Abstract
Exosomes are released by cells as self-contained vesicles with an intact lipid bilayer that encapsulates a small portion of the parent cell. Exosomes have been studied widely as information-rich sources of potential biomarkers that can reveal cellular physiology. We suggest that quantification is essential to understand basic biological relationships between exosomes and their parent cells and hence the underlying interpretation of exosome signals. The number of methods for quantifying exosomes has expanded as interest in exosomes has increased. However, a consensus on proper quantification has not developed, making each study difficult to compare to another. Overcoming this ad hoc approach will require widely available standards that have been adequately characterized, and multiple comparative studies across platforms. We outline the current status of these technical approaches and our view of how they can become more coherent.
Keywords: Exosomes, Nanosight, Flow Cytometry, Tunable Resistive Pulse Sensing
Introduction
Exosomes are small vesicular structures, ranging from 40-150 nm in diameter, that are released by almost all cell types. Exosomes are important because they contain a wide range of cargo that is not typically thought to be released by viable cells including transcription factors, cell-surface receptors, cytosolic and nuclear proteins, microRNA and mRNA. Hence, exosomes represent a novel source of biomarkers for the state of the cells from which they are derived.
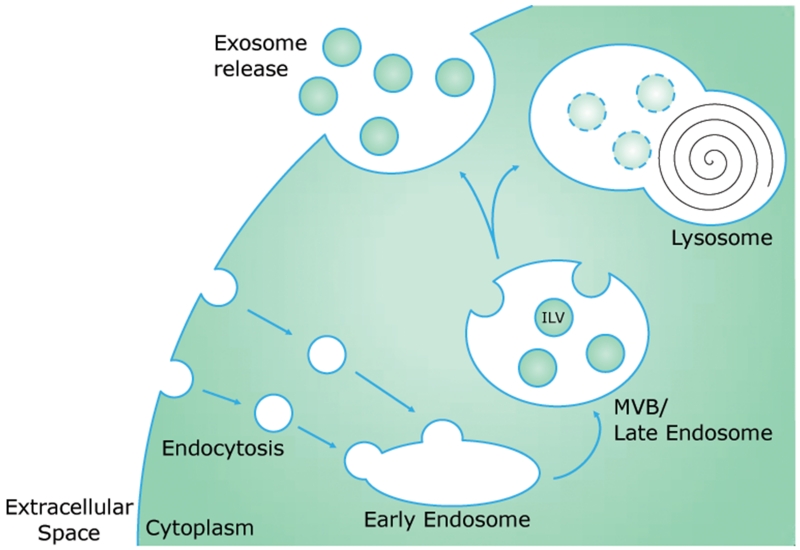
Exosomes are formed as part of the endosomal pathway (Figure 1), as opposed to other classes of EVs, which are formed by direct outward budding from the plasma membrane. Plasma membrane-derived endocytosis results in the formation of endocytic vesicles inside the cell, each of which contains extracellular fluid and has an inside-out membrane orientation. Endocytic vesicles can then fuse with each other to form an endosome. As the endosome grows and matures, it becomes a multivesicular body (MVB). During this maturation process, the endosome/MVB outer membrane deforms inward, generating intraluminal vesicles inside of the endosome/MVB, each of which contains cytosolic contents and now has a right side-out membrane orientation. The MVB then follows one of two paths: it either fuses with a lysosome to degrade and recycle its contents (endolysosomal degradative pathway), or it is routed to the cell surface (exosome release). When the MVB fuses with the plasma membrane, its intraluminal vesicles are released into the extracellular space as exosomes (Thery et al., 2002).
Figure 1.
Exosomes are formed via the endosomal pathway. Endocytic vesicles result from invagination of the plasma membrane and can fuse to form early endosomes. During maturation from early to late endosomes, also called multivesicular bodies (MVBs), invaginations in the endosomal membrane form intraluminal vesicles (ILVs). The late endosome/MVB then either fuses with a lysosome to degrade its contents, or fuses with the plasma membrane and releases the ILVs into the extracellular space as exosomes.
