Abstract
The centrosome organizes microtubule arrays within animal cells and comprises two centrioles surrounded by an amorphous protein mass called the pericentriolar material (PCM). Despite the importance of centrosomes as microtubule-organizing centers, the mechanism and regulation of PCM assembly are not well understood. In C. elegans, PCM assembly requires the coiled-coil protein SPD-5. Here we found that recombinant SPD-5 could polymerize to form micrometer-sized porous networks in vitro. Network assembly was accelerated by two conserved regulators that control PCM assembly in vivo, Polo-like kinase-1 and SPD-2/Cep192. Only the assembled SPD-5 networks, and not unassembled SPD-5 protein, functioned as a scaffold for other PCM proteins. Thus, PCM size and binding capacity emerge from the regulated polymerization of one coiled-coil protein to form a porous network.
One Sentence Summary
Centrosome assembly in C. elegans involves self-assembly of an interconnected, micron-scale network of proteins.
Pericentriolar material (PCM), a matrix of proteins that regulates microtubule nucleation, anchoring, and dynamics (1, 2), assembles around centrioles to form centrosomes that contribute to cell division. The PCM scaffold is thought to form via the assembly of coiled-coil proteins (pericentrin and Cdk5RAP2/centrosomin in vertebrates and Drosophila, and SPD-5 in C. elegans (3–8)). PCM expansion during mitotic entry requires SPD-2/Cep192 and is regulated by mitotic kinases, including the Polo family kinase Plk1 (9–13). However, the mechanism of PCM assembly remains unclear, partly due to the lack of an in vitro system.
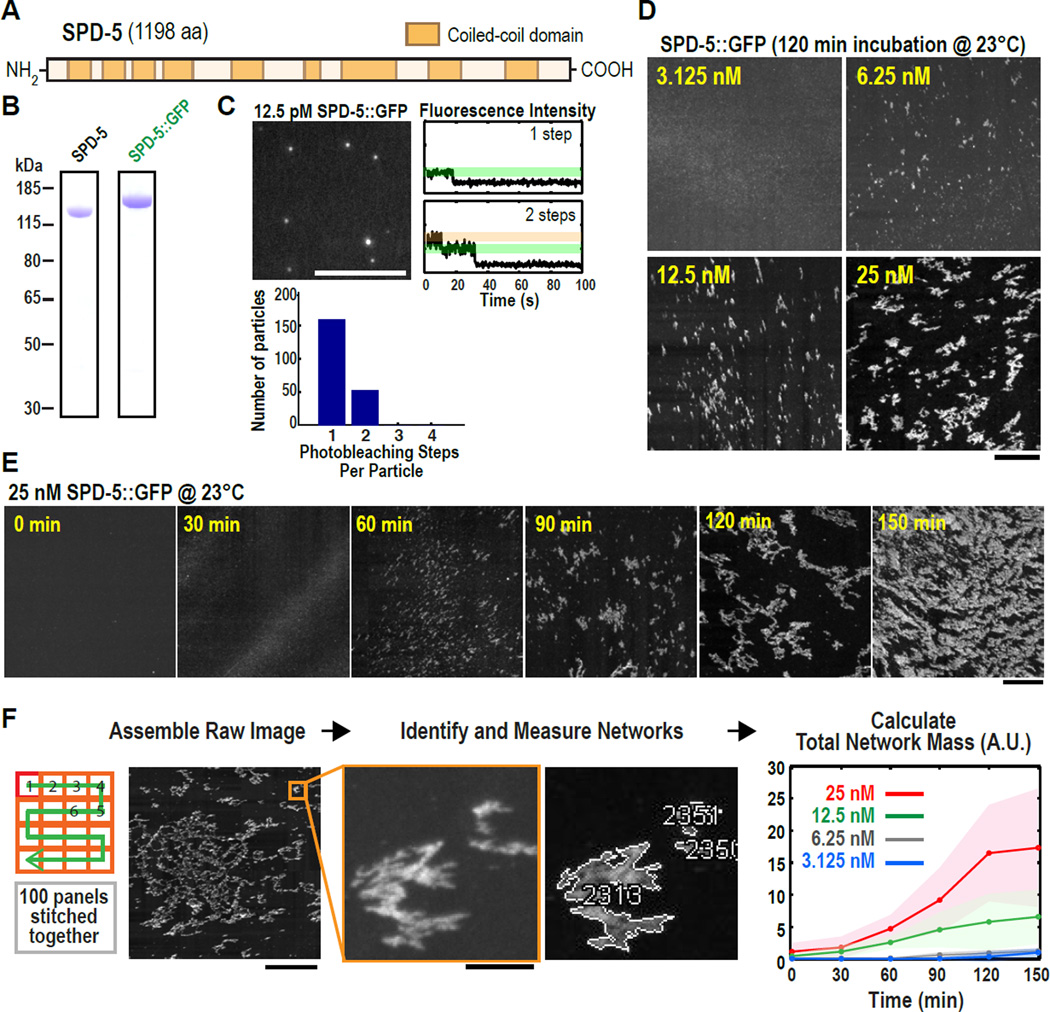
In C. elegans, PCM assembly requires SPD-5, a protein with 9 predicted coiled-coil domains (Fig. 1A) (7). Because coiled-coil domains often mediate homo-oligomerization (14), we hypothesized that SPD-5 self-association underlies PCM assembly. To test this idea, we purified full-length, GFP-labeled SPD-5 (SPD-5::GFP) from baculovirus-infected insect cells (Fig. 1A,B; Fig. S1A shows all proteins used in this study). Using total internal reflection microscopy to count GFP moieties