Abstract
The clinical significance of circulating cell-free DNA (cfDNA) integrity as diagnostic and surveillance biomarker in hepatocellular carcinoma (HCC) was investigated and compared to that of alpha fetoprotein (AFP). Liver cancer patients had lower cfDNA integrity than those with benign diseases (P = 0.0167) and healthy individuals (P = 0.0025). Patients with HCC and non-HCC liver cancers (P = 0.7356), and patients with benign diseases and healthy individuals (P = 0.9138) had comparable cfDNA integrity respectively. cfDNA integrity increased after hepatectomy in cancer patients (P = 0.0003). The AUCs for detecting HCC by cfDNA integrity and AFP were 0.705 (P = 0.005) and 0.605 (P = 0.156), respectively. We found cfDNA integrity decreased in HCC patients and has the potential as promising biomarker for HCC diagnosis and treatment surveillance.
Keywords: circulating cell-free DNA, cfDNA, integrity, hepatocellular carcinoma.
Introduction
Hepatocellular carcinoma (HCC) ranks the third among all cancer-related deaths 1. Its onset is usually asymptomatic at early stage and thus many HCC patients were found late and as a result, tumor recurrence would more likely to occur after treatment, which jointly lead to the poor prognosis of HCC 2. Alpha fetoprotein (AFP) is the most commonly used biomarker for detecting and monitoring HCC, but the sensitivity and specificity of AFP are not satisfactory 3. Thus, identification of biomarkers with better working performance and reliability for early disease detection and timely recurrence prediction is of importance in HCC management.
Circulating cell-free DNA (cfDNA) has been demonstrated to be a promising biomarker 4. Qualifying the concentration and size distribution cfDNA could be easily and reproducibly performed. The concentration of cfDNA in cancer patients was reported higher than that of healthy individuals 5, 6; however, controversies on cfDNA level as biomarker for disease diagnosis were continuously on the rise since it appeared difficult to reach a consensus considering the various pre-analytical factors influencing cfDNA extraction and quantification 7. On the contrary, the size distribution of cfDNA, cfDNA integrity, was vulnerable to processing conditions. cfDNA integrity was measured as the ratio of longer to shorter DNA fragments, based on the hypothesis that cfDNAs released from apoptotic cells (normal blood cells) are uniformly truncated into fragments around 200 bp while cfDNAs released by tumor cells originate from both apoptotic and necrotic source, which are shorter and might thus increase the proportion of shorter DNA copies and lead to decreased cfDNA integrity 8. To this end, cfDNA has the potential for cancer prediction and prognostication 9, 10.
The primary aim of this study was thus to investigate whether the size distribution of cfDNA in patients with liver cancers, mainly HCC, was different from that of patients with benign liver diseases and healthy individuals. In addition, we wanted to know the association of cfDNA integrity with clinicopathological parameters in HCC patients and the dynamical change of cfDNA integrity before and after curative hepatectomy. Finally, we sought to evaluate the diagnostic value of cfDNA integrity in differentiating patients with HCC from healthy individuals, in comparison with AFP level. The results presented here preliminarily testified to the value of cfDNA integrity as a biomarker for HCC.
Materials and Methods
Patient enrollment and blood collection
Patients treated at Liver Surgery Department, Zhongshan Hospital, Fudan University were enrolled. The patients were diagnosed with either liver malignancies or benign liver diseases, without any forms of treatment prior to surgery. The healthy controls were randomly selected from individuals who underwent regular physical examination at our hospital and their health conditions were confirmed individually to ensure that none had acute or chronic diseases at the time of blood sampling. Ten milliliter venous blood was collected from each patient preoperatively and healthy individual, both at morning on an empty stomach using EDTA coated tubes (BD). For enrolled patients, another 10 ml venous blood was taken 5 days postoperatively if possible. All patients and healthy individuals gave their written informed consents and the study was approved by the Institutional Ethics Review Board of Zhongshan Hospital, Fudan University.
cfDNA extraction
Procedures for cfDNA extraction was performed as previously recommended 11. The plasma samples were prepared by centrifugation of blood at 3,000 rpm for 10 min followed by another centrifugation of the supernatant at 14,000 rpm for 10 min. Then the plasma was stored in 1ml aliquots at -80 °C until use. The separation and storage of plasma samples were performed within 3 hours of blood collection at 4 °C. cfDNA was extracted from 1 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN), according to the manufacturer's instructions.
