Abstract
Ocean acidification is expected to have dramatic impacts on oceanic ecosystems, yet surprisingly few studies currently examine long‐term adaptive and plastic responses of marine invertebrates to pCO 2 stress. Here, we exposed populations of the common copepod Pseudocalanus acuspes to three pCO 2 regimes (400, 900, and 1550 μatm) for two generations, after which we conducted a reciprocal transplant experiment. A de novo transcriptome was assembled, annotated, and gene expression data revealed that genes involved in RNA transcription were strongly down‐regulated in populations with long‐term exposure to a high pCO 2 environment, even after transplantation back to control levels. In addition, 747 000 SNPs were identified, out of which 1513 showed consistent changes in nucleotide frequency between replicates of control and high pCO 2 populations. Functions involving RNA transcription and ribosomal function, as well as ion transport and oxidative phosphorylation, were highly overrepresented. We thus conclude that pCO 2 stress appears to impose selection in copepods on RNA synthesis and translation, possibly modulated by helicase expression. Using a physiological hypothesis‐testing strategy to mine gene expression data, we herein increase the power to detect cellular targets of ocean acidification. This novel approach seems promising for future studies of effects of environmental changes in ecologically important nonmodel organisms.
Keywords: acclimation, adaptation, evolution, gene expression, ocean acidification, Pseudocalanus, transcription, transgenerational effects, translation
Introduction
Anthropogenic emissions of CO2 have increased the global pCO2 from 280 ppm at pre‐industrial times to the present day 400 ppm (IPCC 2013). About one‐third of emitted CO2 is absorbed by the world's oceans (Sabine et al. 2004). Dissolution of CO2 into surface water forms H2CO3, carbonic acid, which quickly dissociates into bicarbonate (HCO3−) ions as well as hydrogen (H+) ions, lowering the seawater pH (ocean acidification, OA) (Doney et al. 2009). These changes, which are predicted to persist for thousands of years to come, will expose marine animals to dramatically changed chemical conditions, and negative effects are predicted for many species and ecosystems (Wittman and Pörtner 2013).
Calanoid copepods constitute approximately 80% of the global zooplankton biomass (Mauchline 1998). Many fish species depend on copepods for prey during their larval life (Last 1980), and fish stock recruitments can vary closely with copepod biomass (Beaugrand et al. 2003; Castonguay et al. 2008). Pseudocalanus is widely distributed in temperate and Arctic seas (Aarbakke et al. 2011). They are intensely preyed upon (Ohman 1986; Thor et al. 2008), may at times contribute more than 1/3 of the total zooplankton biomass in boreal and Arctic waters (Lischka and Hagen 2005; Thor et al. 2005), and thus constitute important prey items for many fish species.
Tolerance of calanoid copepods to OA has been assessed in a number of studies to date, and many have found them to be remarkably resilient (Weydmann et al. 2012; Pedersen et al. 2013). However, most studies have focused on species such as Calanus finmarchius (Mayor et al. 2007, 2012; Hildebrandt et al. 2014; Pedersen et al. 2014b), which undergo seasonal diapause (metabolic depression) during which extracellular pH can drop to pH 5 (Schruender et al. 2013). So, these species could already be adapted to tolerate exposure to low pH conditions. Other studies have shown sensitivity even in species otherwise known to be resilient to large changes in water chemistry (Calliari et al. 2008; Cripps et al. 2014). Also, importantly, most are short‐term studies (e.g., Kurihara et al. 2004; Watanabe et al. 2006; Lewis et al. 2013; Engström‐Öst et al. 2014), mostly on adult females (Cripps et al. 2014), so they do not assess transgenerational plasticity or adaptation to low pH (but see Kurihara and Ishimatsu 2008; Pedersen et al. 2014a). Thor and Dupont (2015) recently conducted a multigeneration study, finding negative effects of short‐term (three weeks) pH stress on fecundity and metabolism in Pseudocalanus acuspes. These effects did not decrease after two generations in elevated pCO2 within the present range of natural variability (900 μatm), but in a higher pCO2 deviating from the present range of variability (1550 μatm), authors observed a transgenerational buffering effect decreasing negative effects to only half of those observed in acute pH stress treatments. Reciprocal transplant tests showed that this buffering was caused either by transgenerational plasticity (e.g., epigenetic changes in gene expression) or adaptive evolution, or a combination of both (Thor and Dupont 2015).
The actual mechanism by which OA affects marine organisms has been studied on several different levels. On the whole‐organism level, energy budgets are critical to consider, as in affected animals a greater fraction of the energy budget may be diverted to costs for maintenance, repair, and homeostasis (Pörtner et al. 2004; Stumpp et al. 2012a; Stumpp et al. 2013). On a cellular level, maintaining homeostasis is of utmost importance for cellular function, including mitochondrial function through the electron transport chain (Cortassa et al. 2009), protein folding (Dobson 2003), and cytoskeleton organization (Squirrell et al. 2001). Effects of low pH could lead to increased energy demand for proton pump action maintaining homeostasis (Stumpp et al. 2012b; Pan et al. 2015; Jager et al. 2016). Thus, it could be beneficial for cells to be able to down‐regulate certain functions (e.g., cell division) in order to make more energy available for core function such as maintenance of homeostasis, the end result being slower growth but a higher ability to function in a stressful environment (Stumpp et al. 2011).
At the molecular level, two processes allow for buffering of negative impacts of pH stress: acclimation and adaptation (Calosi et al. 2013; Reusch 2014). Acclimation can be a short‐term, reversible process within an individual, or a transgenerational development of different reaction norms due to for example maternal effects (e.g., egg quality) or epigenetic changes affecting gene expression responses to stress (Riebesell and Gattuso 2014; Magozzi and Calosi 2015). Adaptation on the other hand is a slower process that depends on heritable genetic variation in traits associated with tolerance for natural selection to act upon (Munday et al. 2013; Stillman and Paganini 2015). In the event of rapid environmental change, adaptation from standing genetic variation allows for rapid response (Hermisson and Pennings 2005). This is likely to occur in large populations that experience long‐term environmental fluctuations on a regular basis and has been shown to be common in the marine environment (Johannesson et al. 2010; Feulner et al. 2013; Pespeni et al. 2013; De Wit et al. 2014; Gosset et al. 2014). In many cases, this genetic variation exist as low‐frequency alleles that are neutral (or nearly neutral) in the background environment, but as they become adaptive they can quickly increase in frequency over a few generations, allowing the population to evolve their tolerance limit beyond that possible by nongenetic change (i.e., acclimation). In the most beneficial of cases, it has been hypothesized that a combination of nongenetic short‐term changes can combine with longer‐term genetic changes to facilitate evolution of tolerance limits (Stillman 2003; Ghalambor et al. 2007; Sunday et al. 2014).