in SPD-5::GFP particles by photobleaching revealed that SPD-5::GFP initially exists as monomers and dimers (Fig. 1C), consistent with in vivo data (15). To determine if SPD-5 could form larger assemblies, we shifted SPD-5::GFP from ice to 23°C and observed samples using widefield fluorescence microscopy. After 120 minutes, 25 nM SPD-5::GFP assembled into dense structures spanning several microns that we term “networks” (Fig. 1D); similar assemblies were observed by phase contrast microscopy using untagged SPD-5 (Fig. S1B). Network formation decreased with lower initial SPD-5::GFP concentrations (Fig. 1D). Temporal analysis using 25 nM SPD-5::GFP revealed single puncta that grew into extended meshworks (Fig. 1E); separate networks could also be observed coalescing during assembly (Fig. S1C). SPD-5 networks readily dissolved after dilution or mechanical disruption, indicating that network formation is reversible (Fig. S1D).
Fig. 1. Recombinant SPD-5 molecules polymerize into expansive networks over time in a concentration-dependent manner in vitro.
(A) SPD-5 sequence with coiled-coil domains predicted by the MARCOIL algorithm (31) at 90% threshold.
(B) Coomassie-stained gels depicting purified full-length SPD-5 and SPD-5::GFP.
(C) 12.5 pM SPD-5::GFP was squashed under a cover slip and imaged continuously by total internal reflection microscopy. The intensity of each fluorescent spot was measured over time and photobleaching steps per spot were counted (n = 210). Representative photobleaching profiles are shown. Scale bar, 5 µm.
(D) Visualization of different concentrations of SPD-5::GFP after 120 min. Scale bar, 25 µm.
(E) 25 nM SPD-5::GFP was incubated at 23°C, then squashed under a coverslip at 30 min intervals. Scale bar, 25 µm.
(F) Flowchart of automated SPD-5::GFP network quantification. For each sample, 100 images were collected, stitched together, a threshold was applied, and the networks were identified and measured. The graph plots the integrated intensity of all networks (Total Network Mass) per sample at each time point (n= 4–10; mean with 95% confidence intervals). Scale bars, 25 µm (first panel), 2.5 µm (inset).