In our search for novel urine biomarkers for acute kidney injury, we tested whether exosomes could contain miRNA, and found that at least two abundant kidney miRNAs were detectable under normal conditions in urine exosomes. Furthermore, when rats were subjected to ischemia/reperfusion injury, a classic acute kidney injury animal model, we found that the urine exosomal levels of one of these miRNAs (miR-192) increased as much as 60-fold compared to control levels (Zhou et al., 2008). In contrast, a wide variety of stimuli change cellular miRNA levels by only two- to three-fold across a broad range of cell types and tissues. Two main hypotheses emerged that could explain this discrepancy: a) the number of miRNA molecules per exosome changes, or b) the total number of exosomes released containing this specific miRNA increases. In either case, the number of exosomes in urine would need to be determined to further our fundamental understanding of the biology underlying our biomarker observations.
The importance of quantifying exosomes is illustrated by three groups, examining the stoichiometry of miRNAs per exosome. Stevanato et al (Stevanato et al., 2016) observed a stoichiometry of 12-60 molecules of a specific miRNA per vesicle in exosomes derived from an immortalized neural stem cell line. Akers et al. (Akers et al., 2013; Akers et al., 2016) isolated extracellular vesicles, primarily exosomes, from cerebrospinal fluid (CSF) samples and then determined the number of exosomes by Nanosight Nanoparticle Tracking Analysis and the total content of specific miRNAs by qRT-PCR. The stoichiometry varied widely, ranging from 10−5 to ~10 miRNA molecules per vesicle. Chevillet et al. (Chevillet et al., 2014) similarly found a range of 10−5 to 10−1 miRNA molecules per vesicle in exosomes derived from plasma, seminal fluids, cultured dendritic cells, a mast cell leukemia cell line, and an ovarian cancer cell line. The wide range of stoichiometry suggests that not only are exosomes diverse in terms of their miRNA content, but also that the presence or absence of a small percentage of exosomes containing a single molecule of miRNA can skew the final value. This is particularly true in our case, as the urine exosome population is very heterogeneous, comprised of exosomes derived from the numerous epithelial cell types that line the nephron as well as the rest of the genitourinary tract. Additionally, understanding changes in exosome release is further complicated by the variation in urine composition throughout the day. The kidneys can alter urine osmolarity across almost two orders of magnitude, making it challenging to understand exosomal biomarker concentrations, when the normalization method uses volume as a denominator (per ml). Efforts to normalize to urine markers that are excreted at a more constant rate, such as creatinine (per mg creatinine), mitigates against some variability. Unfortunately these markers/denominators can also vary during non-steady states (such as kidney injury). When urine collection can be timed, an excretion rate (per hour or per day) does not need to be normalized, and two biomarker excretion rates can be properly compared, even if their concentrations are very different.
Techniques for Exosome Quantification
There are a variety of techniques available that are currently used for exosome quantification, but there is a lack of consensus in the field and there is a need for more rigorous head-to-head comparisons of the different techniques.
Nanosight Nanoparticle Tracking Analysis
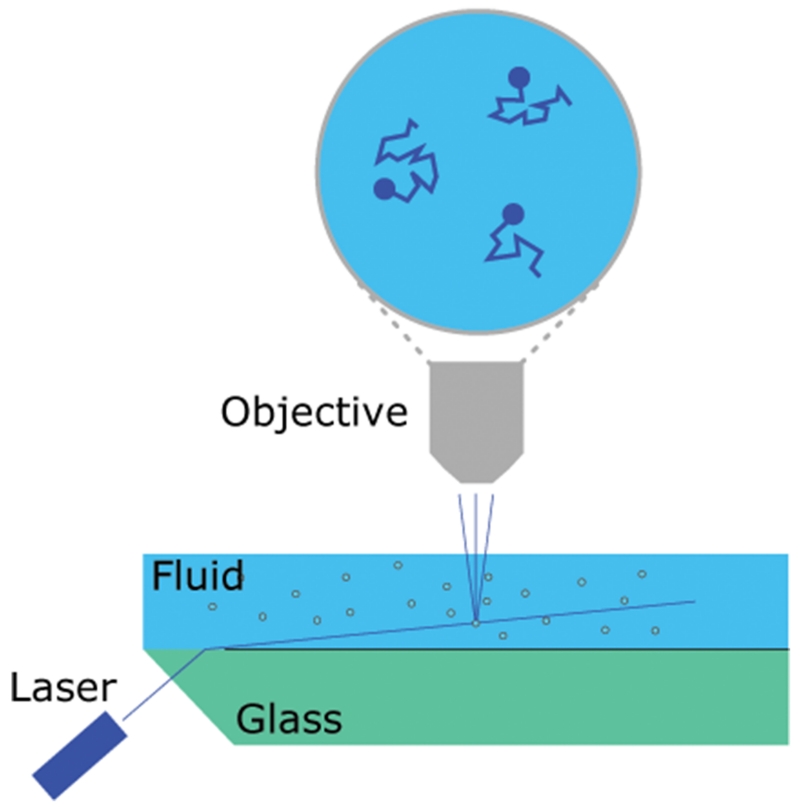