This study is an examination of the molecular response of the copepod P. acuspes used in the experimental setup of Thor and Dupont (2015). In short, copepods were kept for two generations in one of three different pCO2 environments and then reciprocally transplanted. These reciprocal transplants tests indicated that while observed changes in fecundity were caused solely by phenotypic plasticity (i.e., acclimation) at the intermediate pCO2 (900 μatm), a transgenerational physiological buffering effect was observed at the highest pCO2 (1550 μatm). For the study presented here, we hypothesized that expression patterns of involved genes should follow these observations. To investigate this, we used an mRNA‐Seq approach, sequencing pools of individual copepods. We assembled and annotated a transcriptome using available arthropod sequences, and then searched for genes exhibiting changes in expression similar to the changes in fecundity. We also scanned all expressed sequences for single nucleotide polymorphism (SNP) frequency changes associated with exposure to the highest pCO2 treatment, to infer loci potentially under selection pressure. Finally, we searched for nonrandom functional annotations within genes exhibiting interesting expression patterns and changes in SNP frequencies. This combined approach allowed us to gain an increased understanding of the cellular targets of OA and of the relative importance of acclimation and adaptation. Using a physiological hypothesis‐testing strategy to mine gene expression data for co‐expression patterns, rather than traditional differential expression analyses, it was possible to increase the power to detect cellular functional targets of ocean acidification. This novel approach seems promising for future studies of effects of environmental changes in ecologically important nonmodel organisms, where long generation times and lack of replication is a constant issue.
Materials and methods
Experimental setup
Pseudocalanus spp. specimens were collected in the Gullmar fjord in the spring of 2013 (58°16′N, 11°26′E) using a 200 μm WP‐2 plankton net, after which they were kept in culture at the Sven Lovén Centre for Marine Sciences—Kristineberg in Fiskebäckskil, Sweden at 5°C. Species identity was confirmed through PCR with species‐specific primers: DNA from a pool of 100 indiv. was extracted and separated from RNA and proteins using TriZol reagent (Invitrogen). Primers used were for P. minutus PsCOI_1561F/COI_1931R; for P. acuspes PsCOI_1561F/COI_2060R (both described in Gudmundsdottir 2008); and for P. elongatus Pseud‐E 225‐27F/Pseud‐E 345‐22R (Grabbert et al. 2010). The PCR program used for P. minutus and P. acuspes was as follows: 94°C (45 s), 47°C (1 min), 72°C (1 min 30 s) for 40 cycles, and 72°C for 3 min; and the program used for P. elongatus was 94°C (1 min), 62°C (1 min), and 75°C (2 min) for 31 cycles.
After verifying presence of only P. acuspes, 200 adults (F0 generation) were transferred into each of three different pCO2 treatments: Control (400 μatm pCO2), Medium (900 μatm pCO2), and High (1550 μatm pCO2), with two replicates of each for a total of six laboratory populations. The laboratory populations were grown in 40‐L tanks with filtered seawater with a 12 h/12 h light/dark cycle at 5°C for 137 days, until the F2 generation reached maturity. pH was dynamically controlled using pH computers (Aqua Medic, Germany), applying the CO2 immediately next to the air flow. pH electrodes were placed inside the streams of bubbles. Total scale pH and total alkalinity were measured once a week, using a Metrohm 827 pH meter and by titration of 25 mL water in a SI Analytics Titroline potentiometric titrator (Riebesell et al. 2010), respectively, after which pCO2 was calculated in CO2sys version 1.4 (Lewis and Wallace 1998). For food, Rhodomonas baltica were pumped intermittently into the tanks to achieve satiating concentrations. Concentrations were measured every two days with an Elzone 5380 electronic particle counter. Water was changed in all tanks every two weeks by siphoning out water from a large 50‐μm sieve inserted into the tanks, then transferring the animals into clean tanks. Generation sorting (Both between the F0 and F1 generation and the F1–F2 generation) was achieved in a similar fashion during water change by manually removing all adult individuals after two weeks of egg production.
At maturation, the F2 adults were reciprocally transplanted: the 400 μatm laboratory populations (n = 2) were divided into thirds and placed in 400 μatm pCO2, 900 μatm pCO2, and 1550 μatm pCO2 conditions. The 900 μatm (n = 2) and 1550 μatm (n = 2) laboratory populations were split in halves: half placed back in their original pCO2 and half moved into 400 μatm pCO2 conditions. After three weeks, fecundity was measured (Thor and Dupont 2015), after which the animals were placed in RNAlater (Ambion, Foster City, CA, USA) at 4°C for 24 h, then frozen at −20°C for genetic analyses.
Bioinformatic analysis and transcriptome assembly
RNA from 14 pools (seven treatments * two populations) of adult F2 copepods was extracted using TriZol reagent (Invitrogen, Carlsbad, CA, USA) (Table 1). Total RNA concentrations were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). For each pool, 1 μg of total RNA was used as input to Illumina TruSeq RNA sample kit v2 (Illumina, San Diego, CA, USA), following the kit standard protocol except in the final PCR step, where only 12 cycles were used rather than the recommended 15 to reduce the amount of duplicate sequences. Concentrations and fragment size distributions of the cDNA libraries were examined using a high‐sensitivity QuBit 2.0 fluorometric assay (Life Technologies, Carlsbad, CA, USA) and a TapeStation 2200 (Agilent, Santa Clara, CA, USA), respectively, after which libraries were combined equimolarly into three pools of four barcoded libraries each and one consisting of two libraries. Finally, all pools were diluted to 2 nm for sequencing. Sequencing was performed in February 2014 at the Genomics Core Facility of the University of Gothenburg, Sweden, in an Illumina NextSeq 500 sequencing machine with 50 bp read length, paired‐end sequencing.
Table 1.
Sample size, number of reads, and alignment results from the 14 samples used in the study
| Sample ID | Population | Sample size (n copepods) | n reads | % of reads aligning uniquely |
|---|---|---|---|---|
| 1 | 400A | 37 | 19 233 392 | 25.8 |
| 2 | 400B | 68 | 22 531 143 | 23.1 |
| 3 | 900A | 31 | 17 582 146 | 13.7 |
| 4 | 900B | 50 | 13 269 154 | 9.5 |
| 5 | 1550A | 39 | 21 055 372 | 22 |
| 6 | 1550B | 49 | 21 006 096 | 24.1 |
| 7 | 400–900A | 43 | 19 261 833 | 21.5 |
| 8 | 400–900B | 58 | 20 218 284 | 25 |
| 9 | 400–1550A | 38 | 20 271 884 | 16.4 |
| 10 | 400–1550B | 28 | 22 877 758 | 23.4 |
| 11 | 900–400A | 74 | 24 166 899 | 25.7 |
| 12 | 900–400B | 76 | 20 619 393 | 25.7 |
| 13 | 1550–400A | 68 | 12 020 876 | 8.7 |
| 14 | 1550–400B | 57 | 24 491 817 | 23.4 |
The raw sequence data (Table 1) were processed on the University of Gothenburg computer cluster ‘Albiorix’. First, low‐quality (Q < 20) ends were trimmed, and adapter sequences were removed. Remaining sequence data were assembled into a transcriptome using Trinity (Grabherr et al. 2011) version r2013_08_14. Assembled contigs (n = 207 302) were annotated by 1. BLASTx against a BLAST database consisting of all arthropod sequences from the NCBI nr database (April 1, 2014); 2. BLASTx to the curated SwissProt database, using 10−5 as e‐value cutoff. From these searches, top hits were extracted as well as the nr top hit when omitting ‘putative’ and ‘hypothetical’ hits (as very often the descriptions of these hits are not very useful). In addition, GO and KEGG terms were extracted from the SwissProt BLAST results. To be conservative, all un‐annotated contigs were removed from the dataset, as these could potentially consist of contaminant sequences, for a final list of 69 555 annotated contigs. The quality trimmed sequence data were then aligned against the newly created transcriptome, keeping only reads aligning uniquely to one location.