cfDNA integrity analysis
The integrity of cfDNA was determined by qPCR as previously reported 12. Two primer sets (Sangon Biotech) were used to amplify ALU sequence: the primer set for 115 bp ALU amplicon was: forward 5'-CCTGAGGTCAGGAGTTCGAG-3' and reverse 5'-CCCGAGTAGCTGG GATTACA-3'; the primer set for 247 bp ALU amplicon was: forward 5'-GTGGCTCACGCCTGTAATC-3' and reverse 5'-CAGGCTGGAGTGCAGTGG-3'. The qPCR was done in a final volume of 20 μL on the ABI 7500 (Applied Biosystems) in triplicate and each reaction mixture contained 2 μL cfDNA template, 0.5 μL of each forward and reverse primer (10 μM, ALU115 or ALU247), 10 μL 2x SYBR Green (Takara), 0.5 μL Rox Dye II, and 6.5 μL RNase-free water. The reaction condition was 95 oC for 10 s, followed by 40 cycles of 95 oC for 5 s, and annealing at 60 oC for 34 s. The 115-bp ALU amplicon represented the total amount of DNA fragments including both short and long copies whereas the 247-bp ALU amplicon only reflected the amount of long DNA fragments. The amount of ALU 115 and ALU 247 DNA fragments was determined by comparing the CT values of each sample against the calibration curve created by performing qPCR with serial diluted cDNA (100 ng to 0.01 pg; EASY Dilution, Takara). cfDNA integrity was calculated as the relation of ALU 247 to ALU 115 according to the methods of Umetani et al 12.
Statistical analysis
Statistical analysis was performed using SPSS 18.0 (IBM). The values of cfDNA integrity between two groups were expressed as means ± SD and compared using unpaired Student's t test. To evaluate the diagnostic utility of the cfDNA integrity, the area under the curve (AUC) of receiver-operating characteristic (ROC) curve was calculated. The Youden index was calculated as sensitivity-(1-specificty), which were the values of y and x axis in ROC. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy were calculated accordingly using the cutoff value when Youden index was maximal. All P values were two-sided and P < 0.05 was considered statistically significant.
Results
Patient characteristics
Totally, 69 patients with liver malignancies were enrolled in this study and all patients successfully underwent hepatectomy with curative intention; among them, 53 patients had HCCs while the other 16 patients had non-HCC liver cancers including intrahepatic cholangiocarcinoma (ICC, 10 cases) and metastatic liver cancer (6 cases). Among patients with HCCs, the most of them were HBV positive (44/53, 83.0%); 25 patients had large HCC (≥ 5 cm); 27 patients had preoperative AFP levels above 20 ng/ml; 45 patients were staged BCLC (Barcelona Clinic Liver Cancer staging classification) 13 A and 8 patients were staged B. Besides, 15 patients with benign liver diseases, including hepatic hemangioma, hepatic cyst, and focal nodular hyperplasia, and 22 healthy individuals were included as controls.
Measurement of cfDNA integrity and its correlation with clinicopathological parameters
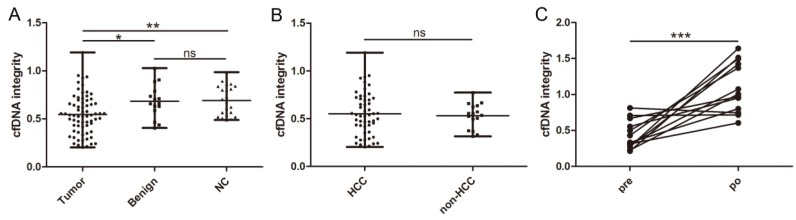
cfDNA was successfully extracted from all collected samples. We first compared the cfDNA integrity of the three different groups. In patients with malignant liver cancers, the mean cfDNA integrity was 0.545 ± 0.205 (range, 0.203-1.191), which was statistically lower than that of patients with benign liver disease (0.683 ± 0.175; range, 0.406-1.028; P = 0.0167, Figure 1A) and healthy individuals (0.692 ± 0.155; range, 0.488-0.986; P = 0.0025, Figure 1A). In contrast, the cfDNA integrity between patients with benign liver diseases and healthy individuals were comparable (P = 0.9138, Figure 1A). Similarly, we did not find any difference between the cfDNA integrity of HCC patients and patients with ICC or metastatic liver cancer (0.550 ± 0.224 vs 0.531 ± 0.131, P = 0.7356, Figure 1B). Among the patients with HCCs, we did not observe any correlation between cfDNA integrity and clinicopathological factors including HBV infection, liver cirrhosis, AFP, alanine aminotransferase, aspartate aminotransferase, tumor number, tumor size, tumor necrosis, encapsulation, and BCLC stage (Table 1).