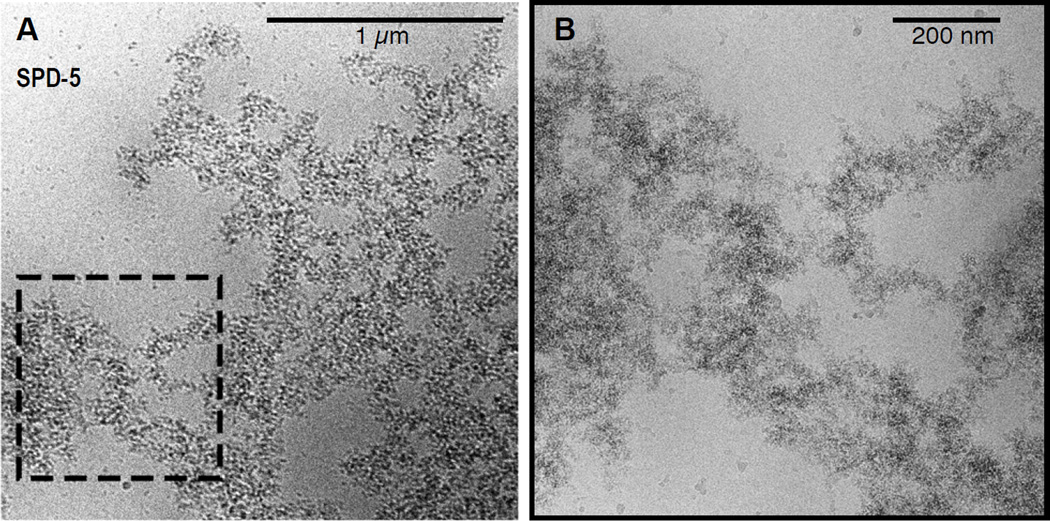
We calculated total SPD-5 network mass per sample by summing the integrated fluorescence intensity of all SPD-5::GFP networks (up to 200,000 per sample) in the imaged area (Fig. 1F). We also calculated the size distribution of the networks in each sample (Fig. S1E). The kinetics of SPD-5::GFP network formation fit a sigmoidal curve typical of growing polymers, including a characteristic lag time and plateau (Fig. 1F). Thus, SPD-5 alone can polymerize in vitro in a concentration and time-dependent manner to form extended networks that reach sizes comparable to fully expanded PCM in vivo (11). Low magnification cryo electron microscopy of SPD-5 networks revealed dense, amorphous assemblies interspersed with pores of various sizes (Fig. 2A); the assemblies lacked any distinct organization at higher magnification (Fig. 2B).
Fig. 2. High-resolution imaging of SPD-5 networks with Cryo-electron microscopy.
(A) Cryo-electron microscopy image of untagged SPD-5.
(B) Higher magnification view.
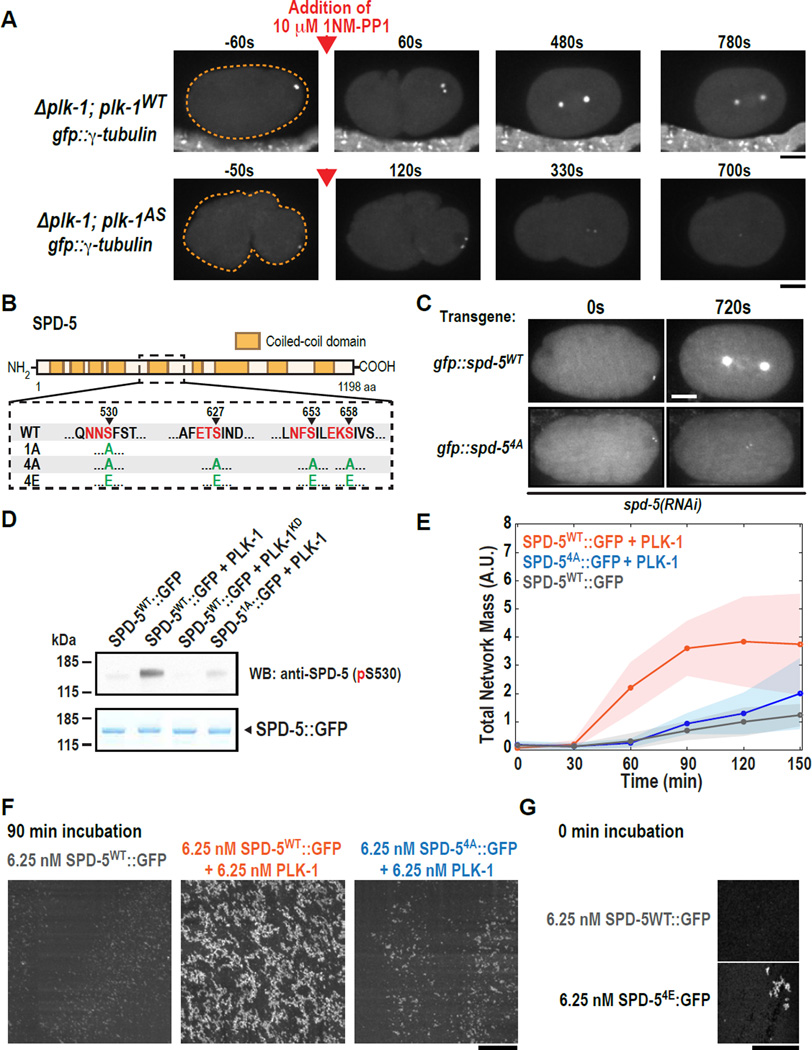
In vertebrate cells, Plk1 is required for the expansion and maintenance of the mitotic PCM (12, 13, 16–19). To determine if C. elegans PLK-1 has a similar regulatory role, we constructed strains deleted for the endogenous plk1 gene that also expressed either wild-type PLK-1 (PLK-1WT) or an analog-sensitive PLK-1 mutant (PLK-1AS) that can be inhibited by the drug 1NM-PP1 (20) (Fig. S2A). Monitoring recruitment of the PCM marker GFP::γ-tubulin revealed that addition of 1NM-PP1 in S-phase abolished mitotic GFP::γ-tubulin accumulation in embryos expressing PLK-1AS but not PLK-1WT (Fig. 3A). Addition of 1NM-PP1 to embryos arrested in mitosis also led to loss of GFP::γ-tubulin only in embryos expressing PLK-1AS (Fig. S2B). Thus, PLK-1 activity is required for both assembly and maintenance of the mitotic PCM.