Gene expression analysis
In cases where different sequences (‘seq’) of the same Trinity component (‘comp’) had the same annotation, it was concluded that they most likely represented different isoforms of the same gene. Thus, counts of different isoforms were combined into 28 879 ‘Unigene’ counts (mean n reads = 4.254 Mreads/sample, SD 1.645 Mreads). To compare gene expression levels between samples, counts were scale normalized in the DESeq package in R (Anders and Huber 2010). In addition, genes with greater standard deviation than mean and/or at least one sample with zero counts were excluded. A hypothesis‐testing method was employed to search for genes exhibiting a similar expression pattern as the observed changes in fecundity (see Figure 1 in Thor and Dupont 2015) using analysis of covariance (ancova using the SAS software (SAS Institute, Cary, NC, USA); the Shapiro–Wilk test was used to check that the data were normally distributed). Specifically, we scanned the expression data for genes showing no significant (P > 0.05) difference in neither slope nor elevation (value of gene expression at the midpoint between the two treatments) of the linear regressions of the gene expression changes between the 400→900 and 900→400 μatm of pCO2 transplants (indicating phenotypic plasticity), AND:
significantly different slopes between the 400→1550 and 1550→400 μatm pCO2 transplant regression lines (‘Hypothesis 1’, i.e., transgenerational development of different phenotypic plasticity; Fig. 1A)
significantly different elevation between the 400→1550 and 1550→400 μatm pCO2 transplant regression lines (‘Hypothesis 2’, i.e., transgenerational development of different phenotype; Fig. 1B)
significantly different slopes and elevation between the 400→1550 and 1550→400 μatm pCO2 transplant regression lines (‘Hypothesis 3’, i.e., transgenerational development of both different phenotype and plasticity; Fig. 1C).
Figure 1.
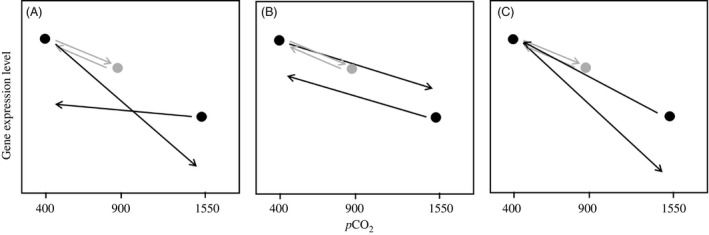
The three hypotheses used to examine the gene expression data. (A) Hypothesis 1: Different slopes but equal elevation interpreted as a transgenerational development of different reaction norms (i.e., transgenerational development of different phenotypic plasticity) at 1550 μatm pCO 2; (B) Hypothesis 2: Equal slopes but different elevation interpreted as a transgenerational development of different phenotype (i.e., adaptation) at 1550 μatm pCO 2; (C) Hypothesis 3: Different slopes and elevation. All three hypotheses assumed pure phenotypic plasticity between 400 and 900 μatm pCO 2 as depicted by the gray arrows.
The lists of genes matching these three hypotheses were tested for nonrandom distribution of functions using a GO enrichment analysis with the online software called ‘Gene Ontology Enrichment Analysis Software Toolkit’ (GOEAST) (http://omicslab.genetics.ac.cn/GOEAST/) (Zheng and Wang 2008) in the ‘Custom Microarray’ setting, using the recommended settings (Hypergeometric tests with FDR under dependency).
As ‘Helicase activity’ was indicated as strongly overrepresented in the differential expression dataset matching Hypothesis 2 (see Results), we scanned the transcriptome for other genes exhibiting the same expression pattern, the idea being that anything with identical expression may be part of the same gene regulatory network. Means of the two replicates were calculated, and the logical test was designed as follows: (900→400 > 400→400 AND 400→400 > 1550→400 AND 400→900 > 900→900 AND 400→1550 > 1550→1550 AND 400→400 > 400→900 AND 400→1550 > 400→900 AND 400→900 > 1550→400 AND 400→900 > 1550→1550) (Fig. 2). The resulting list of genes passing these conditions was then tested for functional enrichment as described above.
Figure 2.
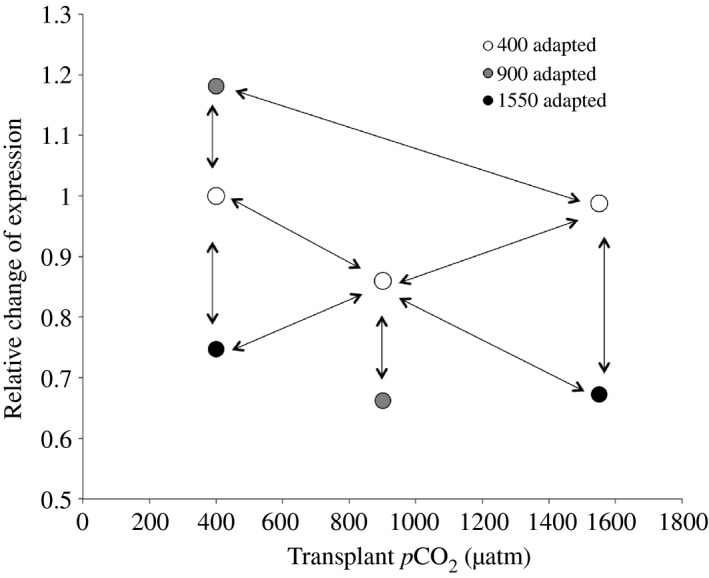
General expression pattern exhibited by contigs associated with helicase activity, used to scan the data for co‐expression patterns. Arrows indicate the logical rules used in the test.
Allele frequency changes
The PoPoolation2 pipeline (Kofler et al. 2011) and scripts were used to analyze allele frequency changes in the pools, using the trimmed data files and only annotated contigs from the transcriptome assembly (although keeping isoforms separate, nCONTIGS = 69 555) (https://code.google.com/p/popoolation2/wiki/Tutorial), employing the SAMtools (Li et al. 2009) mpileup command to calculate allele frequencies at all sites for the 14 pools, then following with the Cochran–Mantel–Haenszel (CMH) test for testing for consistent and significant changes in allele frequency between the replicate treatments of 400 μatm pCO2 and 1550 μatm pCO2. A total of 747 423 variant sites identified by mpileup in the previous step were used for this test. We considered the 400→1550 μatm transplants as replicates of the 400 μatm treatments, and the 1550→400 μatm transplants as replicates of the 1550 μatm treatment, thus arriving at four replicates for the CMH test. As no mortality had occurred during the transplant, we assumed that allele frequencies had not changed during this time. A GO category functional enrichment test was conducted for the resulting gene list using GOEAST, as described above. In addition, gene expression levels between treatments were compared in these genes as well, as described above.
Results
Transcriptome assembly
The Pseudocalanus acuspes transcriptome assembly initially consisted of 207 302 contigs, with an N50 (contig length at which 50% of the assembly consists of contigs the same length or longer) of 851 bp, and a GC content of 51.1%. After removing contigs that could not be annotated, the remaining 69 555 contigs (Appendix S1) had an N50 of 1236 bp and a GC content of 53.3%, reflecting that shorter contigs are less likely to be annotated. Information about nr arthropod and SwissProt top hits, as well as GO and KEGG terms, is given in Appendix S2.