Figure 1.
Distribution and dynamic of cfDNA integrity. A, Comparison of cfDNA integrity among patients with malignant liver tumors, benign liver diseases, and healthy individuals; B, comparison of cfDNA integrity between patients with HCC and non-HCC liver cancers; C, comparison of cfDNA integrity before and after hepatectomy in patients with liver cancers (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, non-significant).
Table 1.
Comparison of cfDNA integrity in HCC patients with different clinicopathological parameters.
| Parameters | Number | cfDNA integrity | P Value |
|---|---|---|---|
| HBsAg | |||
| Positive | 44 | 0.556 ± 0.224 | 0. 720 |
| Negative | 9 | 0.526 ± 0.236 | |
| Cirrhosis | |||
| Yes | 45 | 0.552 ± 0.225 | 0.920 |
| No | 8 | 0.543 ± 0.230 | |
| AFP | |||
| > 20 ng/ml | 27 | 0.586 ± 0.216 | 0.243 |
| < 20 ng/ml | 26 | 0.514 ± 0.230 | |
| ALT | |||
| > 50 u/L | 14 | 0.534 ± 0.215 | 0.750 |
| < 50 u/L | 39 | 0.556 ± 0.229 | |
| AST | |||
| > 40 u/L | 19 | 0.503 ± 0.184 | 0.250 |
| < 40 u/L | 34 | 0.577 ± 0.242 | |
| Tumor No. | |||
| Single | 42 | 0.554 ± 0.228 | 0.834 |
| Multiple | 11 | 0.538 ± 0.215 | |
| Tumor size | |||
| Large | 25 | 0.539 ± 0.188 | 0.731 |
| Small | 28 | 0.561 ± 0.254 | |
| Edmonson grade | |||
| I+II | 35 | 0.572 ± 0.214 | 0.340 |
| III+IV | 18 | 0.509 ± 0.242 | |
| Necrosis | |||
| Yes | 16 | 0.576 ± 0.257 | 0.593 |
| No | 37 | 0.540 ± 0.211 | |
| Encapsulation | |||
| Yes | 31 | 0.572 ± 0.253 | 0.419 |
| No | 22 | 0.521 ± 0.177 | |
| Vascular invasion | |||
| Yes | 19 | 0.504 ± 0.195 | 0.265 |
| No | 34 | 0.576 ± 0.237 | |
| BCLC stage | |||
| A | 45 | 0.553 ± 0.223 | 0.853 |
| B | 8 | 0.537 ± 0.243 |
In 13 patients with malignant liver cancers, a second blood sample was available 5 days after curative hepatectomy and we found the level of cfDNA integrity closely correlated with the disease status since it significantly increased after the tumor was removed (0.431 ± 0.199 vs 1.100 ± 0.348, P = 0.0003, Figure 1C).
Differential ability of cfDNA integrity as biomarker for HCC
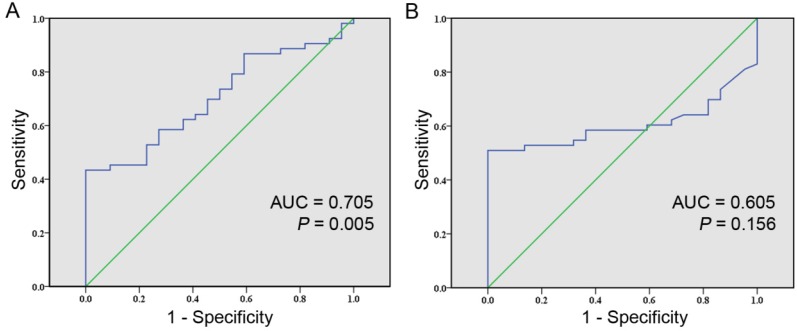
At last, we evaluated the ability of cfDNA integrity in differing HCC patients from healthy individuals. The ROC curves were generated separately for cfDNA integrity and AFP levels with AUC calculated accordingly. The AUC of distinguishing HCC patients from healthy individuals by cfDNA integrity was 0.705 (95% CI: 0.588-0.822; P = 0.005, Figure 2A). The Youden index was maximal when the cfDNA integrity was 0.488 and by using this cutoff value, the sensitivity was 43.4% with the specificity being 100%, the accuracy was 60.0%, the PPV was 100% and the NPV was 42.3% (Table 2). The AUC of distinguishing HCC patients from healthy individuals by AFP level was 0.605 (95% CI: 0.480-0.729; P = 0.156, Figure 2B). Practically, the cutoff value of AFP level was set at 20 ng/ml, at which level the sensitivity was 50.9% at a specificity of 100%, the accuracy was 65.3%, the PPV was 100% and the NPV was 45.8% (Table 2). Furthermore, we evaluated the diagnostic value of cfDNA integrity in combination with AFP level and the sensitivity, specificity, accuracy, PPV, and NPV were 79.2%, 100%, 85.3%, 100%, and 66.7% respectively (Table 2).
