Abstract
Abnormal expression of microRNAs plays important role in tumor metastasis. Migration and invasion of cancer cells accord for the metastasis and deterioration of breast cancer. However, the regulatory role of microRNAs in the invasion and migration of breast cancer cells has not completely understood yet. Here we found that microRNA-34c (miR-34c) was significantly downregulated in metastatic tissue of breast cancer. In vitro study showed that miR-34c negatively regulated GIT1 protein expression by binding to the 3'UTR of GIT1 mRNA. Consistently, GIT1 protein expression was found upregulated significantly in metastatic breast cancer. Moreover, miR-34c overexpression suppressed the expression of GIT1 protein, and this effect was restored by AMO-miR-34c in breast cancer cells. Overexpression of miR-34c suppressed cell migration and invasion in both MCF-7 and MDA-MD-231 breast cancer cells. Furthermore, knockdown of endogenous GIT1 expression reduced the migration and invasion of both two breast cancer cells. Collectively, miR-34c downregulation in breast cancer cells resulted in the upregulation of GIT1, which in turn enhanced the migration and invasion of breast cancer. This study highlights molecular mechanism of migration and invasion of breast cancer cells.
Keywords: miR-34c, breast cancer, GIT1, metastasis.
Introduction
Nowadays, breast cancer has been the most common cancer among females all over the world, and over 1.6 million cases are newly diagnosed per year 1. Metastasis of breast cancer is a significant life-threatening transformation and remains the leading cause of breast cancer-associated deaths 2. Intensive efforts have been focused on understanding cancer metastasis, including the molecular mechanisms of breast cancer metastasis, as well as the novel targets in suppressing cancer metastasis 3-4. Cell migration and invasion are the key steps in breast cancer metastasis. However, the precise mechanisms of breast cancer cell migration and invasion has not been elucidated yet.
G protein-coupled receptor kinase interacting protein 1 (GIT1) was originally identified as an ADP ribosylation factor GTPase-activating protein (ARF-GAP) that binds to G-protein-coupled receptor kinases (GRKs) and regulates membrane trafficking 5. Recently, GIT1 was shown to play a critical role in regulating focal adhesion, cell migration and lamellipodia formation 6. Structural analysis of GIT1 consists of multiple domains, including coiled-coil, Spa homology, focal adhesion targeting (FAT) domain of focal adhesion kinase (FAK), FAT-Homology (FAH), paxillin and Hic-5 LD3-binding domains 7-8. These structural properties of GIT1 contribute to tumor metastasis, including cell migration, invasion, focal adhesion and angiogenesis 9-11. microRNAs (miRNAs), a large class of small noncoding RNAs with approximately 22 nucleotides in length, have emerged as new important mediators of cancer development and treatment 12. They have been shown to regulate specific oncogenes, and participate in the processes of tumor growth and cancer metastasis. Also, miRNAs were promised to be key targets in the diagnosis, prognosis and therapy of cancers 13-14. In the present study, miR-34c was screened out by microarray from mouse with breast cancer metastasis and presented a powerful role in regulating GIT1 expression, which mediated the suppression of cells migration and invasion in breast cancer cells.
Material and Methods
Cell culture
Human breast adenocarcinoma cell line Michigan Cancer Foundation-7 (MCF-7) and MDA-MB-231 was cultivated in Dulbecco's Modified Eagle's Medium-High Glucose (Hyclone, South Logan, Utah, USA) with 4 mM L-glutamine, 10% FBS (Hyclone, South Logan, Utah, USA), and 100 U/mL penicillin/streptomycin (Hyclone, South Logan, Utah, USA). Cells were grown in a humidified atmosphere of 5% CO2 in air at 37 oC and passaging with 0.05% trysin-EDTA every 3-4 days. MCF-7 and MDA-MB-231 cells were transfected with 50 nM miR-34c, miR-34c inhibitor (AMO-miR-34c) or NC miRNAs (Guangzhou RiboBio Co., Ltd. Guangzhou, China) with Lipofectamine™ RNAiMAX (Life Technologies, Grand Island, NY, USA). 48 hours after transfection, cells were collected for further total RNA or protein extraction.
siRNA transfection
siRNA technology was preformed to silence the expression of GIT1 in MCF-7 and MDA-MB-231 cells. si-GIT1 (Santa Cruz Biotechnology, Dallas, Texas, USA) at the concentration of 100 nM or the negative control siRNA (si-Control) were delivered by LipofectamineTM RNAiMAX in 6-well plates cells, and then was further incubated for 48 hours at 37 °C in an incubator. All the transfections were repeated more than three times independently.
Quantitative Reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol (Life Technologies, Carlsbad, CA, USA) and miRNeasy mini kit (QIAGEN, Valencia, CA, USA) according to manufacturer's instructions, which efficiently recovered all RNA species, including miRNAs. RNA quality and quantity was measured using NanoDrop spectrophotometer (ND-1000, NanoDrop Technologies) and RNA integrity was determined by gel electrophoresis.
Total RNA was extracted from tumor tissue and cell line using TRIzol reagents according to the manufacturer's instructions. cDNA was assessed with SYBR Green PCR Master Mix (TOYOBO) by ABI PRISM® 7500 Sequence Detection System (Applied Biosystems). The U6 and 18S were used as internal standards for miRNA and mRNA respectively. The primer pairs are listed in Table 1. We determined the appropriate cycle threshold (Ct) using the automatic baseline determination feature and analyzed data by relative quantitative analysis method. The mean Ct values (± SEM) of three independent experiments are presented.
Table 1.