Nanosight Nanoparticle Tracking Analysis (Malvern Instruments, Figure 2) provides a combined analysis of Brownian motion via light scattering to count and size nanoparticles in a liquid suspension (Dragovic et al., 2011; Filipe et al., 2010; Gardiner et al., 2013; Oosthuyzen et al., 2013)). Particles in suspension are passed through a flow chamber and are illuminated using a laser source. The light scatter produced from this is recorded using a video camera. The instrument is able to account for net flow, allowing for the addition of a syringe pump to the system. The use of a syringe pump improves measurement quality due to the significantly larger quantity of unique particles analyzed. Nanosight NTA is currently considered to be the best method available for exosome quantification because it does not rely on detection of a specific marker and because the exosomes can be suspended in a wide range of solutions. Despite this, there are still important limitations to using Nanosight NTA for exosome quantification. Nanosight instruments have a somewhat limited dynamic range for particle concentration measurements of ~107-109 particles/mL, and it is important that the sample be diluted to a concentration within that range to obtain accurate and reproducible size distribution and concentration data. There is also the potential for some variability based on the choice of camera level and detection threshold settings used, making it important to ensure that users are properly trained on how to use the instrument and NTA software (Filipe et al., 2010)). Additionally, there are inconsistencies in the Nanosight protocols used in different laboratories that can further complicate comparisons. The most critical decision is whether or not to use a syringe pump with the instrument. The syringe pump slowly pushes particles through the chamber which allows for the visualization and analysis of more unique particles in the sample compared to one static field of view. Although many Nanosight users do not use a syringe pump, we have found that it greatly improves the quality and reproducibility of Nanosight data.
Figure 2.
Schematic representation of the Nanosight NTA system.
A laser beam refracts at the glass/liquid interface illuminating a plane within the flow chamber. Particles in the solution scatter light that is detected by a microscope objective and video camera focused on the illuminated plane. Due to Brownian motion the position of each particle changes between frames. Small particles will move further than large particles, enabling the distance moved to be used to calculate the particle size. A size distribution can be developed by studying multiple particle tracks.
Tunable Resistive Pulse Sensing
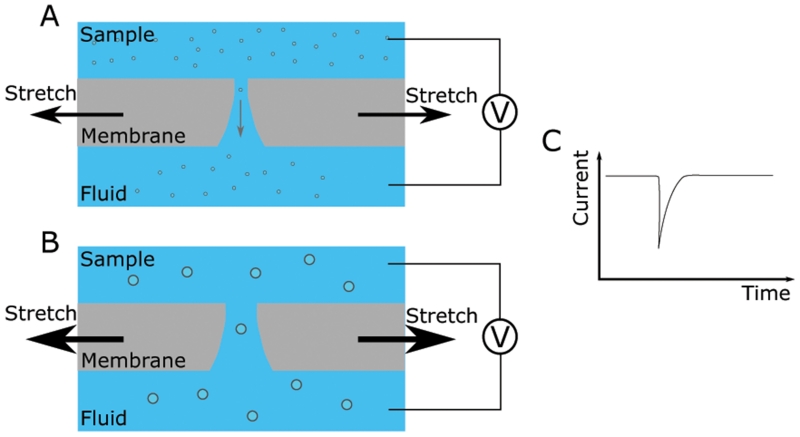
Tunable resistive pulse sensing (TRPS) (Izon Science) detects the passage of individual particles through a pore in a membrane (Figure 3), and operates similar to a Coulter counter. A pore is created in a membrane and a voltage is applied across the membrane creating a current through the pore (Blundell et al., 2015). A sample is applied to one side of the membrane and individual particles pass through the pore driven by a pressure difference and the voltage. As the particles have a higher resistance than the electrolyte they momentarily reduce the current passing through the pore. This can be detected providing both concentration and size information. The concentration is calculated from the frequency of events; the particle size is calculated from the drop in current. The membrane used is elastic and can be stretched to alter the pore size. By tuning the size of the pore the sensitivity and accuracy of the technique can be optimized for every sample. Momentarily dilating the pore or reversing the pressure differential across the membrane can be used to clear any blockages. Changes in the pressure and voltage applied across the membrane can also be used to detect particle charge, although to date, particle charge has not been reported for exosomes (Maas et al., 2014). Drawbacks to using TRPS for exosome quantification include issues with pore clogging and instrument sensitivity. It has been shown that a significant portion of smaller exosomes can fall below the detection threshold for TRPS when compared to EM quantification, an observation that may explain variability in TRPS-determined exosome data in the literature (Anderson et al., 2015). Additionally, TRPS analysis is limited to counting and sizing all particles within a given sample, while other techniques provide the ability to discriminate between particle subpopulations using antibody binding and fluorescence.