Gene expression
Out of the 28 879 Unigenes, 15 850 remained after discarding ones with greater variance than mean and/or at least one sample with 0 counts after scaling normalization (Appendix S3). Out of these, 40.0% (6345 contigs) had a mean count across all samples >100, 41.5% (6570 contigs) had a mean count between 20 and 100, and 18.5% (2935 contigs) had a mean count < 20. While overall there was a large amount of variability between samples, both between and within treatments (see Figure S1), the ancova analysis identified 684 genes matching Hypothesis 1, 686 genes matching Hypothesis 2 and 26 genes matching Hypothesis 3 (see Fig. 1 for all hypotheses). The lists of genes matching Hypotheses 1 and 3 contained a random distribution of GO terms compared to the full transcriptomic dataset. However, the genes matching Hypothesis 2 were significantly enriched for a cascade of functions involving Helicase expression (Fig. 3). There were 32 genes involved in this functional category, all exhibiting the same pattern of decreased expression after spending two generations at 1550 μatm pCO2 (mean log twofold change −0.42; SD 0.20), even after being transplanted back into 400 μatm (mean log twofold change −0.42; SD 0.25) (Fig. 4). This reduction in expression, although not as severe, was also seen in both the short‐ and long‐term 900 μatm pCO2 treatments, but the populations that had been located in 900 μatm for two generations increased their expression back to the same expression level (or even slightly higher, although not significant) as the 400 μatm natives, when transplanted back into 400 μatm.
Figure 3.
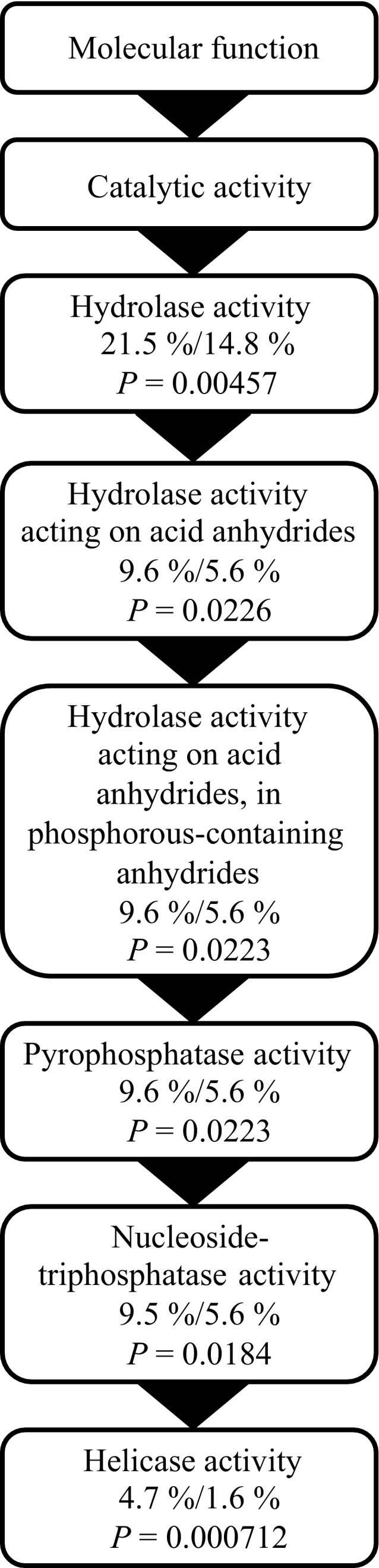
All Gene Ontology terms significantly enriched in the set of 686 contigs matching Hypothesis 2: Transgenerational development of different phenotype. Percentages are given as % contigs with GO term in list/% contigs with GO term in transcriptome, along with false‐discovery rate corrected P‐values.
Figure 4.
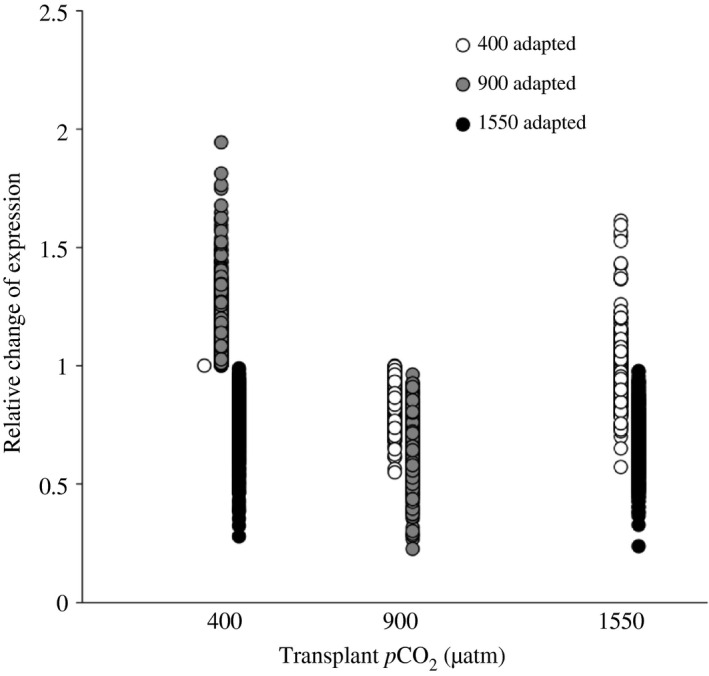
Expression of the 32 contigs associated with helicase activity. Expression levels are given as relative to the expression level in the control treatment.
Finally, we could also identify 321 additional contigs exhibiting the same expression pattern as the helicase contigs (Appendix S4), almost all of which are involved in RNA metabolism or DNA replication/repair (Fig. 5; Figure S2).
Figure 5.
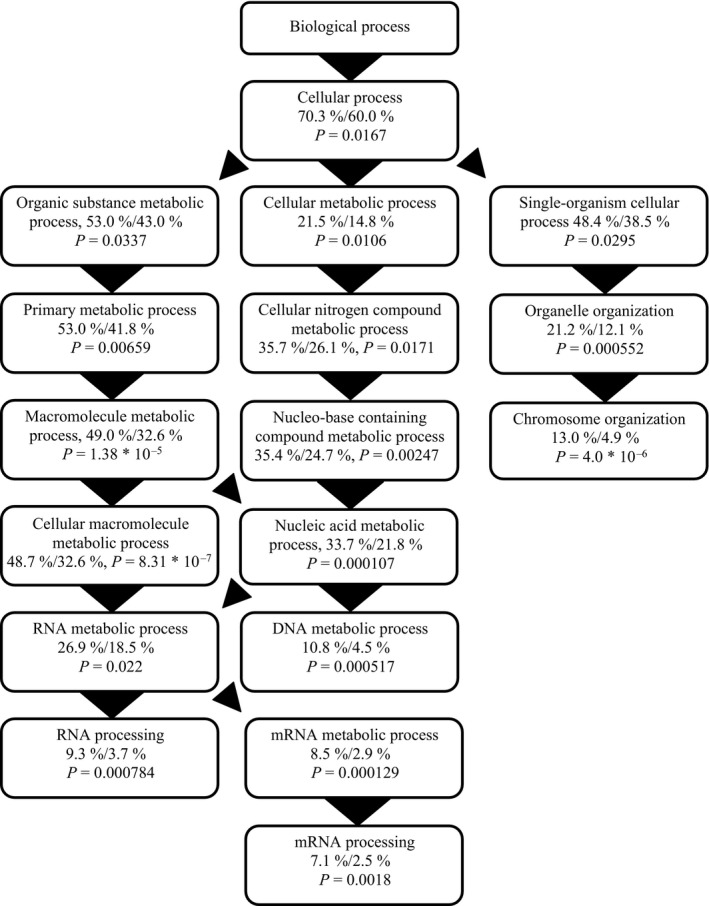
The most important Gene Ontology terms significantly enriched in the set of 353 contigs having the same expression pattern as the contigs involved in Helicase activity (for the full set, see Figure S2). Percentages are given as % contigs with GO term in list/% contigs with GO term in transcriptome, along with false‐discovery rate corrected P‐values.