Primers for qRT-PCR and luciferase assay.
| Primer | Primer Sequence (5'-3') | |
|---|---|---|
| mmu-miR-34c | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGGCAATCAGCTAAC |
| Forward | ACACTCCAGCTGGGAGGCAGTGTAGTTAGCTGAT | |
| mmu-miR-93 | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGCTACCTGCACG |
| Forward | ACACTCCAGCTGGGCAAAGTGCTGTTCGTGCAGG | |
| mmu-miR-21 | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGTCAACATCAGTC |
| Forward | ACACTCCAGCTGGGTAGCTTATCAGACTGATGTT | |
| Mmu-miR-27a | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGGCGGAACTTAG |
| Forward | ACACTCCAGCTGGG TTCACAGTGGCTAAGTTCC | |
| mmu-miR-375 | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGTCACGCGAGCCG |
| Forward | ACACTCCAGCTGGGTTTGTTCGTTCGGCTCGCGT | |
| miRNA | Reverse | CTCAACTGGTGTCGTGGA |
| U6 | RT | AACGCTTCACGAATTTGCGT |
| Forward | CTCGCTTCGGCAGCACA | |
| Reverse | AACGCTTCACGAATTTGCGT | |
| GIT1 | Forward | GCCGACAGTGACTATGAGAACA |
| Reverse | CCCGCAACAGTTCCTGAAT | |
| 18S | Forward | CCTGGATACCGCAGCTAGGA |
| Reverse | GCGGCGCAATACGAATGCCCC | |
| GIT1-luc | Forward | (XhoI)ccgctcgagCCTCTCTCCCCACACCCTCACC |
| Reverse | (NotI)ataagaatgcggccgcTTAACAGCTCATGGTCACTTCTTTATTTT | |
| mutGIT1-luc | Forward | CGGGTAGCCAGTTTTGGTCCTGCCACGACGGAGCCTGATCCCTCCCCACTCTCGCCCC |
| Reverse | GGGGCGAGAGTGGGGAGGGATCAGGCTCCGTCGTGGCAGGACCAAAACTGGCTACCCG |
Note: miRNA Reverse primer as a common reverse primer used for all miRNAs detected by qRT-PCR in our study. XhoI and NotI are name of restriction enzyme.
miRNA microarray
Total RNA of four tumor tissue samples of mouse model in each group was randomly selected. After RNA isolation from the samples, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) was used according to the manufacturer's guideline for miRNA labeling. Microarray analysis was completed with the 6th generation of miRCURYTM LNA Array (v.16.0) (Exiqon, Inc. Woburn, MA, USA), which contains more than 1891 capture probes, covering all human, mouse and rat microRNAs annotated in miRBase 16.0. Slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA), and images were imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs that intensities >50 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the median normalization. After normalization, differentially expressed miRNAs were identified through Fold Change filtering. Hierarchical clustering was performed using MEV software (version 4.6, TIGR).
Immunocytochemistry
MCF-7 cells was cultured in chamber slides (Invitrogen, Life Technologies, Carlsbad, CA, USA) and 4% paraformaldehyde fixed for 30 mins in room temperature. After quenching endogenous peroxidase activity with 3% H2O2, the sections were blocked with 10% non-immune goat serum and then incubated overnight with primary anti-GIT1 antibody (Epitomics, Burlingame, CA, USA) (1:200), anti-Paxillin antibody (Epitomics, Burlingame, CA, USA) (1:200) at 4 oC, and then incubate slides with second antibodies for 40 mins in room temperature. The reaction was stopped by rinsing sections with 0.01 M phosphate-buffered saline (PBS). Negative controls were incubated with serum instead of primary antibody under the same conditions. The signals were detected with DAB substrate (ChemMateTM DAKO Envision TM Detection Kit).
Transwell migration and invasion assays
Transwell migration and invasion assays were carried out using 24-well Transwell chambers with 8 μm size polytrylene membranes (BD Biosciences, San Jose, CA, USA). MCF-7 and MDA-MB-231 cells were starved for 24 hours, and 1×105 cells were seeded on the top chamber. The bottom chambers were filled with 10% FBS in DMEM. For cell migration assay, cells were placed in the upper chamber with 100 μl serum-free medium. For the invasion assay, cells were placed in the upper chamber with 400 μl prechilled Matrigel (BD Biosciences, USA). 24 hours later, migration and invasion activity of MCF-7 and MDA-MB-231 cells were determined by counting cells positive with crystal violet staining after fixed with 4% paraformaldehyde for 20 mins.
Western blot
Tumor tissues were homogenized in lysis buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton and 1× complete protease inhibitor cocktail), and spined extract 10 mins at 14,000g in 4 oC centrifuge. Cells were lysed in lysis buffer, and sonicated briefly, centrifuged extract 15 mins at 14,000g in 4 oC centrifuge. Total protein samples (80 μg) were analyzed by 8% SDS-PAGE gel. The protein was ransferred to polyvinylidene difluoride (PVDF) membranes by a wet blotting procedure (100 V, 120 mins, 4oC). After blocked with 5% blocking buffer, the membranes incubated with primary antibodies at 4oC overnight using the following concentration: anti-GIT1 antibody (Epitomics, Burlingame, CA, USA) (1:1000), anti-Paxillin antibody (Epitomics, Burlingame, CA, USA) (1:1000) and GAPDH (1:3000), followed by alkaline phosphatase conjugated secondary antibody. Visualization of bound antibody was achieved with film (Kodak, Rochester, NY USA) by using 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) (Beyotime Institute of Biotechnology, Jiangsu, China) at several time points (2 mins, 10 mins and 2 hr). We chose the optimal conditions of GIT1/GAPDH signal image respectively and presented here. GAPDH acted as the internal control for normalization of gene expression data.