Figure 2.
Comparison of the diagnostic values between cfDNA integrity and AFP level for HCC using ROC curve. A, ROC curve of cfDNA integrity for detecting HCC (AUC, 0.705; 95% CI , 0.588-0.822; P = 0.005); B, ROC curve of AFP level for detecting HCC (AUC, 0.605; 95% CI , 0.480-0.729; P = 0.156).
Table 2.
Diagnostic value of cfDNA integrity and AFP level for HCC patients.
| Factor | Sensitivity | Specificity | Accuracy | PPV | NPV |
|---|---|---|---|---|---|
| cfDNA integrity | 43.4% (23/53) | 100% (22/22) | 60.0% (45/75) | 100% (23/23) | 42.3% (22/52) |
| AFP level | 50.9% (27/53) | 100% (22/22) | 65.3% (49/75) | 100% (27/27) | 45.8% (22/48) |
| cfDNA integrity + AFP level | 79.2% (42/53) | 100% (22/22) | 85.3% (64/75) | 100% (42/42) | 66.7% (22/33) |
Discussions
In this study, we evaluated cfDNA integrity in individuals with liver cancers, benign liver diseases, and healthy controls. Lower cfDNA integrity was observed in patients with liver cancers while patients with benign liver diseases and healthy individuals had comparable cfDNA integrity. In HCC patients, cfDNA integrity had non-inferior performance in diagnosing HCCs compared with AFP. What's important, the sharp increase of cfDNA integrity after hepatectomy suggested its close relevance with disease status, thus qualifying it as a potential surveillance biomarker.
cfDNA is increasingly being valued in cancer since part of cfDNAs is originated from tumor cells (circulating tumor DNA, ctDNA) 14. ctDNA is highly informative and taken as liquid biopsy to profile the genome of cancer 15; however, the high cost of next generation sequencing makes it less accessible 16. cfDNA concentration has been reported as diagnostic and/or prognostic biomarker in breast cancer 17, colorectal cancer 18, non-small cell lung cancer 19, pancreatic malignancies 20, and even HCC 21; but one should note that cfDNA concentration could be easily biased by preanalytical procedures: different cfDNA extraction kits/methods have distinct DNA recovery efficiencies 22, large plasma volume extraction with low volume of dissolution buffer increases the final concentration 23, cell lysis leads to increased DNA concentration 24 while prolonged storage decreases 25, and variances among quantitation modalities 26 make it less credible to make comparison between studies. Therefore, the cfDNA concentration among groups were not compared on purpose in this study.
In contrast, cfDNA integrity is less affected by much influential factors. Since cfDNA could be extracted from serum and plasma, its integrity also changed accordingly. In this study, we used plasma to extract cfDNA. Compared to serum, plasma is better for cfDNA extraction since genomic DNA from peripheral blood mononuclear cells would be released due to cell lysis during blood coagulation, leading to increased long DNA copies of serum cfDNA and thus elevated integrity 11. Herein, the blood was preserved in anticoagulant EDTA coated tubes for further plasma separation and EDTA could inhibit DNases to protect cfDNA from ex vivo degradation 27. Although we observed outcomes different from the results of two former studies by El-Shazly et al 28 and Chen et al 29, it should be noticed that they extracted cfDNA from serum and the cfDNA integrity might then be increased and our results were consistent with a recent study which reported that short DNA copies positively correlated with the fractional concentration of tumor-derived DNA in plasma 30.