Allele frequency changes
The SAMtools mpileup algorithm identified 747 423 polymorphic sites within the P. acuspes transcriptome. Within these, the CMH test identified 1517 SNPs and located in 303 contigs that were showing consistent and significant allele frequency changes among the biological replicates, after Bonferroni multiple‐test correction (P > 0.05) (Appendices S5 and S6). These genes were strongly enriched for functions involving protein translation (GO:0006412, P = 4.43*10−56), proton transport (GO:0015985, P = 7.93*10−7), mitotic spindle elongat‐ion (GO:0000022, P = 5.24*10−7), and cellular respiration (mostly mitochondrial genes) (GO:0006754, P = 1.08*10−5) (see Figure S3 for all enriched terms). No significant gene expression changes between transplant treatments were seen in these genes, however.
Discussion
In this study, we identified 303 genes that could be involved in adaptive evolution to pCO2 stress. This list included genes that encode for most of the proteins involved in ribosome formation and a large fraction of all mitochondrial genes. As all mitochondrial genes are inherited as one unit, it is difficult to speculate on which of these could be the ultimate target of natural selection, although it is interesting that also rather conserved sequences such as cytochrome oxidase subunit I also exhibit differences between treatments. Rather, it can be concluded that the mitochondrial function of oxidative phosphorylation was a target of natural selection. Respiration rates were higher at 900 μC, which was interpreted as a result of metabolic expenses due to an increased allocation of resources in order to acclimate to the new environment (Thor and Dupont 2015). Respiration rates were higher at 900 μatm as compared to 400 μatm and were correlated with a decrease in fecundity. As metabolic expenses vary depending on energy allocation to egg production in copepods (Thor 2002; Thor et al. 2002), Thor and Dupont (2015) interpreted this as a reallocation of resources caused by increased energy costs in high pCO2. On the contrary, at 1550 μatm respiration rates were similar to the ones at 400 μatm, with transplant tests suggesting that this could have arisen as a result of adaptation (Thor and Dupont 2015) due to differential mortality in the high pCO2 treatment. It therefore seems plausible that selection could be acting on maintenance of efficient oxidative phosphorylation at high pCO2 levels (Cortassa et al. 2009; Beaufort et al. 2011), to maintain energy production levels. We did not observe any increase in mitochondrial gene expression levels among treatments, which might have been expected if an increased energy demand had required more ATP to be produced. However, mitochondrial energy production can be modulated in more ways than expression levels (e.g., mtDNA copy number, transcription rate, mtRNA turnover, translation, mitochondrial fission/fusion). Other genes of interest showing allele frequency changes between treatments include Ferritin, which is associated with oxidative stress, known to be induced by high pCO2 levels in oysters (Tomanek et al. 2011), and also several subunits of the proteasome, which has been shown to evolve in response to high pCO2 stress in Sea Urchins (Pespeni et al. 2013).
A large fraction of the genes showing changes in allele frequencies between the high and low pCO2 treatments were mitochondrial, and must thus be linked as the mitochondrial genome is inherited maternally without recombination in copepods. Thus, the population of P. acuspes used in this experiment must have contained at least two mitochondrial lineages in order for this type of evolution to be observed. It is not uncommon for large marine invertebrate populations to show considerable variation in mitochondrial sequence (e.g., Silberman et al. 1994; Meyer and Paulay 2005), so this could be considered a realistic experimental situation. The rather rapid transfer of individuals (gradual increase over three days) from low to high pCO2 at the initiation of the experiment can be criticized as a less than realistic situation (although a frequent experimental practice), as ocean acidification is a slow and gradual process that will take place over a several hundred year period. By showing that P. acuspes has the potential to buffer even this rapid change, however, it is likely that they can do so also over longer time scales.
While genetic changes seem to be concentrated around the translational mechanism (ribosome formation) and mitochondrial functions, the gene expression data suggest transgenerational changes in RNA transcription and potentially DNA replication through changes in helicase activity. Interestingly, while the ancova approach used to identify genes with expression patterns matching the observed changes in fecundity only used ‘difference in expression elevation’ (between acute versus multigeneration treatment in 1550 μatm) as a criterion without considering the direction of the change (up‐ or down‐regulation), all of the 32 contigs associated with helicase activity showed the same pattern (Fig. 4): Strong down‐regulation after two generations at 1550 μatm pCO2, with no compensation when transplanted back into 400 μatm pCO2. At the intermediate 900 μatm pCO2, a decrease in expression (although not as strong as at 1550 μatm pCO2) can be seen at both acute and transgenerational scales, but upon reintroduction into 400 μatm pCO2 expression re‐attained 400 μatm levels. These changes in expression could be associated with changes in energy allocation to different functions.
All contigs associated with helicase activity consistently showed identical expression patterns (Fig. 2) despite being identified though ancova by the rather vague ‘Difference in elevation’ approach (‘Hypothesis 2’). Thus, it was of interest to scan the gene expression data for contigs showing the same pattern, to potentially identify other genes part of the same regulatory network. This search identified 353 contigs (including the helicase contigs) involved in RNA transcription and DNA replication (Fig. 5, Appendix S4). We cannot distinguish which cellular functions are the primary targets of this consistent down‐regulation of transcription/replication, but rather conclude that this can play a role in changes in energy allocation to different functions.
Using both the gene expression and the SNP frequency change data, a picture emerges on the cellular response and adaptive potential of P. acuspes to pCO2 stress. The high pCO2 level induces a transgenerational change in helicase activity [either through natural selection in a control region (Wray 2007), or nongenetic changes (Goldberg et al. 2007)], which is used to modify energy allocation. Helicase is involved in the separation of double‐stranded DNA and is a regulator of both RNA transcription and DNA replication, so changes in helicase expression would have significant effects on certain cellular functions, depending on the type of helicase involved. For example, reducing costs associated to DNA replication/cellular division, thereby reducing egg production as observed, would allow energy to be allocated to maintenance of homeostasis through ion pumps. Interestingly, a similar pattern has been observed in sea urchins, where gonadal tissue was used as an energy source during acclimation to pH changes (Dupont et al. 2013). At the same time, there is a selective advantage (either through selective mortality or perhaps more likely through larval development) for certain ribosomal structures and mitochondria that are better suited to a low‐energy metabolic mode.