Luciferase assays
We synthesized fragments of GIT1 3'UTR containing the exact target site for miR-34c or the mutated target site using the Taq PCR amplification. The primer pairs are listed in Table 1. We insert the GIT1 3'UTR fragments into the multiple cloning sites (SgfI and XhoI) of the psiCHECKTM-2 luciferase miRNA expression reporter vector (Promega, Madison, WI, USA). The HEK293T cells (2×104 pre well) were transfected with miR-34c (50 nM) or AMO-miR-34c (50 nM) and 0.5 μg plasmid by Lipofectamine™ RNAiMAX, according to the manufacturer's instructions. Luciferase activities were measured 48 hours after transfection with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) on a luminometer (GloMax, Promega).
Statistical analysis
Student's t-test was used to compare the difference between two groups. Statistical comparisons among multiple groups were performed by Tukey's post hoc test and One-Way ANOVA. All data are presented as Mean ± SEM. All of the figures were plotted using GraphPad Prism (GraphPad Software Inc., San Diego, USA). P<0.05 was considered as significantly different.
Results
miR-34c downregulated in metastatic breast cancer
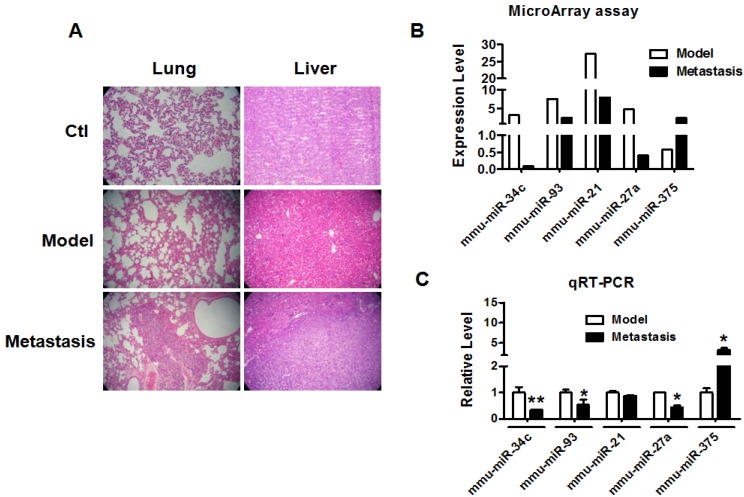
We established metastatic breast cancer model by subcutaneous injection of 4T1 breast cancer cells into BALB/c female mouse as described previously 15. When tumor size reached over 100 mm3, the mice were divided into model group and metastasis group. The mice of metastasis group were normally raised one more month than mice of model group. H&E staining of lung and liver tissues was used to determine the metastasis of breast cancer in each group. As shown in Figure 1A, the lung and liver metastasis of breast cancer were confirmed in metastasis group but not model group. Microarray and qRT-PCR profiling identified miRNAs differentially expressed in the tumor tissues of model group and metastasis group. As illustrated in Figure 1B, miR-34c, miR-93, miR-21 and miR-27a were downregulated by >50%, whereas miR-375 was upregulated by more than 2 fold in metastasis group compared with model group. Furthermore, qRT-PCR verification showed that miR-34c, miR-93 and miR-27a were decreased and miR-375 was increased significantly in metastasis group (Figure 1C). Particularly, miR-34c demonstrated the most significantly change in metastasis group, which suggested that miR-34c is involved in the process of breast cancer metastasis.
Figure 1.
miR-34c downregulated in breast cancer metastasis. A. H&E staining of mouse lung and liver tissues of each group (40 ×). Multiple metastatic tumors were observed in lung and liver of metastasis group compare with model group. B. The miRNAs expression profile of breast tumors between model group and metastasis group. C. Quantitative Real-Time PCR verification of the miRNA expression profile in breast tumors of model group and metastasis group. * P<0.05 vs. model group, ** P<0.01 vs. model group. n=3 independent samples for each group.
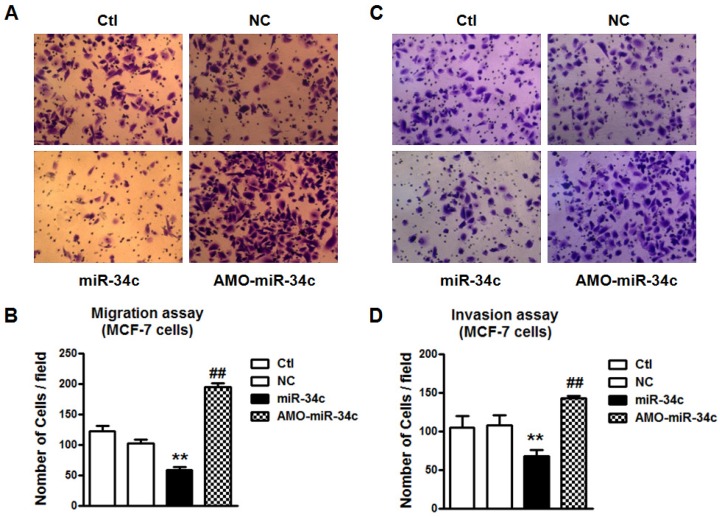
Overexpression of miR-34c suppressed cell migration and invasion in MCF-7 cells
To verify the role of miR-34c in breast cancer metastasis, we performed the migration and invasion assay. Overexpression of miR-34c suppressed cells migration and invasion significantly in MCF-7 cells (p<0.01), and the effects were reversed substantially by co-transfection with AMO-miR-34c (Figure 2). It indicated that miR-34c suppressed cell migration and invasion of MCF-7 breast cancer cells.
Figure 2.