Fig. 3. PLK-1 phosphorylation of SPD-5 drives PCM assembly in vivo and SPD-5 polymerization in vitro.
(A) plk-1WT (n=9) and plk-1AS (n=13) embryos expressing the PCM marker GFP::γ-tubulin were visualized by fluorescence confocal microscopy (orange dashed line is embryo outline). 10 µM 1-NM-PP1 (PLK-1AS inhibitor) was added to permeabilized embryos prior to mitotic entry (red arrow).
(B) Diagram of phospho-epitopes on SPD-5 and different mutant constructs. Canonical PLK-1 consensus motifs (32, 33) are indicated in red. The arrowheads indicate the phosphorylated residue in each motif. The complete set of phosphorylation sites identified by MS/MS is included in Table S3.
(C) Centrosome size was visualized in embryos expressing RNAi-resistant GFP::SPD-5WT or GFP::SPD-54A. Images are sum intensity projections from z-stacks. Scale bar, 25 µm. See also Fig. S3.
(D) In vitro kinase assay. SPD-5WT::GFP was incubated with buffer alone, PLK-1WT, or a kinase dead version of PLK-1 (PLK-1KD). Phosphorylation at serine 530 was detected by western blot using a phospho-specific antibody.
(E) Kinetics of 6.25 nM SPD-5WT::GFP network formation in vitro. Values represent mean with 95% confidence intervals (n= 8–14).
(F) Representative images of SPD-5::GFP networks from (E). Scale bar, 25 µm.
(G) A phosphomitetic version of SPD-5 (SPD-54E::GFP) already formed networks at t = 0 min, bypassing the need for incubation at room temperature. Scale bar, 25 µm.
