålberg E., Laporte J. Adult course in dynamin 2 dominant centronuclear myopathy with neonatal onset. Neuromuscul. Disord. 2010;20:53–56. doi: 10.1016/j.nmd.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Mim C., Cui H., Gawronski-Salerno J.A., Frost A., Lyman E., Voth G.A., Unger V.M. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto O.A., Martins C.D.A., Carvalho M., Chadi G., Seitz K.W., Souza A., Oliveira B., Reed U.C., Laporte J., Zanoteli E. DNM2 mutations in a cohort of sporadic patients with centronuclear myopathy. Genet. Mol. Biol. 2015;151:147–151. doi: 10.1590/S1415-4757382220140238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S., Schmid S.L. Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J. Biol. Chem. 2013;288:25119–25128. doi: 10.1074/jbc.M113.490474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot A.-S., Toussaint A., Tosch V., Kretz C., Wallgren-Pettersson C., Iwarsson E., Kingston H., Garnier J.-M., Biancalana V., Oldfors A., Mandel J.-L., Laporte J. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- Owen D., Wigge P., Vallis Y., Moore J., Evans P., McMahon H.T. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 1998;17:5273–5285. doi: 10.1093/emboj/17.18.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.M., Zoncu R., Lucast L., De Camilli P., Toomre D. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19332–19337. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J.G., Evans P.R., McMahon H.T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Picas L., Viaud J., Schauer K., Vanni S., Hnia K., Fraisier V., Roux A., Bassereau P., Gaits-Iacovoni F., Payrastre B., Laporte J., Manneville J.-B., Goud B. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 2014;5:5647. doi: 10.1038/ncomms6647. [DOI] [PubMed] [Google Scholar]
- Raimondi A., Ferguson S.M., Lou X., Armbruster M., Paradise S., Giovedi S., Messa M., Kono N., Takasaki J., Cappello V., O’Toole E., Ryan T.A., De Camilli P. Overlapping role of dynamin isoforms in synaptic vesicle endocytosis. Neuron. 2011;70:1100–1114. doi: 10.1016/j.neuron.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjaun A.R., McPherson P.S. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J. Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- Razzaq A., Robinson I.M., Mcmahon H.T., Skepper J.N., Su Y., Zelhof A.C., Jackson A.P., Gay N.J., Kane C.J.O. Amphiphysin is necessary for organization of the excitation–contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold T.F., Faelber K., Plattner N., Posor Y., Ketel K., Curth U., Schlegel J., Haucke V., Daumke O., Eschenburg S., Anand R., Manstein D.J., Noe F. Crystal structure of the dynamin tetramer. Nature. 2015 doi: 10.1038/nature14880. [DOI] [PubMed] [Google Scholar]
- Roux A., Koster G., Lenz M., Sorre B., Manneville J.-B., Nassoy P., Bassereau P. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer B., Hnia K., Gavriilidis C., Tronchère H., Tosch V., Laporte J. The myotubularin–amphiphysin 2 complex in membrane tubulation and centronuclear myopathies. EMBO Rep. 2013;14:907–915. doi: 10.1038/embor.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D., Elliott K.J., Wechsler-Reya R., Prendergast G.C. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- Segrest J.P., Jackson R.L., Morrisett J.D., Gotto A.M. A molecular theory of lipid—protein interactions in the plasma lipoproteins. FEBS Lett. 1974;38:247–253. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- Sidiropoulos P.N.M., Miehe M., Bock T., Tinelli E., Oertli C.I., Kuner R., Meijer D., Wollscheid B., Niemann A., Suter U. Dynamin 2 mutations in Charcot-Marie-Tooth neuropathy highlight the importance of clathrin-mediated endocytosis in myelination. Brain. 2012;135:1395–1411. doi: 10.1093/brain/aws061. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Shurland D., Newman-smith E.D., Pishvaee B., Bliek A.M., Van Der A model for dynamin self-assembly based on binding between three different protein domains. Biochemistry. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Smith L.L., Gupta V.A., Beggs A.H. Bridging integrator 1 (Bin1) deficiency in zebrafish results in centronuclear myopathy. Hum. Mol. Genet. 2014;23:3566–3578. doi: 10.1093/hmg/ddu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B., Callan-Jones A., Manzi J., Goud B., Prost J., Bassereau P., Roux A. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc. Natl. Acad. Sci. U.S.A. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susman R.D., Quijano-Roy S., Yang N., Webster R., Clarke N.F., Dowling J., Kennerson M., Nicholson G., Biancalana V., Ilkovski B., Flanigan K.M., Arbuckle S., Malladi C., Robinson P., Vucic S., Mayer M., Romero N.B., Urtizberea J.A., García-Bragado F., Guicheney P., Bitoun M., Carlier R.-Y., North K.N. Expanding the clinical, pathological and MRI phenotype of DNM2-related centronuclear myopathy. Neuromuscul. Disord. 2010;20:229–237. doi: 10.1016/j.nmd.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Takei K., Slepnev V.I., Haucke V., De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Cowling B.S., Hnia K., Mohr M., Oldfors A., Schwab Y., Yis U., Maisonobe T., Stojkovic T., Wallgren-Pettersson C., Laugel V., Echaniz-Laguna A., Mandel J.L., Nishino I., Laporte J. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- Tsutsui K., Maeda Y., Tsutsui K., Seki S., Tokunaga A. CDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem. Biophys. Res. Commun. 1997;236:178–183. doi: 10.1006/bbrc.1997.6927. [DOI] [PubMed] [Google Scholar]
- Wang L., Barylko B., Byers C., Ross J.A., Jameson D.M., Albanesi J.P. Dynamin 2 mutants linked to centronuclear myopathies form abnormally stable polymers. J. Biol. Chem. 2010;285:22753–22757. doi: 10.1074/jbc.C110.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock D.E., Hinshaw J.E., Schmid S.L. Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R., Sakamuro D., Zhang J., Duhadaway J., Prendergast G.C. Structural Analysis of the Human BIN1 Gene: evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem. 1997;272:31453–31458. doi: 10.1074/jbc.272.50.31453. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R.J., Elliott K.J., Prendergast G.C. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell. Biol. 1998;18:566–575. doi: 10.1128/mcb.18.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Baumgart T. BIN1 membrane curvature sensing and generation show autoinhibition regulated by downstream ligands and PI(4,5)P2. Biochemistry. 2014;53:7297–7309. doi: 10.1021/bi501082r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Shi Z., Baumgart T. Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization. PLoS One. 2014;9:e93060. doi: 10.1371/journal.pone.0093060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Kinuta M., Abe T., Liang S., Araki K., Cremona O., Di Paolo G., Moriyama Y., Yasuda T., De Camilli P., Takei K. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 2004;23:3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]