Overexpression of miR-34c suppressed cell migration and invasion of MCF-7 cells. A. Migration assay of MCF-7 cells after treatment with miR-34c, AMO-miR-34c and scramble miRNA (negative control, NC) for 48 hours. B. Quantitative analysis of cell migration in MCF-7 cells of each group. C. Invasion assay of MCF-7 cells after transfection of miR-34c, AMO-miR-34c and NC for 48 hours. D. Quantitative analysis of cell invasion in MCF-7 cells of each group. ** P<0.01 vs. NC group. ## P<0.01 vs. miR-34c group. n=6 independent batches of cells for each group.
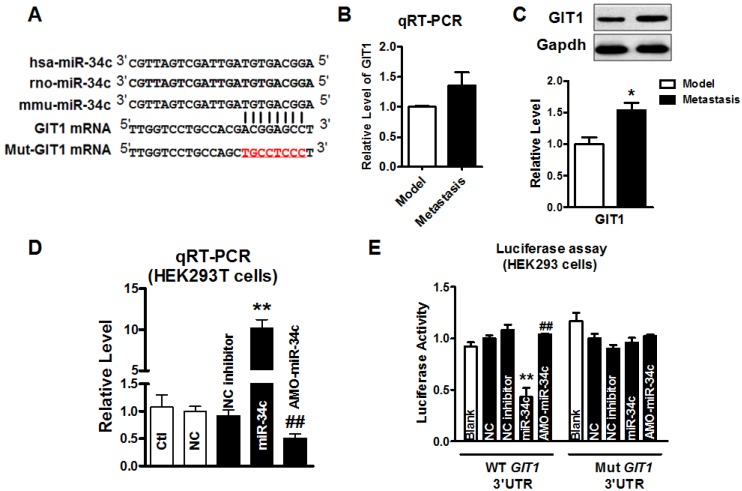
miR-34c targeting at GIT1
We further explored the mechanism underlying the function of miR-34c in suppressing migration and invasion of MCF-7 cells. Computational prediction using TargetScan and miRnada predicted GIT1 gene which encodes GIT1 protein in tumor cells, as a potential target of miR-34c (Figure 3A). Then, we employed qRT-PCR and western blot to determine GIT1 mRNA and protein expression in tumor tissues from model group and metastasis group. As shown in Figure 3B and 3C, the protein level of GIT1 was upregulated significantly, but mRNA level did not change significantly in tumor tissue of metastasis group compared with that in model group. Furthermore, we performed the luciferase assay to determine whether miR-34c targets at GIT1 via binding to GIT1 mRNA 3'UTR. After verified that miR-34c was successfully transfected into HEK293T cells by qRT-PCR (Figure 3D), luciferase assay was carried out. Overexpression miR-34c (50 nM) induced a substantial reduction of luciferase activity of constructs carrying fragment of WT GIT1 3'UTR to 43% (P<0.01) compared with NC group, and this effect was reversed by AMO-miR-34c transfection (Figure 3E). Moreover, transfection of miR-34c or AMO-miR-34c did not affect the luciferase reporter activity with binding site mutation in GIT1 mRNA 3'UTR.
Figure 3.
miR-34c targeting at GIT1. A. The seed sequence of miR-34c is conserved among human, rat and mouse species. Computational prediction of binding site of miR-34c in GIT1 mRNA 3'UTR. 8 nucleotides of miR-34c binding site in GIT1 mRNA 3'UTR was mutated. (red) B. The mRNA level of GIT1 expression showed the increase tendency in metastasis group, but not significantly larger than in tumor of model group. C. GIT1 protein expression was upregulated significantly in metastasis group compared with model group. * P<0.05 vs. model group. n=3 independent samples for each group. D. Transfection efficiency of miR-34c in HEK293T cells was verified by qRT-PCR. E. Luciferase assay in HEK293T cells for post-transcriptional inhibition of GIT1 by miR-34c. WT GIT1 3'UTR luciferase reporter contained the full length of GIT1 3'UTR including miR-34c binding site. Mut GIT1 3'UTR luciferase reporter vector was inserted with mutant 8 mucleotides of miR-34c binding site in full length of GIT1 3'UTR. ** P<0.01 vs. NC group. ## P<0.01 vs. miR-34c group. n=3 independent batches of cells for each group.
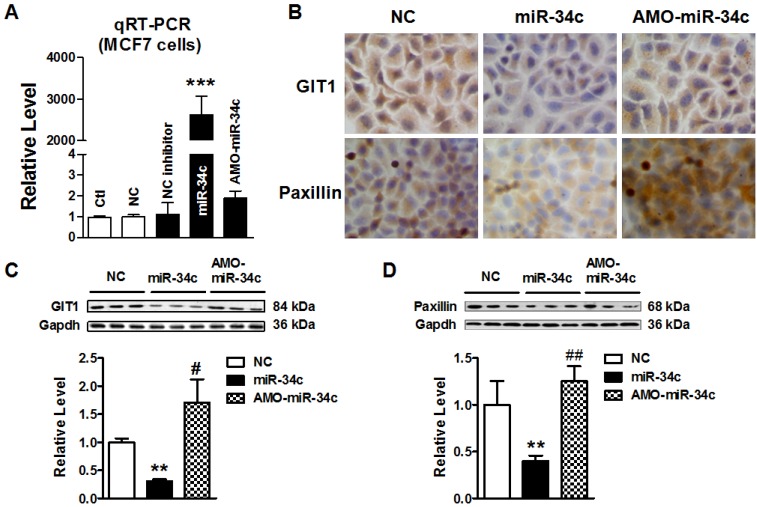
miR-34c suppressed GIT1 protein expression in MCF-7 cells
To further test that miR-34c regulates GIT1 expression in MCF-7 cells, we transfected miR-34c mimics and its inhibitor AMO-miR-34c in MCF-7 cells for 48 hr. Firstly; we identify the efficiency of microRNA transfection in MCF-7 cells. As shown in Figure 4A, miR-34c was effectively overexpressed, and AMO-miR-34c successfully suppressed endogenous miR-34c in MCF-7 cells. Then, immunocytochemistry assay showed that GIT1 protein expression was significantly reduced in cytoplasm of MCF-7 cells after transfected with miR-34c. Previous study has reported that inhibition of GIT1 resulted in protein degradation and reduction of paxillin in cancers 16. In this study, we also found that Paxillin was downregulated in MCF-7 cells with miR-34c overexpression. Inversely, AMO-miR-34c transfection induced the increase of GIT1 and Paxillin protein expression (Figure 4B). Consistent with immunocytochemistry assay results, the protein level of GIT1 expression was decreased significantly in MCF-7 cells after treatment with miR-34c mimics, and these changes were reversed by treatment with AMO-miR-34c. Similar results were obtained in Paxillin protein expression (Figure 4C and D). These results demonstrated miR-34c targeting at GIT1, and also affected expression of Paxillin, GIT1 related gene, in breast cancer cells, which indicated miR-34c potentially participates in the process of tumor metastasis.