PLK-1 could control PCM assembly by phosphorylating SPD-5 to promote its polymerization. Consistent with this hypothesis, centrosomal SPD-5 accumulation was impaired in plk-1(RNAi) embryos (Fig. S2C). Immunoprecipitation of SPD-5 from worm embryo extracts followed by mass spectrometry identified 19 phosphorylation sites (Table S3). Among others, three central residues (S530, S627 and S658) were identified as likely to be PLK-1 target sites (Fig. 3B, Table S2; (21)). Residue S653 was also predicted to be a PLK-1 phosphorylation site, but we could not confirm its phosphorylation status. To test the importance of these residues, we generated RNAi-resistant transgenes encoding GFP fusions with wild-type SPD-5 (GFP::SPD-5WT) or with a mutant in which S530, S627, S653 and S658 were mutated to alanine (GFP::SPD-54A) (Fig. S3A). Both transgenes were expressed at similar levels (Fig. S3B). All subsequent experiments were performed after RNAi silencing of endogenous spd-5 to ensure that the RNAi-resistant transgenes were the sole source of SPD-5 (Fig. S3C).
In embryos expressing GFP::SPD-5WT, centrioles entering the embryo during fertilization rapidly acquired a small shell of GFP::SPD-5WT and the PCM expanded as cells progressed into mitosis (Fig. 3C). In embryos expressing GFP::SPD-54A, centrioles acquired a small shell of GFP::SPD-54A, but the PCM failed to expand (Fig. 3C, S3D–G), similar to the plk-1(RNAi) phenotype (Fig. S2C). γ-tubulin recruitment was also impaired and centrosomes failed to separate (Fig. S3F,G). Consistent with an essential role in vivo, PLK-1 could also phosphorylate SPD-5 in vitro. An antibody raised against phosphorylated S530 recognized SPD-5 after incubation with PLK-1 and ATP, but not if S530 was mutated to alanine (SPD-51A), or if a kinase dead version of PLK-1 (PLK-1KD) was used (Fig. 3D). Additionally, a pan-phospho-specific antibody recognized previously de-phosphorylated SPD-5 only after incubation with PLK-1 and ATP (Fig. S5A). To investigate the relative importance of the four PLK-1 phosphorylation sites, we mutated S530, S627, S653 and S658 to alanine individually. Only the S658A mutant appreciably reduced PCM assembly (Fig. S4). A double S653A S658A mutant reduced PCM assembly to an extent comparable to the 4A mutant (Fig. S4). Thus, S658 is the most important site in this region and phosphorylation of S653 can partially compensate when S658 cannot be phosphorylated. Phosphorylations on S530 and S627 are not essential, but could also have redundant roles. Although these results suggest that phosphorylation of SPD-5 on these central sites controls mitotic PCM expansion, they do not exclude the possibility that a second PLK-1-based mechanism governs γ-tubulin maintenance.
Our results suggest that phosphorylation of SPD-5 by PLK-1 is required for PCM expansion in vivo. We therefore wanted to determine if phosphorylation by PLK-1 would also promote SPD-5::GFP network formation in vitro. For all subsequent growth experiments, an initial concentration of 6.25 nM SPD-5::GFP was used. In the absence of PLK-1, SPD-5WT::GFP networks appeared after 90 min. Addition of 6.25 nM PLK-1 and 0.2 mM ATP enhanced network formation, such that SPD-5 networks appeared after 30 min (Fig. 3E,F). This stimulation was abrogated when the 4A mutant (SPD-54A::GFP) was used (Fig. 3E,F). SPD-5WT::GFP and SPD-54A::GFP polymerized similarly in the absence of PLK-1, indicating that, although insensitive to the effects of PLK-1, SPD-54A::GFP is otherwise functional (Fig. S5B). While SPD-54A::GFP formed networks at later time points in vitro, centrosomes did not expand in cells expressing the 4A mutant in vivo (Fig. 3C). This could be due to the short duration of the C. elegans embryo cell cycle. A SPD-5 mutant harboring four phospho-mimetic mutations (SPD-54E::GFP; Fig. 3B) already formed networks at t=0 min, suggesting that phosphorylation at these four residues is sufficient to catalyze SPD-5 polymerization (Fig. 3G). Thus, PLK-1 enhances SPD-5 polymerization through direct phosphorylation in vitro.