Figure 3.
Schematic representation of the TRPS instrument.
A voltage is applied across a stretchable membrane containing a pore (A). The dynamic range of the instrument can be tuned to match the particle properties by stretching the membrane to change the pore diameter (B). The current across the membrane is monitored and will drop as particles pass through the pore, momentarily increasing its electrical resistance (C). The number of events can be used to calculate the concentration and the magnitude of the event used to develop a size distribution.
Vesicle Flow Cytometry
Flow cytometry detects particles suspended in a fluid by their interaction with a laser beam as they flow through a detection cell. A sheath fluid is used to spatially confine particles in the center of the detection cell. As particles pass through the laser beam they scatter light, and if appropriate fluorophores are present, the particles also fluoresce. Flow cytometry instruments used for the detection of exosomes have either been commercial instruments originally designed for cells then modified to lower the detection threshold (Groot Kormelink et al., 2016; van der Vlist et al., 2012), or custom instruments (Pospichalova et al., 2015; Yang et al., 2009). Modifications include smaller probe volumes, reduced flow rates and changes to the optics and detectors used. Detection of scattered light requires the highest sensitivity. Although forward and side scatter is widely used for the analysis of cells the intensity is proportional to the sixth power of the particle diameter, meaning that for very small particles such as exosomes the signal intensity is very close to the background noise. Fluorescence can provide a stronger signal and has been used on modified commercial instruments. A limitation of flow cytometry is the insensitivity in the smallest size range, which may potentially skew size distributions towards larger particles. Even microvesicles, which are larger than exosomes, can have a “swarming” effect, where a threshold concentration of particles is required to see any signal, which at the low end of detection is not linear with respect to concentration (Erdbrugger et al., 2014). Determining size is also challenging, as differences in the optical properties of any standard will alter the relationship between particle size and signal intensity. Analysis by side scatter requires a standard of similar refractive index to exosomes, and for fluorescence an equivalent number of bound fluorophores is needed.
Surface Plasmon Resonance
Unlike the above techniques, surface plasmon resonance (SPR) based approaches for quantification of exosomes have not yet been developed commercially. SPR can detect exosome binding to a surface by the changes induced in the optical properties of the surface (Im et al., 2015; Zhu et al., 2014). The requirement for binding potentially compromises the determination of particle concentration with existing reports using immuno-affinity for CD63, a widely expressed, but not necessarily universal exosome marker. There are conflicting reports in the literature, with at least one claiming CD63 as the most universal outward facing immune-affinity target on exosomes (Kowal et al., 2016), and at least one report claiming that other markers work better (Salih et al., 2016). Thus, the detector will miss any exosomes not displaying CD63. A combination of multiple markers could improve detection, and SPR may yet find its niche in quantifying exosome subpopulations or individual proteins present in exosomes, rather than in determining the number or concentration of exosomes.