Figure 4.
miR-34c suppressed GIT1 protein expression in MCF-7 cells. A. The transfection efficiency of miR-34c in MCF-7 cells was identified by qRT-PCR. B. Immunocytochemistry assay showed the GIT1 and Paxillin protein expression in cytoplasm of MCF-7 cells in different groups. Both GIT1 and Paxillin protein expression were suppressed by miR-34c transfection, and these effects were restored by AMO-miR-34c treatment in MCF-7 cells. C. Western blot detected GIT1 protein expression in MCF-7 cells of miR-34c, AMO-miR-34c and NC groups. D. The protein level of Paxillin in MCF-7 cells of miR-34c, AMO-miR-34c and NC groups detected by western blot technique. ** P<0.01 vs. NC group. *** P<0.001 vs. NC group. # P<0.05 vs. miR-34c group. ## P<0.01 vs. miR-34c group. n=3 independent batches of cells for each group.
Knockdown of GIT1 restored cell migration and invasion in MCF-7 cells
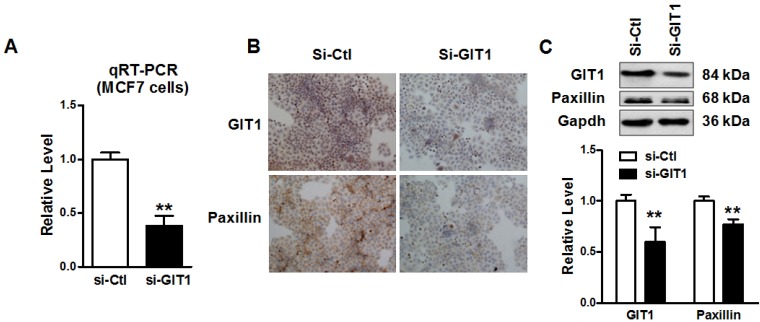
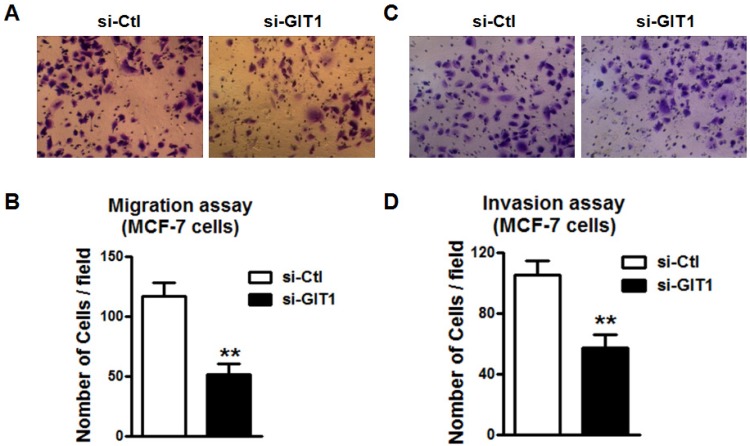
To assess the role of GIT1 in cancer metastasis, we knockdowned endogenous GIT1 expression by small interfering RNAs (siRNAs) transfection in MCF-7 cells. After transfected with si-GIT1 in MCF-7 cells for 48 hours, the mRNA level of GIT1 was significantly suppressed (Figure 5A) and the protein levels of both GIT1 and Paxillin expression were significantly decreased by immunocytochemistry and western blot as well (Figure 5B and C). It suggested that siRNA si-GIT1 effectively suppressed endogenous GIT1 protein expression. Consistently, si-GIT1 also inhibited cell migration and invasion significantly in MCF-7 cells (Figure 6). Thus, knockdown of GIT1 suppressed cell migration and invasion of MCF-7 breast cancer cells.
Figure 5.
Knockdown of endogenous GIT1 expression in MCF-7 cells. A. GIT1 mRNA level was significantly reduced by si-GIT1 transfection in MCF-7 cells for 48 hours. B. Immunocytochemistry assay showed that GIT1 and Paxillin protein expression was reduced in MCF-7 cells. C. Western blot assay demonstrated that si-GIT1 successfully suppressed endogenous GIT1 protein expression, and Paxillin protein expression was downregulated with GIT1 protein expression as well. ** P<0.01 vs. si-Ctl group. n=3 independent batches of cells for each group.
Figure 6.