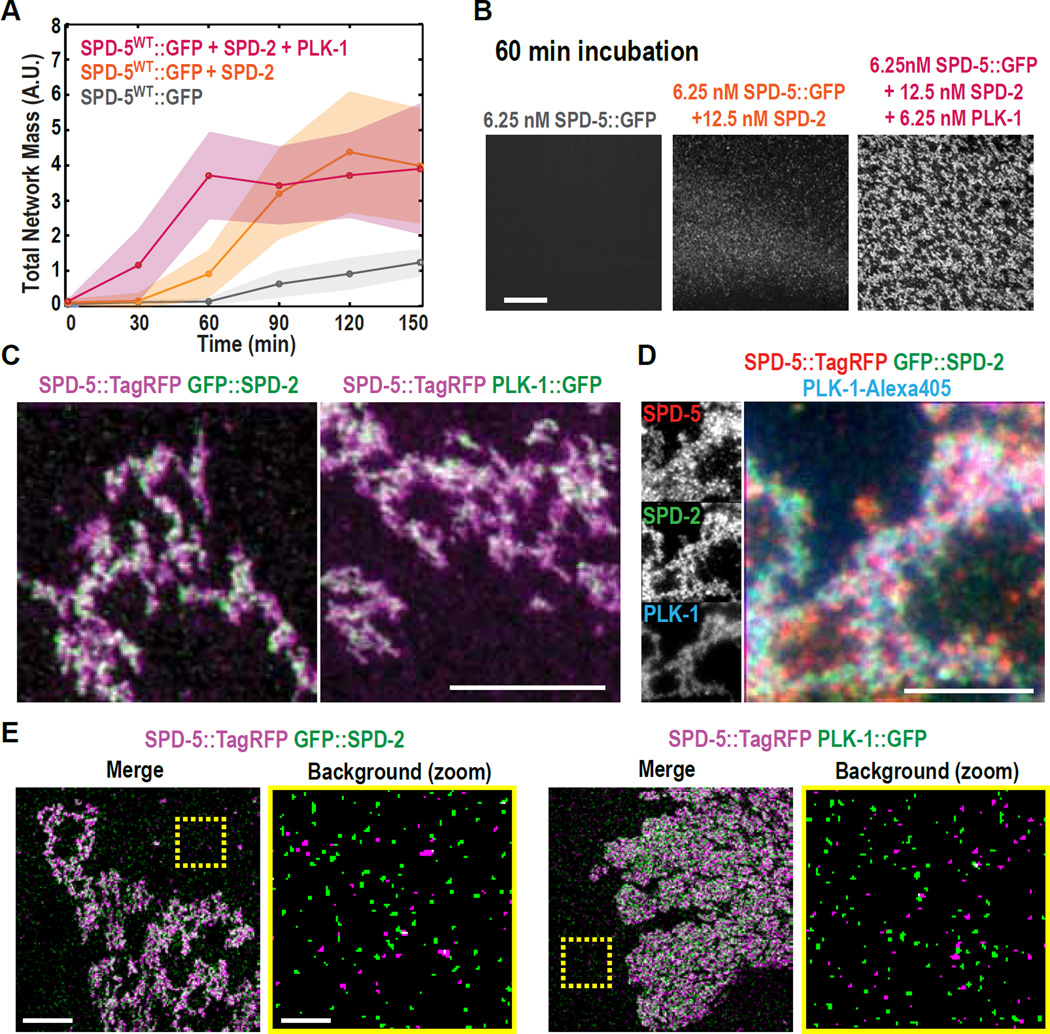
SPD-2/Cep192 has a conserved role in PCM assembly (9, 10, 22–25), yet its precise role remains unclear. We therefore addressed the role of C. elegans SPD-2 in PCM formation using our in vitro system. Addition of 12.5 nM purified full-length SPD-2 enhanced SPD-5::GFP network assembly, such that networks were seen after 60 min (Fig. 4A,B), indicating that SPD-2 by itself can promote SPD-5 polymerization. Simultaneous addition of 6.25 nM PLK-1 and 12.5 nM SPD-2 enhanced SPD-5::GFP network formation beyond what was observed by adding either PLK-1 or SPD-2 alone (Figs. 4A, S6A). Thus, SPD-2 and PLK-1 cooperatively enhance SPD-5 polymerization in vitro, consistent with the essential roles for both proteins in PCM expansion in vivo.