Electron Microscopy
As the first method used to identify exosomes, electron microscopy (EM) serves as a gold standard for verifying the quality of exosome preparations. Although EM has been used to count exosomes, this method is not as commonly used for several reasons, primarily the labor-intensive, low-throughput nature of the technique. Standard EM methods are likely to cause exosome loss during dehydration and embedding (Thery et al., 2006). Thus, EM-based quantification is likely to significantly underestimate exosome numbers (Akers et al., 2016). Additionally, there is a lack of studies quantifying the yield of the EM sample preparation process, and there is little information on the variability in yield for this process (Chairoungdua et al., 2010; Fernandez-Llama et al., 2010).
Discussion
Developing a consensus on methods and standards for exosome quantification will be important in bringing prospective exosomal biomarkers closer to clinical use. Independently verified NIST-certified nanosphere standards are currently used to calibrate instruments to measure particle size properly. Concentration standards do not yet exist but are needed to calibrate exosome counting techniques, as significant differences have been reported in comparative studies (Akers et al., 2016; van der Pol et al., 2014). Currently, the only option is to use the sizing nanosphere standards back-calculated to an approximate particle concentration based on density. There are significant limitations to this approach, as the calculated value has only two significant figures due to the density data provided by the manufacturer of the nanospheres. This is particularly important when for some of these techniques, it is necessary to perform a 1,000,000-fold dilution of a standard in order to have a particle concentration within an instrument’s dynamic range, a process that can introduce significant additional variability into the measured concentration. In the absence of an independently verified concentration standard, sharing an exosome preparation would be an easy way to normalize measurements and reproducibility between laboratories, but this still does not establish absolute quantitation. Ideally, a set of certified concentration standards should cover the dynamic ranges of each of these techniques and consist of particles with a size range similar to exosomes. Their composition should match the physical properties exosomes as closely as possible, as differences between standard and sample can introduce artifact, such as the impact of refractive index on flow cytometry (Chandler et al., 2011) or Nanosight NTA (Filipe et al., 2010) measurements. The development of a particle concentration standard would also allow for the use of the standard addition method (Bader, 1980; Miller and Miller, 1988) to verify instrument readings.
Other ways to normalize exosomal biomarkers have been proposed. Webber et al. propose a reasonable method for normalization that involves using total protein measurements (Webber and Clayton, 2013), but this would require establishing the “normal” total protein content for each source of exosomes. This approach would be difficult in urine, where liquid phase protein concentration can vary over 3 orders of magnitude (normal to nephrotic syndrome). We have published urine exosomal biomarker data normalized to urine creatinine, a marker of kidney function (Zhou et al., 2013). Urine exosomal biomarkers have also been normalized by using relative Tamm Horsfall protein measurements by western blot (Fernandez-Llama et al., 2010). Additionally, for exosome studies based on cell-culture systems, groups have used cell counts as a normalizing factor, which is less than ideal. We think that normalization based on exosome number provides the most direct comparison and represents the most generalizable option.
Direct quantification of exosomes is a relatively new field, and there is currently no consensus on either an optimal approach or how to properly compare results using different approaches. Ultimately, these techniques require continued study identifying their reproducibility, dynamic range, sensitivity, specificity, and the appropriate protocols to maintain their performance between laboratories.