Cell migration and invasion in MCF-7 cells was restored by knockdown of endogenous GIT1 expression. A. Cell migration of MCF-7 cells was inhibited by knockdown of endogenous GIT1 protein by si-GIT1. B. Quantitative analysis of cell migration of MCF-7 cells in si-Ctl and si-GIT1 groups. C. Cell invasion assay of MCF-7 cells was restored by si-GIT1. D. Quantitative analysis of cell invasion in MCF-7 cells of each group. ** P<0.01 vs. si-Ctl group. n=6 independent batches of cells for each group.
miR-34c regulated the migration and invasion of MDA-MB-231 cells
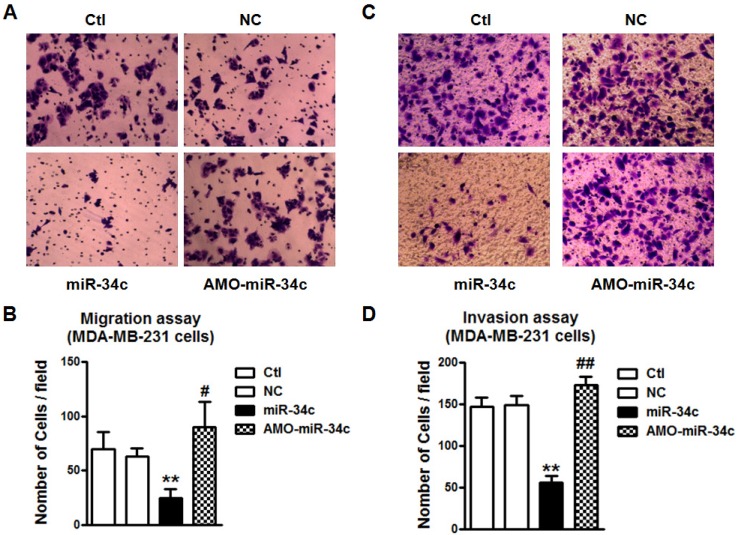
We also tested the regulatory effects of miR-34c on the migration and invasion of MDA-MB-231 cells. We found that overexpression of miR-34c using miR-34c mimics inhibited the migration of MDA-MB-231 cells, but the suppression of endogenous miR-34c promoted MDA-MB-231 migration (Figure 7A and B). The MDA-MB-231 cell invasion was also markedly inhibited after transfection of miR-34c, and enhanced by treatment with AMO-miR-34c (Figure 7C and D). It suggests that miR-34c also plays regulatory roles in the migration and invasion of MDA-MB-231 cells.
Figure 7.
miR-34c suppresses cell migration and invasion of MDA-MB-231 cells. A. After treatment with miR-34c mimics, AMO-miR-34c and NC in MDA-MB-231 cells for 48 hours, migration assay was performed. B. Quantitative analysis of cell migration in MDA-MB-231 cells treated with miR-34c mimics, AMO-miR-34c and NC. C. The representative pictures of MDA-MB-231 invasion after transfection of miR-34c, AMO-miR-34c and NC for 48 hours. D. Statistical analysis of MDA-MB-231 cell invasion of each group. ** P<0.01 vs. NC group. # P<0.01 vs. miR-34c group. ## P<0.01 vs. miR-34c group. n=6 independent batches of cells for each group.
GIT1 mediated the inhibition of MDA-MB-231 cell migration and invasion by miR-34c
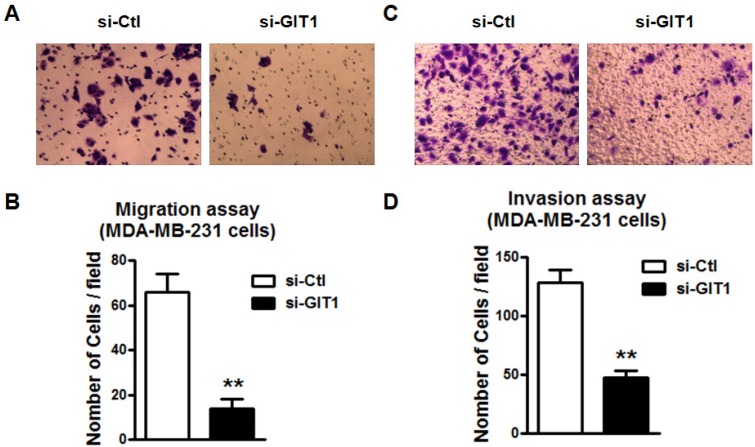
Further study was performed to study if GIT1 is also involved in miR-34c-induced the inhibition of migration and invasion of MDA-MB-231 cells. Similarly, si-GIT1 transfection led to the suppression of cell migration of MDA-MB-231 (Figure 8A and B). The invasion of MDA-MB-231 cells was also suppressed by si-GIT1 (Figure 8C and D). This implicates that GIT1 as the target of miR-34c plays important roles in MDA-MB-231 cell migration and invasion.
Figure 8.
GIT1 regulated cell migration and invasion of MDA-MB-231 cells. A and B. si-GIT1 suppressed cell migration of MDA-MB-231 cells. C and D. Cell invasion assay was preformed to study the effects of si-GIT1 on invasion of MDA-MB-231 cells. ** P<0.01 vs. si-Ctl group. n=6 independent batches of cells for each group.
Discussion
In present study, we found that miR-34c was aberrantly downregulated in breast tumor of mice with lung and liver metastasis, accompanied by increased expression of GIT1. Consistently, loss of function of miR-34c promoted the migration and invasion of both MCF-7 cell and MDA-MB-231 breast cancer cells. Inversely, gain of function of miR-34c did the opposite. We further identified that knockdown of GIT1 led to the inhibition of cell migration and invasion of breast cancer cells. The study therefore provides the evidence that miR-34c plays regulatory roles in breast cancer cell migration and invasion.