Fig. 4. SPD-2 and PLK-1 independently bind to SPD-5::GFP networks and cooperatively stimulate network formation in vitro.
(A) Kinetics of 6.25 nM SPD-5::GFP network formation in the presence of 12.5 nM SPD-2 and/or 6.25 nM PLK-1. Values represent mean with 95% confidence intervals (n= 8–14).
(B) Representative images of SPD-5::GFP networks from (A) after 60 min. Scale bar, 25 µm.
(C) Dual color images of SPD-5::TagRFP networks assembled in the presence of GFP::SPD-2 or PLK-1::GFP. Scale bar, 5 µm. See also Figure S6.
(D) Triple color images of SPD-5::TagRFP networks assembled in the presence of GFP::SPD-2 and PLK-1-Alexa405. Scale bar, 5 µm.
(E) Co-localization analysis using structured illumination microscopy. For each image pair, the right panel depicts a zoomed-in image of the non-network-assembled SPD-5::TagRFP particles selected from the area bounded by the yellow box. Scale bar, 5 µm (left panel), 1 µm (zoomed in, right panel).
In vivo, loss of SPD-5 abolishes recruitment of all known PCM proteins, suggesting that SPD-5 forms the structural framework of the PCM (7). To test if in vitro assembled SPD-5 networks could serve as scaffolds for the recruitment of other PCM proteins, we observed SPD-5::TagRFP networks assembled in the presence of fluorescently-labeled PLK-1 and SPD-2. PLK-1::GFP and GFP::SPD-2 were both recruited to SPD-5::TagRFP networks independently of each other (Fig. 4C). Purified γ-tubulin and four non-centrosomal proteins were not recruited to SPD-5::TagRFP networks (Fig. S6B,C), demonstrating that the association of PLK-1 and SPD-2 with SPD-5 networks is specific. Alexa405-labeled PLK-1 and GFP::SPD-2 were simultaneously recruited to the same networks (Fig. 4D). Thus, PLK-1 and SPD-2 can exist concomitantly on SPD-5 networks and their localization is not interdependent.
Structured illumination microscopy revealed that non-network-associated SPD-5::TagRFP particles did not co-localize with either PLK-1::GFP or GFP::SPD-2 (Fig. 4E), suggesting that only the network-assembled form of SPD-5 can bind to PLK-1 and SPD-2. To further test this possibility, we determined the amount of SPD-2 or PLK-1 that co-precipitated with beads coated with non-assembled SPD-5. As expected, we did not detect enrichment in the amount of SPD-2 or PLK-1 pulled down by the SPD-5-coated beads as compared to the control beads (Fig. S6D). Thus, SPD-5 networks, rather than unassembled SPD-5 molecules, serve as scaffolds for the recruitment of SPD-2 and PLK-1 in vitro. Indeed, a recent study shows that SPD-5, SPD-2, and PLK-1 do not interact prior to their incorporation into the PCM in living embryos (15). Yeast-two-hybrid analysis has shown that a SPD-5 fragment can interact with SPD-2 (26). In light of our findings, it is possible that this interaction domain is blocked when SPD-5 is monomeric, but becomes accessible when SPD-5 molecules assemble into a network.