From the results of this study, we cannot conclude which specific physiological functions are down‐regulated and what the long‐term fitness consequences might be (apart from the apparent decrease in fecundity). For example, elevated pCO2 can lead to modulation of the immune‐response and lead to a reduced ability to fight against pathogens (e.g., Asplund et al. 2014), and reduced levels of genetic variability due to one selective factor might impede the population's ability to adapt to additional stressors (Pistevos et al. 2011). Thus, an exciting field for future studies lies within investigating consequences of the observed ‘adaptation’ in the context of multiple environmental drivers (Dupont and Pörtner 2013). Despite these issues, this study demonstrates the great ability that marine invertebrates have to adapt from standing genetic variation. In most cases, these small organisms harbor large amounts of genetic diversity and have large population sizes and short generation times (Hellberg et al. 2002), so adaptation from standing genetic variation will surely have a large role in the maintenance of ecosystem stability in an unstable future environment.
Presently, the long‐term consequences of ocean acidification are difficult to predict. Even between closely related taxa, short‐term responses vary considerably (see, e.g., Kroeker et al. 2010), and very little is known about the effects of evolutionary change on a global scale. However, by understanding the effects of OA from a cellular perspective over a longer timeframe in ecologically important species (such as copepods), we might be able to build predictive models of global ecosystem changes in the future. To do this, much more data will be needed from a variety of different organisms, but as sequencing methods become more available and easier to use and standards for experimental design are being improved, this goal becomes more and more achievable in the years to come. In addition, by testing gene expression data against specific hypotheses generated by physiological data, we can gain power in detecting the cellular mechanisms involved in adaptation and acclimation to OA, and we predict that future studies increasingly will adopt this type of approach rather than the more exploratory differential gene expression analyses used to date.
Data archiving statement
All raw Illumina reads have been submitted to the NCBI Short Read Archive (SRA) (Bioproject SRP063962). The transcriptome assembly and annotation, as well as gene expression count data, are available as online supporting material (Appendices S1–S3).
Supporting information
Appendix S1. De novo transcriptome assembly of P. acuspes.
Appendix S2. Annotation data for all assembly contigs.
Appendix S3. Scale‐normalized count data.
Appendix S4. List of contigs exhibiting the same expression pattern as the helicase‐annotated contigs.
Appendix S5. List of SNPs with significant results from the CMH‐test.
Appendix S6. Annotation of contigs containing SNPs with significant CMH‐test results.
Figure S1 PCA plot of the 2 most informative dimensions of the gene expression data.
Figure S2. GO terms significantly overrepresented in the list of contigs matching the expression pattern observed in the helicase‐annotated contigs (Appendix S4).
Figure S3 GO terms significantly overrepresented in the list of contigs containing SNPs with significant CMH‐test results (Appendix S6).
Acknowledgements
Sam Dupont is funded by the Centre for Marine Evolutionary Biology, CeMEB (http://www.cemeb.science.gu.se/) and supported by a Linnaeus grant from the Swedish Research Councils. Peter Thor is funded by the Fram Centre Flagship programme ‘Ocean acidification and ecosystem effects in Northern waters’. Sequencing was funded by grant #2252279/E40 from the Norwegian Research Council, and Pierre De Wit was funded by the Marcus and Amalia Wallenberg Foundation. We would also like to thank the Genomics Core Facility platform at the Sahlgrenska Academy, University of Gothenburg., for providing excellent service and input on technical details. In addition, we would like to thank the technical staff at the Sven Lovén Centre for Marine Sciences at Kristineberg for help with managing the laboratory populations, and Ron Burton (Scripps Institution of Oceanography) for input on copepod cellular biology.
Literature cited
- Aarbakke, O. N. S. , Bucklin A., Halsband C., and Norrbin F. 2011. Discovery of Pseudocalanus moultoni (Frost, 1989) in Northeast Atlantic waters based on mitochondrial COI sequence variation. Journal of Plankton Research 33:1487–1495. [Google Scholar]
- Anders, S. , and Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asplund, M. , Baden S., Russ S., Ellis R., Gong N., and Hernroth B. 2014. Ocean acidification and host‐pathogen interactions: blue mussels, Mytilus edulis, encountering Vibrio tubiaschii . Environmental Microbiology 16:1029–1039. [DOI] [PubMed] [Google Scholar]
- Beaufort, L. , Probert I., de Garidel‐Thoron T., Bendif E. M., Ruiz‐Pino D., Metzl N., Goyet C. et al. 2011. Sensitivity of coccolithophores to carbonate chemistry and ocean acidification. Nature 476:80–83. [DOI] [PubMed] [Google Scholar]
- Beaugrand, G. , Brander K. M., Lindley J. A., Souissi S., and Reid P. C. 2003. Plankton effect on cod recruitment in the North Sea. Nature 426:661–664. [DOI] [PubMed] [Google Scholar]
- Calliari, D. , Andersen Borg M. C., Thor P., Gorokhova E., and Tiselius P. 2008. Instantaneous salinity reductions affect the survival and feeding rates of the co‐occurring copepods Acartia tonsa Dana and A. clausi Giesbrecht differently. Journal of Experimental Marine Biology and Ecology 362:18–25. [Google Scholar]
- Calosi, P. , Rastrick S. P. S., Lombardi C., de Guzman H. J., Davidson L., Jahnke M., Giangrande A. et al. 2013. Adaptation and acclimatization to ocean acidification in marine ectotherms: an in situ transplant experiment with polychaetes at a shallow CO2 vent system. Royal Society Philosophical Transactions Biological Sciences 368:20120444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay, M. , Plourde S., Robert D., Runge J. A., and Fortier L. 2008. Copepod production drives recruitment in a marine fish. Canadian Journal of Fisheries and Aquatic Sciences 65:1528–1531. [Google Scholar]
- Cortassa, S. , O'Rourke B., Winslow R. L., and Aon M. A. 2009. Control and regulation of mitochondrial energetics in an integrated model of cardiomyocyte function. Biophysical Journal 96:2466–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps, G. , Lindeque P., and Flynn K. J. 2014. Have we been underestimating the effects of ocean acidification in zooplankton? Global Change Biology 20:3377–3385. [DOI] [PubMed] [Google Scholar]
- De Wit, P. , Rogers‐Bennett L., Kudela R. M., and Palumbi S. R. 2014. Forensic genomics as a novel tool for identifying the causes of mass mortality events. Nature Communications 5:3652. [DOI] [PubMed] [Google Scholar]
- Dobson, C. M. 2003. Protein folding and misfolding. Nature 426:884–890. [DOI] [PubMed] [Google Scholar]
- Doney, S. C. , Fabry V. J., Feely R. A., and Kleypas J. A. 2009. Ocean acidification: the other CO2 problem. Annual Review of Marine Science 1:169–192. [DOI] [PubMed] [Google Scholar]
- Dupont, S. , and Pörtner H. 2013. Get ready for ocean acidification. Nature 498:429. [DOI] [PubMed] [Google Scholar]