Due to the high morbidity and mortality following breast cancer metastasis, it is urgently needed to look for new molecules for the early diagnosis and treatment of breast cancer metastasis. A large body of evidence has revealed that miRNAs act as oncogenes or tumor suppressor in process of tumor development and progression, by affecting tumor cell proliferation, apoptosis, migration and invasion 17-18. Therefore, miRNAs potentially function as new promising targets for cancer diagnosis and therapeutics 19. It has been shown that miRNAs are frequently associated with chromosomal instability and amplification in multiple human cancers 17. Nowadays, many miRNAs have been found correlated with breast cancer. It was reported by Wang et al. that miR-320a inhibits the proliferation and invasion of breast cancer by targeting RAB11A 18. Li et al. reported that miR-762 promotes the proliferation and invasion of breast cancer MCF-7 cells by targeting IRF7 expression 20. However, whether miRNAs takes part in the development of breast cancer has not been reported yet.
In the in vivo study, we found that miR-34c, miR-93, miR-21 and miR-27a were significantly decreased in metastatic breast tumors. Furthermore, qRT-PCR was verified that miR-34c is the most significantly decreased compared with other miRNAs. Actually, previous studies have reported that miR-34c was downregulated in the tissues of non-small cell lung cancer, nasopharyngeal carcinoma, and prostate cancer cells 21-23. It indicated that miR-34c might participate in metastasis of breast cancer. To further verify the function of miR-34c in this process, we forced expression miR-34c in MCF-7 and MDA-MB-231 cells in in vitro study and found that migration and invasion of breast cancer cells were significantly inhibited by transfection with miR-34c. Conversely, silencing of miR-34c by AMO-miR-34c promoted the migration and invasion of both MCF-7 and MDA-MB-231 cells. This result is in consistent with the previous findings on the antitumor effects of miR-34c 24.
Then, we further explored the molecular mechanisms of miR-34c in breast cancer metastasis. It has been widely reported that GIT1 was associated with tumor cell migration, invasion as well as angiogenesis 10, 16, 25. As a key factor of tumor metastasis, GIT1 protein level was significantly increased while its mRNA level was only elevated slightly in metastasis mouse model, which suggested that the expression of GIT1 is posttranscriptionally regulated in the process of breast cancer metastasis. By using two microRNA database software (TargetScan, miRnada), we found that miR-34c was conserved in multiple species (Human, mouse and rat), and predicted that GIT1 is one of the targets of miR-34c. The result of luciferase assay proved that miR-34c targeted GIT1 mRNA 3'UTR and negatively regulated GIT1 protein expression. Paxillin is a signal transduction adaptor protein, and functions to adhering cells to the extracellular matrix. The N-terminal region of paxillin is rich in protein-protein interaction sites. The protein that may bind to paxillin includes focal adhesion kinase (FAK)-Src complex 26, and GIT1/βPIX/pTyr-PAK1 complex, which regulate breast cancer cell motility and invasion 8, 27. Here we found that overexpression miR-34c induced GIT1 downregulation, accompanied by Paxillin downregulation in MCF-7 cells, which indicated miR-34c potentially participated in the migration and invasion of breast cancer cell through GIT1/Paxillin pathway. It was shown that GIT1 can interact with paxillin, which increased instability of paxillin and prevented it from protein degradation in cells 16. In this study, we found that knockdown of GIT1 by siRNA induced the downregulation of both GIT1 protein and Paxillin protein, which restored cell migration as well as invasion in breast cancer cells. It strongly supported that GIT1/Paxillin is responsible for the migration and invasion of breast cancer cells, which is partially consistent with current reports 16.
At last, the limitation of this study is lacking of observation in clinical patients of metastatic breast cancer. It has reported that high level of GIT1 is correlated with advanced stages and lymph node metastasis of breast cancer 16. Therefore, the level of miR-34c in human patients with metastatic breast cancer will be examined in the future study.
Our present study showed that miR-34c regulates cell migration and invasion of breast cancer cells through targeting GIT1 signaling pathway. It suggests that miR-34c has the potential to become a new diagnostic biomarker for breast cancer metastasis, and miR-34c-GIT1 is a novel microRNA pathway for migration and invasion of breast cancer. This study provided a new strategy for treatment of breast cancer.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81101997/H1622), Foundation of Educational Commission of Heilongjiang Province (12521280), Heilongjiang Postdoctoral Fund (LBH-Z11085), China Postdoctoral Science Foundation (2011M500694, 2014T70368), Specialized Research Fund for the Doctoral Program of Higher Education of China for new teacher (20112307120019), Natural Science Foundation of Heilongjiang Province (H2016052) and the funding of The Affiliated Tumor Hospital of Harbin Medical University (JJZ2010-14).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. doi:10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Dow KH, Ferrell BR, Leigh S, Ly J, Gulasekaram P. An evaluation of the quality of life among long-term survivors of breast cancer. Breast Cancer Res Treat. 1996;39:261–73. doi: 10.1007/BF01806154. [DOI] [PubMed] [Google Scholar]