In summary, we have developed an in vitro system for studying regulated PCM assembly. The coiled-coil protein SPD-5 can polymerize into interconnected, porous networks that specifically recruit downstream PCM proteins such as PLK-1, SPD-2, and ZYG-9 (chTOG homolog) (Fig. S6B,C). Thus, SPD-5 networks are likely to represent the structural framework of the PCM. PLK-1 kinase directly phosphorylates SPD-5 to promote its polymerization and mitotic PCM expansion, and SPD-2 makes an additional independent contribution to PCM assembly. Consistent with our findings for C. elegans SPD-5, PCM formation in Drosophila requires SPD-2 and Polo kinase phosphorylation at multiple sites on centrosomin (Cdk5RAP2) (27, 28). Like SPD-5, centrosomin is a large coiled-coil protein that recruits downstream PCM proteins (7, 27–30). Thus, SPD-2/Polo kinase-driven polymerization of a scaffolding component is a conserved mechanism for PCM assembly.
Supplementary Material
Acknowledgments
We thank the Protein Expression & Purification (Dresden, Martinsried) and Light Microscopy (Dresden) facilities, M. Braun, H. Petzold, B. Ferreira Gomes, M. Podolski, and A. Hibbel for experimental help; and L. Pelletier for helpful comments. This project was funded by the Max Planck Society and the European Commission's 7th Framework Programme grant (FP7-HEALTH-2009-241548/MitoSys) to A.H. and an NIH grant (R01-GM074207) to K.O. K.O. received salary and additional support from the Ludwig Institute for Cancer Research. J.S.A. and J.B. were funded in part by the EU FP7 PROSPECTS network grant (HEALTH-F4-2008-201648). J.B.W. was supported by Alexander von Humboldt and EMBO fellowships. J.M. was supported by EMBO and HFSP fellowships. E.Z. was supported by a Deutsche Forschungsgemeinschaft fellowship. The supplemental materials contain additional data.
Footnotes
Supporting Online Material:
Materials and Methods
Figs. S1–S6
Tables S1–S3
Data Set D1
References (33–42)
References and Notes
- 1.Nigg EA, Raff JW. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Woodruff JB, Wueseke O, Hyman AA. Phil. Trans. R. Soc. B. 2014;369:1–10. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dictenberg JB, et al. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mol. Biol. Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchman JJ, et al. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 6.Conduit PT, et al. Curr. Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Hamill DR, Severson AF, Carter JC, Bowerman B. Dev. Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelletier L, et al. Curr. Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Dev. Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 11.Decker M, et al. Curr. Biol. 2011;21:1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Lane HA, Nigg EA. J. Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haren L, Stearns T, Lüders J. PLoS ONE. 2009;4(6):e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 15.Wueseke O, et al. Mol. Biol. Cell. 2014;25:2984–2992. doi: 10.1091/mbc.E13-09-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumara I, et al. Curr. Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Lénárt P, et al. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 18.Santamaria A, et al. Mol. Biol. Cell. 2007;18:4024–4036. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K, Rhee K. J. Cell Biol. 2011;195:1093–1101. doi: 10.1083/jcb.201106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop AC, et al. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, et al. Brief. Bioinformatics. 2013;14:344–360. doi: 10.1093/bib/bbs041. [DOI] [PubMed] [Google Scholar]
- 22.Dix CI, Raff JW. Curr. Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Curr. Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Ferreria MA, et al. Curr. Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhu F, et al. Curr. Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 26.Boxem M, et al. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conduit PT, et al. Elife. 2014;3:e03399. doi: 10.7554/eLife.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conduit PT, et al. Dev. Cell. 2014;28:659–669. doi: 10.1016/j.devcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megraw TL, Li K, Kao LR, Kaufman TC. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- 30.Lucas EP, Raff JW. J. Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delorenzi M, Speed T. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 32.Santamaria A, et al. Mol. Cell. Proteomics. 2010;10(1) doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosstessner-Hain K, et al. Mol. Cell. Proteomics. 2011;10(11) doi: 10.1074/mcp.M111.008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stiernagle T. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frøkjær-Jensen C, et al. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lettman MM, et al. Dev. Cell. 2013;25:284–298. doi: 10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmons L, Fire A. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 38.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Genome Biol. 2001;2(1):research0002–research0002.10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindelin J, et al. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho A, et al. PLoS ONE. 2011;6(9):e24656. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamath RS, et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 42.Cox J, Mann M. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.