Whereas cfDNA integrity significantly decreased in patients with liver malignancies, patients with benign liver diseases and healthy individuals had comparable size distribution. Increased necrosis within tumor resulted in highly fragmented DNA copies released into circulation 31 and improved sensitivity of ctDNA measurement using short PCR amplicons also indicated tumor derived copies were much shorter 32. This is supported by the finding that cfDNA integrity sharply increased after liver cancers being removed. The comparable cfDNA integrity between patients with benign liver diseases and healthy individuals again supported the notion that necrosis caused by malignant pathology increased short DNA fragments and cfDNA in normal human (possibly including patients with benign diseases) plasma is derived from apoptotic cells 33. We did not find any correlation between cfDNA integrity with clinicopathological factors whereas in the previous studies 28, 29, increased cfDNA integrity was significantly associated with large tumor size, vascular invasion etc. Possible explanations include different cfDNA source and relative small sample size. Since it remains largely unknown how clinicopathological factors influence cfDNA release and subsequent integrity, further cohort and experimental studies are needed. The similar cfDNA integrity between HCC and non-HCC liver cancers indicates that fragmentation of cfDNA is a frequent event regardless of malignancy type and decreased cfDNA integrity in cancer patients might be a universal phenomenon. However, it also raises another issue that cfDNA integrity might not be taken as a biomarker specifically for HCC but be more appropriate for treatment surveillance in HCC patients.
Although we observed better AUC value of cfDNA integrity, it would be rash to conclude cfDNA integrity outperforms AFP in detecting HCC. One should be aware that the value of cfDNA integrity in distinguishing HCC was made under the premise that individuals were already diagnosed with HCC. Considering cfDNA integrity in other malignancies also differed from normal people 34, it would be impractical to use cfDNA integrity alone to diagnose HCC. Pepes et al. have proposed that the development of biomarkers for early detection of cancer should undergo five phases 35 and till now, AFP is the only biomarker that has finished all five phases and been approved by FDA (Food and Drug Administration), though its sensitivity and specificity need improvement. Thus, it would be long before cfDNA integrity could be used as diagnostic biomarker for HCC and at present, it might be better used to help AFP for diagnosis.
Conclusively, cfDNA is more fragmentized in HCC patients compared with patients with benign liver diseases and healthy individuals. The sharp increase of cfDNA integrity after hepatectomy indicated its close relevance with tumor status and it shows similar working performance with AFP for HCC diagnosis. Further studies are warranted to examine the value of cfDNA integrity in the diagnosis and treatment surveillance of HCC.
Acknowledgments
The authors have declared that no competing interest exists. This study was jointly funded by the National Science Foundation for Distinguished Young Scholars of China (No. 81225019), the National Natural Science Foundation of China (No. 81572823), the National Science and Technology Major Project (No.2013ZX10002007-005), the National Key Research and Development Plan (2016YFC0902400, 2016YFC0902401), and Shanghai Hospital Development Center (SHDC12015104).
References
- 1.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Lee SC, Tan HT, Chung MC. Prognostic biomarkers for prediction of recurrence of hepatocellular carcinoma: current status and future prospects. World J Gastroenterol. 2014;20:3112–3124. doi: 10.3748/wjg.v20.i12.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rich N, Singal AG. Hepatocellular carcinoma tumour markers: current role and expectations. Best Pract Res Clin Gastroenterol. 2014;28:843–853. doi: 10.1016/j.bpg.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 4.Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci. 2013;14:18925–18958. doi: 10.3390/ijms140918925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shukla D, Kale AD, Hallikerimath S, Yerramalla V, Subbiah V. Can quantifying free-circulating DNA be a diagnostic and prognostic marker in oral epithelial dysplasia and oral squamous cell carcinoma? J Oral Maxillofac Surg. 2013;71:414–418. doi: 10.1016/j.joms.2012.04.039. [DOI] [PubMed] [Google Scholar]
- 6.Kienel A, Porres D, Heidenreich A, Pfister D. cfDNA as a Prognostic Marker of Response to Taxane Based Chemotherapy in Patients with Prostate Cancer. J Urol; 2015. [DOI] [PubMed] [Google Scholar]