- Dupont, S. , Dorey N., Stumpp M., Melzner F., and Thorndyke M. 2013. Long‐term and trans‐life‐cycle effects of exposure to ocean acidification in the green sea urchin Strongylocentrotus droebachiensis. Marine Biology 160:1835–1843. [Google Scholar]
- Engström‐Öst, J. , Holmborn T., Brutemark A., Hogfors H., Vehmaa A., and Gorokhova E. 2014. The effects of short‐term pH decrease on the reproductive output of the copepod Acartia bifilosa – a laboratory study. Marine and Freshwater Behaviour and Physiology 47:173–183. [Google Scholar]
- Feulner, P. G. D. , Chain F. J. J., Panchal M., Eizaguirre C., Kalbe M., Lenz T. L., Mundry M. et al. 2013. Genome‐wide patterns of standing genetic variation in a marine population of three‐spined sticklebacks. Molecular Ecology 22:635–649. [DOI] [PubMed] [Google Scholar]
- Ghalambor, C. , McKay J., Carroll S., and Reznick D. 2007. Adaptive versus non‐adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology 21:394–407. [Google Scholar]
- Goldberg, A. D. , Allis C. D., and Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128:635–638. [DOI] [PubMed] [Google Scholar]
- Gosset, C. C. , Do Nascimento J., Auge M.‐T., and Bierne N. 2014. Evidence for adaptation from standing genetic variation on an antimicrobial peptide gene in the mussel Mytilus edulis . Molecular Ecology 23:3000–3012. [DOI] [PubMed] [Google Scholar]
- Grabbert, S. , Renz J., Hirche H.‐J., and Bucklin A. 2010. Species‐specific PCR discrimination of species of the calanoid copepod Pseudocalanus, P. acuspes and P. elongatus, in the Baltic and North Seas. Hydrobiologia 652:289–297. [Google Scholar]
- Grabherr, M. G. , Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., et al. 2011. Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir, R. 2008. Pseudocalanus in Svalbard Waters: Identification and Distribution Patterns of two Sibling Copepod Species, Department of Aquatic Bio Sciences, University of Tromsø, Norwegian College of Fishery Science Master's thesis, http://munin.uit.no/handle/10037/1656. [Google Scholar]
- Hellberg, M. E. , Burton R. S., Neigel J. E., and Palumbi S. R. 2002. Genetic assessment of connectivity among marine populations. Bulletin of Marine Science 70:273–290. [Google Scholar]
- Hermisson, J. , and Pennings P. 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169:2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt, N. , Niehoff B., and Sartoris F. J. 2014. Long‐term effects of elevated CO2 and temperature on the Arctic calanoid copepods Calanus glacialis and C. hyperboreus . Marine Pollution Bulletin 80:59–70. [DOI] [PubMed] [Google Scholar]
- IPCC 2013. IPCC Fifth Assessment Report: Climate Change 2013. The Physical Science Basis, Cambridge. [Google Scholar]
- Jager, T. , Ravagnan E., and Dupont S.. 2016. Near‐future ocean acidification impacts maintenance costs in sea‐urchin larvae: identification of stress factors and tipping points using a DEB modelling approach. Journal of Experimental Marine Biology and Ecology 474:11–17. [Google Scholar]
- Johannesson, K. , Panova M., Kemppainen P., Andre C., Rolan‐Alvarez E., and Butlin R. K. 2010. Repeated evolution of reproductive isolation in a marine snail: unveiling mechanisms of speciation. Philosophical Transactions of the Royal Society B‐Biological Sciences 365:1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler, R. , Vinay Pandey R., and Schloetterer C. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool‐Seq). Bioinformatics 27:3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker, K. J. , Kordas R. L., Crim R. N., and Singh G. G. 2010. Meta‐analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecology Letters 13:1419–1434. [DOI] [PubMed] [Google Scholar]
- Kurihara, H. , and Ishimatsu A. 2008. Effects of high CO2 seawater on the copepod (Acartia tsuensis) through all life stages and subsequent generations. Marine Pollution Bulletin 56:1086–1090. [DOI] [PubMed] [Google Scholar]
- Kurihara, H. , Shimode S., and Shirayama Y. 2004. Effects of raised CO2 concentration on the egg production rate and early development of two marine copepods (Acartia steueri and Acartia erythraea). Marine Pollution Bulletin 49:721–727. [DOI] [PubMed] [Google Scholar]
- Last, J. M. 1980. The Food of Twenty Species of Fish Larvae in the West‐Central North Sea. Ministry of Agriculture, Fisheries and Food, Lowestoft (UK). [Google Scholar]
- Lewis, E. , and Wallace D. 1998. CO2SYS. Department of Applied Science, Brookhaven National Laboratory, Upton, NY. [Google Scholar]
- Lewis, C. N. , Brown K. A., Edwards L. A., Cooper G., and Findlay H. S.. 2013. Sensitivity to ocean acidification parallels natural pCO2 gradients experienced by Arctic copepods under winter sea ice. Proceedings of the National Academy of Sciences of the United States of America 110:E4960–E4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G. et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka, S. , and Hagen W. 2005. Life histories of the copepods Pseudocalanus minutus, P. acuspes (Calanoida) and Oithona similis (Cyclopoida) in the Arctic Kongsfjorden (Svalbard). Polar Biology 28:910–921. [Google Scholar]
- Magozzi, S. , and Calosi P. 2015. Integrating metabolic performance, thermal tolerance, and plasticity enables for more accurate predictions on species vulnerability to acute and chronic effects of global warming. Global Change Biology 21:181–194. [DOI] [PubMed] [Google Scholar]
- Mauchline, J. 1998. The biology of calanoid copepods In Blaxter J. H. S., Southward A. J., and Tyler P. A., eds. Advances in Marine Biology. Academic Press, San Diego, CA. [Google Scholar]
- Mayor, D. J. , Matthews C., Cook K., Zuur A. F., and Hay S. 2007. CO2‐induced acidification affects hatching success in Calanus finmarchicus . Marine Ecology Progress Series 350:91–97. [Google Scholar]
- Mayor, D. J. , Everett N. R., and Cook K. B. 2012. End of century ocean warming and acidification effects on reproductive success in a temperate marine copepod. Journal of Plankton Research 34:258–262. [Google Scholar]
- Meyer, C. P. , and Paulay G. 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday, P. L. , Warner R. R., Monro K., Pandolfi J. M., and Marshall D. J. 2013. Predicting evolutionary responses to climate change in the sea. Ecology Letters 16:1488–1500. [DOI] [PubMed] [Google Scholar]
- Ohman, M. D. 1986. Predator‐limited population‐growth of the copepod Pseudocalanus sp. Journal of Plankton Research 8:673–713. [Google Scholar]
- Pan, T.‐C. F. , Applebaum S. L., and Manahan D. T. 2015. Experimental ocean acidification alters the allocation of metabolic energy. Proceedings of the National Academy of Sciences of the United States of America 112:4696–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, S. A. , Hansen B. H., Altin D., and Olsen A. J. 2013. Medium‐term exposure of the North Atlantic copepod Calanus finmarchicus (Gunnerus, 1770) to CO2‐acidified seawater: effects on survival and development. Biogeosciences 10:7481–7491. [Google Scholar]
- Pedersen, S. A. , Håkedal O. J., Salaberria I., Tagliati A., Gustavson L. M., Jenssen B. M., Olsen A. J. et al. 2014a. Multigenerational exposure to ocean acidification during food limitation reveals consequences for copepod scope for growth and vital rates. Environmental Science and Technology 48:12275–12284. [DOI] [PubMed] [Google Scholar]