While much is understood about exosome biogenesis in the normal state, and many studies have examined the relative amounts of exosomal biomarkers in disease versus normal states, absolute quantification has been missing. Rather than inferring that the changes in exosome cargo reflect changes in the parent cell, absolute quantification can directly address the question of whether sequestration of cargo has changed or whether exosome production has changed. Similarly, absolute quantification would allow us to examine the yield of exosomes in biofluids and account for the fate of exosomes once they have been released from their cell of origin.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, Kalinina J, Hua W, Kesari S, Mao Y, Breakefield XO, Hochberg FH, Van Meir EG, Carter BS, Chen CC. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu CC, Hochberg FH, Chen CC, Carter BS. Comparative Analysis of Technologies for Quantifying Extracellular Vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF) PLoS One. 2016;11(2):e0149866. doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W, Lane R, Korbie D, Trau M. Observations of Tunable Resistive Pulse Sensing for Exosome Analysis: Improving System Sensitivity and Stability. Langmuir. 2015;31(23):6577–6587. doi: 10.1021/acs.langmuir.5b01402. [DOI] [PubMed] [Google Scholar]
- Bader M. A Systematic-Approach to Standard Addition Methods in Instrumental Analysis. J Chem Educ. 1980;57(10):703–706. [Google Scholar]
- Blundell ELCJ, Mayne LJ, Billinge ER, Platt M. Emergence of tunable resistive pulse sensing as a biosensor. Anal Methods-UK. 2015;7(17):7055–7066. [Google Scholar]
- Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190(6):1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler WL, Yeung W, Tait JF. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer. J Thromb Haemost. 2011;9(6):1216–1224. doi: 10.1111/j.1538-7836.2011.04283.x. [DOI] [PubMed] [Google Scholar]
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7(6):780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrugger U, Rudy CK, M EE, Dryden KA, Yeager M, Klibanov AL, Lannigan J. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry A. 2014;85(9):756–770. doi: 10.1002/cyto.a.22494. [DOI] [PubMed] [Google Scholar]
- Fernandez-Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T, Knepper MA. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77(8):736–742. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of extracellular vesicles. 2013:2. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot Kormelink T, Arkesteijn GJ, Nauwelaers FA, van den Engh G, Nolte-’t Hoen EN, Wauben MH. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytometry A. 2016;89(2):135–147. doi: 10.1002/cyto.a.22644. [DOI] [PubMed] [Google Scholar]
- Im H, Shao H, Weissleder R, Castro CM, Lee H. Nano-plasmonic exosome diagnostics. Expert review of molecular diagnostics. 2015;15(6):725–733. doi: 10.1586/14737159.2015.1041378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968–977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SL, De Vrij J, Broekman ML. Quantification and size-profiling of extracellular vesicles using tunable resistive pulse sensing. J Vis Exp. 2014;(92):e51623. doi: 10.3791/51623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Miller JN. Statistics for analytical chemistry. E. Horwood ; Halsted Press; Chichester, England New York: 1988. [Google Scholar]
- Oosthuyzen W, Sime NE, Ivy JR, Turtle EJ, Street JM, Pound J, Bath LE, Webb DJ, Gregory CD, Bailey MA, Dear JW. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol. 2013;591(23):5833–5842. doi: 10.1113/jphysiol.2013.264069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. Journal of extracellular vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih M, Fenton RA, Knipscheer J, Janssen JW, Vredenbregt-van den Berg MS, Jenster G, Zietse R, Hoorn EJ. An Immunoassay for Urinary Extracellular Vesicles. Am J Physiol Renal Physiol. 2016 doi: 10.1152/ajprenal.00463.2015. ajprenal 00463 02015. [DOI] [PubMed] [Google Scholar]
- Stevanato L, Thanabalasundaram L, Vysokov N, Sinden JD. Investigation of Content, Stoichiometry and Transfer of miRNA from Human Neural Stem Cell Line Derived Exosomes. PLoS One. 2016;11(1):e0146353. doi: 10.1371/journal.pone.0146353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3 22. [DOI] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, Sturk A, van Leeuwen TG, Nieuwland R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12(7):1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- Webber J, Clayton A. How pure are your vesicles? Journal of extracellular vesicles. 2013:2. doi: 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhu S, Hang W, Wu L, Yan X. Development of an ultrasensitive dual-channel flow cytometer for the individual analysis of nanosized particles and biomolecules. Analytical chemistry. 2009;81(7):2555–2563. doi: 10.1021/ac802464a. [DOI] [PubMed] [Google Scholar]
- Zhou H, Cleary R, Bogaert Y, Leelahavanichkul A. Combination of microRNA192 and microRNA27b from urinary exosomes differentiate between renal tubular damage and glomerular injury. J Am Soc Nephrol. 2008;19:672A. [Google Scholar]
- Zhou H, Kajiyama H, Tsuji T, Hu X, Leelahavanichkul A, Vento S, Frank R, Kopp JB, Trachtman H, Star RA, Yuen PS. Urinary exosomal Wilms’ tumor-1 as a potential biomarker for podocyte injury. Am J Physiol Renal Physiol. 2013;305(4):F553–559. doi: 10.1152/ajprenal.00056.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L, Yang G, Hu Z. Label-free quantitative detection of tumor-derived exosomes through surface plasmon resonance imaging. Analytical chemistry. 2014;86(17):8857–8864. doi: 10.1021/ac5023056. [DOI] [PMC free article] [PubMed] [Google Scholar]