- 3.Jin X, Mu P. Targeting Breast Cancer Metastasis. Breast Cancer (Auckl) 2015;9:23–34. doi: 10.4137/BCBCR.S25460. doi:10.4137/BCBCR.S25460 bcbcr-suppl.1-2015-023 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ursini-Siegel J, Siegel PM. The influence of the pre-metastatic niche on breast cancer metastasis. Cancer Lett; 2015. doi:S0304-3835(15)00683-7 [pii] 10.1016/j.canlet.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–75. doi: 10.1242/jcs.02925. doi:119/8/1469 [pii]10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 6.Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, Yu JS. Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Mol Biol Cell. 2010;21:287–301. doi: 10.1091/mbc.E09-03-0232. doi:E09-03-0232 [pii]10.1091/mbc.E09-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlenker O, Rittinger K. Structures of dimeric GIT1 and trimeric beta-PIX and implications for GIT-PIX complex assembly. J Mol Biol. 2009;386:280–9. doi: 10.1016/j.jmb.2008.12.050. doi:S0022-2836(08)01570-2 [pii]10.1016/j.jmb.2008.12.050. [DOI] [PubMed] [Google Scholar]
- 8.Schmalzigaug R, Garron ML, Roseman JT, Xing Y, Davidson CE, Arold ST. et al. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell Signal. 2007;19:1733–44. doi: 10.1016/j.cellsig.2007.03.010. doi:S0898-6568(07)00096-4 [pii]10.1016/j.cellsig.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang JS, Su CY, Yu WH, Lee WJ, Liu YP, Lai TC. et al. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget. 2015;6:36278–91. doi: 10.18632/oncotarget.5531. doi:5531 [pii]10.18632/oncotarget.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penela P, Nogues L, Mayor F Jr. Role of G protein-coupled receptor kinases in cell migration. Curr Opin Cell Biol. 2014;27:10–7. doi: 10.1016/j.ceb.2013.10.005. doi:S0955-0674(13)00158-0 [pii]10.1016/j.ceb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Majumder S, Sowden MP, Gerber SA, Thomas T, Christie CK, Mohan A. et al. G-protein-coupled receptor-2-interacting protein-1 is required for endothelial cell directional migration and tumor angiogenesis via cortactin-dependent lamellipodia formation. Arterioscler Thromb Vasc Biol. 2014;34:419–26. doi: 10.1161/ATVBAHA.113.302689. doi:ATVBAHA.113.302689 [pii]10.1161/ATVBAHA.113.302689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vescovo VD, Denti MA. microRNA and Lung Cancer. Adv Exp Med Biol. 2015;889:153–77. doi: 10.1007/978-3-319-23730-5_9. doi:10.1007/978-3-319-23730-5_9. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA Processing and Human Cancer. J Clin Med. 2015;4:1651–67. doi: 10.3390/jcm4081651. doi:jcm4081651 [pii]10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansoori B, Mohammadi A, Shirjang S, Baradaran B. Micro-RNAs: The new potential biomarkers in cancer diagnosis, prognosis and cancer therapy. Cell Mol Biol (Noisy-le-grand) 2015;61:1–10. [PubMed] [Google Scholar]
- 15.Tao WY, Liang XS, Liu Y, Wang CY, Pang D. Decrease of let-7f in low-dose metronomic Paclitaxel chemotherapy contributed to upregulation of thrombospondin-1 in breast cancer. Int J Biol Sci. 2015;11:48–58. doi: 10.7150/ijbs.9969. doi:10.7150/ijbs.9969ijbsv11p0048 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY. et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–507. doi: 10.1038/onc.2014.10. doi:onc201410 [pii]10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. doi:nature03702 [pii]10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Yang Z, Wang H, Cao Z, Zhao Y, Gong C. et al. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am J Cancer Res. 2015;5:2719–29. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang V, Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15–23. doi: 10.2165/00063030-200923010-00002. doi:2 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Huang R, Wang L, Hao J, Zhang Q, Ling R. et al. microRNA-762 promotes breast cancer cell proliferation and invasion by targeting IRF7 expression. Cell Prolif. 2015;48:643–9. doi: 10.1111/cpr.12223. doi:10.1111/cpr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Wang X, Li J, Gu K, Lv L, Zhang S. et al. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015;48:582–92. doi: 10.1111/cpr.12201. doi:10.1111/cpr.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YQ, Ren XY, He QM, Xu YF, Tang XR, Sun Y. et al. MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. 2015;6:e1618.. doi: 10.1038/cddis.2014.582. doi:cddis2014582 [pii]10.1038/cddis.2014.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagman Z, Haflidadottir BS, Ansari M, Persson M, Bjartell A, Edsjo A. et al. The tumour suppressor miR-34c targets MET in prostate cancer cells. Br J Cancer. 2013;109:1271–8. doi: 10.1038/bjc.2013.449. doi: 10.1038/bjc.2013.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez JA, Alvarez-Salas LM. Differential effects of miR-34c-3p and miR-34c-5p on SiHa cells proliferation apoptosis, migration and invasion. Biochem Biophys Res Commun. 2011;409:513–9. doi: 10.1016/j.bbrc.2011.05.036. doi:S0006-291X(11)00796-0 [pii]10.1016/j.bbrc.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 25.Pang J, Xu X, Wang X, Majumder S, Wang J, Korshunov VA. et al. G-protein-coupled receptor kinase interacting protein-1 mediates intima formation by regulating vascular smooth muscle proliferation, apoptosis, and migration. Arterioscler Thromb Vasc Biol. 2013;33:999–1005. doi: 10.1161/ATVBAHA.112.300966. doi:ATVBAHA.112.300966 [pii]10.1161/ATVBAHA.112.300966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kratimenos P, Koutroulis I, Marconi D, Syriopoulou V, Delivoria-Papadopoulos M, Chrousos GP. et al. Multi-targeted molecular therapeutic approach in aggressive neuroblastoma: the effect of Focal Adhesion Kinase-Src-Paxillin system. Expert Opin Ther Targets. 2014;18:1395–406. doi: 10.1517/14728222.2014.952280. doi:10.1517/14728222.2014.952280. [DOI] [PubMed] [Google Scholar]
- 27.Hammer A, Diakonova M. Tyrosyl phosphorylated serine-threonine kinase PAK1 is a novel regulator of prolactin-dependent breast cancer cell motility and invasion. Adv Exp Med Biol. 2015;846:97–137. doi: 10.1007/978-3-319-12114-7_5. doi:10.1007/978-3-319-12114-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]