- 7.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A. 2015;112:3178–3179. doi: 10.1073/pnas.1501321112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao TB, Shi W, Shen XJ. et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111:1482–1489. doi: 10.1038/bjc.2014.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iqbal S, Vishnubhatla S, Raina V. et al. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. Springerplus. 2015;4:265. doi: 10.1186/s40064-015-1071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009;404:100–104. doi: 10.1016/j.cca.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Umetani N, Giuliano AE, Hiramatsu SH. et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 13.Llovet JM, Zucman-Rossi J, Pikarsky E. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 14.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 15.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis JM, Heineck DP, Heller MJ. Detecting cancer biomarkers in blood: challenges for new molecular diagnostic and point-of-care tests using cell-free nucleic acids. Expert Rev Mol Diagn. 2015;15:1187–1200. doi: 10.1586/14737159.2015.1069709. [DOI] [PubMed] [Google Scholar]
- 17.Agassi R, Czeiger D, Shaked G. et al. Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am J Clin Pathol. 2015;143:18–24. doi: 10.1309/AJCPI5YHG0OGFAHM. [DOI] [PubMed] [Google Scholar]
- 18.da Silva Filho BF, Gurgel AP, Neto MA. et al. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol. 2013;66:775–778. doi: 10.1136/jclinpath-2013-201521. [DOI] [PubMed] [Google Scholar]
- 19.Nygaard AD, Holdgaard PC, Spindler KL, Pallisgaard N, Jakobsen A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer. 2014;110:363–368. doi: 10.1038/bjc.2013.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High Levels of Cell-Free Circulating Nucleic Acids in Pancreatic Cancer are Associated With Vascular Encasement, Metastasis and Poor Survival. Cancer Invest. 2015;33:78–85. doi: 10.3109/07357907.2014.1001894. [DOI] [PubMed] [Google Scholar]
- 21.Chen K, Zhang H, Zhang LN. et al. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol. 2013;19:3143–3149. doi: 10.3748/wjg.v19.i20.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page K, Guttery DS, Zahra N. et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One. 2013;8:e77963. doi: 10.1371/journal.pone.0077963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazurek AM, Fiszer-Kierzkowska A, Rutkowski T. et al. Optimization of circulating cell-free DNA recovery for KRAS mutation and HPV detection in plasma. Cancer Biomark. 2013;13:385–394. doi: 10.3233/CBM-130371. [DOI] [PubMed] [Google Scholar]
- 24.Page K, Powles T, Slade MJ. et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci. 2006;1075:313–317. doi: 10.1196/annals.1368.042. [DOI] [PubMed] [Google Scholar]
- 25.Sozzi G, Roz L, Conte D. et al. Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst. 2005;97:1848–1850. doi: 10.1093/jnci/dji432. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran K, Speer CG, Fiddy S, Reis IM, Singal R. Free circulating DNA as a biomarker of prostate cancer: comparison of quantitation methods. Anticancer Res. 2013;33:4521–4529. [PubMed] [Google Scholar]
- 27.Barra GB, Santa Rita TH, Vasques JA, Chianca CF, Nery LF, Costa SS. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem; 2015. [DOI] [PubMed] [Google Scholar]
- 28.El-Shazly SF, Eid MA, El-Sourogy HA, Attia GF, Ezzat SA. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis C virus-related hepatocellular carcinoma. Int J Biol Markers. 2010;25:79–86. doi: 10.1177/172460081002500204. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Sun LY, Zheng HQ, Zhang QF, Jin XM. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44:318–324. doi: 10.1097/PAT.0b013e328353a24c. [DOI] [PubMed] [Google Scholar]
- 30.Jiang P, Chan CW, Chan KC, Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang BG, Huang HY, Chen YC. et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- 32.Andersen RF, Spindler KL, Brandslund I, Jakobsen A, Pallisgaard N. Improved sensitivity of circulating tumor DNA measurement using short PCR amplicons. Clin Chim Acta. 2015;439:97–101. doi: 10.1016/j.cca.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki N, Kamataki A, Yamaki J, Homma Y. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta. 2008;387:55–58. doi: 10.1016/j.cca.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Gu G, Ju S. Recent advances in clinical applications of circulating cell-free DNA integrity. Lab Med. 2014;45:6–11. doi: 10.1309/lmkkox6ujzqgw0ea. [DOI] [PubMed] [Google Scholar]
- 35.Pepe MS, Etzioni R, Feng Z. et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]