- Pedersen, S. A. , Våge V. T., Olsen A. J., Hammer K. M., and Altin D. 2014b. Effects of elevated carbon dioxide (CO2) concentrations on early developmental stages of the marine copepod Calanus finmarchicus Gunnerus (Copepoda: Calanoidae). Journal of Toxicology and Environmental Health Part A 77:535–549. [DOI] [PubMed] [Google Scholar]
- Pespeni, M. H. , Sanford E., Gaylord B., Hill T. M., Hosfelt J. D., Jaris H. K., LaVigne M. et al. 2013. Evolutionary change during experimental ocean acidification. Proceedings of the National Academy of Sciences of the United States of America 110:6937–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistevos, J. C. A. , Calosi P., Widdicombe S., and Bishop J. D. D. 2011. Will variation among genetic individuals influence species responses to global climate change? Oikos 120:675–689. [Google Scholar]
- Pörtner, H. O. , Langenbuch M., and Reipschlager A. 2004. Biological impact of elevated ocean CO2 concentrations: lessons from animal physiology and earth history. Journal of Oceanography 60:705–718. [Google Scholar]
- Reusch, T. B. H. 2014. Climate change in the oceans: evolutionary versus phenotypically plastic responses of marine animals and plants. Evolutionary Applications 7:104–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebesell, U. , and Gattuso J.‐P. 2014. Lessons learned from ocean acidification research. Nature Climate Change 5:12–14. [Google Scholar]
- Riebesell, U. , Fabry V. J., Hansson L., and Gattuso J.‐P. 2010. Guide to Best Practices for Ocean Acidification Research and Data Reporting. Publications Office of the European Union, Luxembourg. [Google Scholar]
- Sabine, C. L. , Feely R. A., Gruber N., Key R. M., Lee K., Bullister J. L., Wanninkhof R. et al. 2004. The ocean sink for CO2 . Science 305:367–371. [DOI] [PubMed] [Google Scholar]
- Schruender, S. , Schnack‐Schiel S. B., Auel H., and Sartoris F. J. 2013. Control of diapause by acidic pH and ammonium accumulation in the hemolymph of Antarctic copepods. PLoS ONE 8:e77498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman, J. D. , Sarver S. K., and Walsh P. J. 1994. Mitocondrial DNA variation and population structure in the spiny lobster Panulirus argus . Marine Biology 120:601–608. [Google Scholar]
- Squirrell, J. M. , Lane M., and Bavister B. D. 2001. Altering intracellular pH disrupts development and cellular organization in preimplantation hamster embryos. Biology of Reproduction 64:1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman, J. H. 2003. Acclimation capacity underlies susceptibility to climate change. Science 301:65. [DOI] [PubMed] [Google Scholar]
- Stillman, J. H. , and Paganini A. W. 2015. Biochemical adaptation to ocean acidification. The Journal of Experimental Biology 218:1946–1955. [DOI] [PubMed] [Google Scholar]
- Stumpp, M. , Wren J., Melzner F., Thorndyke M. C., and Dupont S. T. 2011. CO2 induced seawater acidification impacts sea urchin larval development I: elevated metabolic rates decrease scope for growth and induce developmental delay. Comparative Biochemistry and Physiology A‐Molecular and Integrative Physiology 160:331–340. [DOI] [PubMed] [Google Scholar]
- Stumpp, M. , Hu M. Y., Melzner F., Gutowska M. A., Dorey N., Himmerkus N., Holtmann W. C. et al. 2012a. Acidified seawater impacts sea urchin larvae pH regulatory systems relevant for calcification. Proceedings of the National Academy of Sciences of the United States of America 109:18192–18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpp, M. , Truebenbach K., Brennecke D., Hu M. Y., and Melzner F. 2012b. Resource allocation and extracellular acid‐base status in the sea urchin Strongylocentrotus droebachiensis in response to CO2 induced seawater acidification. Aquatic Toxicology 110:194–207. [DOI] [PubMed] [Google Scholar]
- Stumpp, M. , Hu M., Casties I., Saborowski R., Bleich M., Melzner F., and Dupont S. 2013. Digestion in sea urchin larvae impaired under ocean acidification. Nature Climate Change 3:1044–1049. [Google Scholar]
- Sunday, J. M. , Calosi P., Dupont S., Munday P. L., Stillman J. H., and Reusch T. B. H. 2014. Evolution in an acidifying ocean. Trends in Ecology and Evolution 29:117–125. [DOI] [PubMed] [Google Scholar]
- Thor, P. 2002. Specific dynamic action and carbon incorporation in Calanus finmarchicus copepodites and females. Journal of Experimental Marine Biology and Ecology 272:159–169. [DOI] [PubMed] [Google Scholar]
- Thor, P. , and Dupont S. 2015. Transgenerational effects alleviate severe fecundity loss during ocean acidification in a ubiquitous planktonic copepod. Global Change Biology 21:2261–2271. [DOI] [PubMed] [Google Scholar]
- Thor, P. , Cervetto G., Besiktepe S., Ribera‐Maycas E., Tang K. W., and Dam H. G. 2002. Influence of two different green algal diets on specific dynamic action and incorporation of carbon into biochemical fractions in the copepod Acartia tonsa . Journal of Plankton Research 24:293–300. [Google Scholar]
- Thor, P. , Nielsen T. G., Tiselius P., Juul‐Pedersen T., Michel C., Møller E. F., Dahl K. et al. 2005. Post spring bloom community structure of pelagic copepods in the Disko Bay, Western Greenland. Journal of Plankton Research 27:341–356. [Google Scholar]
- Thor, P. , Nielsen T. G., and Tiselius P. 2008. Mortality rates of epipelagic copepods in the post‐spring bloom period in the Disko Bay, Western Greenland. Marine Ecology Progress Series 359:151–160. [Google Scholar]
- Tomanek, L. , Zuzow M. J., Ivanina A. V., Beniash E., and Sokolova I. M. 2011. Proteomic response to elevated pCO2 level in eastern oysters, Crassostrea virginica: evidence for oxidative stress. The Journal of Experimental Biology 214:1836–1844. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. , Yamaguchi A., Ishida H., Harimoto T., Suzuki S., Sekido Y., Ikeda T. et al. 2006. Lethality of increasing CO2 levels on deep‐sea copepods in the western North Pacific. Journal of Oceanography 62:185–196. [Google Scholar]
- Weydmann, A. , Søreide J. E., Kwasniewski S., and Widdicombe S. 2012. Influence of CO2‐induced acidification on the reproduction of a key Arctic copepod Calanus glacialis . Journal of Experimental Marine Biology and Ecology 428:39–42. [Google Scholar]
- Wittman, A. C. , and Pörtner H. O. 2013. Sensitivities of extant animal taxa to ocean acidification. Nature Climate Change 3:995–1001. [Google Scholar]
- Wray, G. A. 2007. The evolutionary significance of cis‐regulatory mutations. Nature Reviews Genetics 8:206–216. [DOI] [PubMed] [Google Scholar]
- Zheng, Q. , and Wang X.‐J. 2008. GOEAST: a web‐based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Research 36:W358–W363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. De novo transcriptome assembly of P. acuspes.
Appendix S2. Annotation data for all assembly contigs.
Appendix S3. Scale‐normalized count data.
Appendix S4. List of contigs exhibiting the same expression pattern as the helicase‐annotated contigs.
Appendix S5. List of SNPs with significant results from the CMH‐test.
Appendix S6. Annotation of contigs containing SNPs with significant CMH‐test results.
Figure S1 PCA plot of the 2 most informative dimensions of the gene expression data.
Figure S2. GO terms significantly overrepresented in the list of contigs matching the expression pattern observed in the helicase‐annotated contigs (Appendix S4).
Figure S3 GO terms significantly overrepresented in the list of contigs containing SNPs with significant CMH‐test results (Appendix S6).


