Abstract
Some adult-onset disorders may be linked to dysregulated embryonic development, yet the mechanisms underlying this association remain poorly understood. Congenital retinal degenerative diseases are blinding disorders characterized by postnatal degeneration of photoreceptors, and affect nearly 2 million individuals worldwide, but ∼50% do not have a known mutation, implicating contributions of epigenetic factors. We found that embryonic deletion of the histone methyltransferase (HMT) Ezh2 from all retinal progenitors resulted in progressive photoreceptor degeneration throughout postnatal life, via derepression of fetal expression of Six1 and its targets. Forced expression of Six1 in the postnatal retina was sufficient to induce photoreceptor degeneration. Ezh2, although enriched in the embryonic retina, was not present in the mature retina; these data reveal an Ezh2-mediated feed-forward pathway that is required for maintaining photoreceptor homeostasis in the adult and suggest novel targets for retinal degeneration therapy.
Postnatal degeneration of photoreceptors underlies the most common causes of irreversible blindness. Rod-cone photoreceptor dystrophy is a group of congenital retinal degenerative disorders characterized by postnatal onset of progressive photoreceptor cell loss1,2, leading to severe visual impairment and deterioration. Molecular genetic studies have revealed the underlying causes in only ∼50% of these patients. Striking characteristics of these diseases are their genetic heterogeneity and large variability in the age of onset, progression, and final visual outcome that can be found even between monozygotic twins3,4,5, suggesting the impact of factors other than genetics on its pathogenesis. Contribution of epigenetically-regulated events in utero toward the adult onset of retinal degeneration is unclear.
Due to minimal cell turnover, the postnatal retina is particularly vulnerable to insults during embryonic development. Epigenetic events mediated by repressive histone methylation regulate the transcriptional programs in proliferating embryonic stem cells and their differentiated, post-mitotic progeny by suppressing transcription in specific fetal genes. These early developmental events may serve as a nexus point of cell proliferation and differentiation that persists into adulthood to allow establishment of stabilized expression of postnatal genes and maintain tissue homeostasis. Perturbation of this process, via inhibition of repressive histone methyltransferase (HMT) activity in embryonic retinal progenitors, may lead to disruption of postnatal homeostasis. Consequently, inhibition of repressive HMT activity could lead to postnatal retinal degeneration through “developmental reprogramming” that stems from aberrant derepression of a subset of normally silenced genes6.
A candidate repressive HMT whose embryonic disruption may interfere with postnatal retinal homeostasis is enhancer of zeste homolog 2 (Ezh2), the enzymatic subunit of the polycomb repressive complex 2 (PRC2). Ezh2 trimethylates histone H3 at lysine 27 (H3K27me3), a repressive histone mark that is known to persist into adulthood7,8. Previous reports have shown that Ezh2/EZH2 is abundant during embryonic development but is absent during the late postnatal period and adulthood in mice and humans9,10,11. Conditional loss-of-function experiments in which an early (E9.5) retinal Cre driver, such as Dkk3-12 or Pax6-αCre, was used to delete retinal Ezh2, has demonstrated a role for this HMT in the proliferation and differentiation of retinal progenitor cells (RPCs)13,14. To circumvent this issue and focus on a role for Ezh2-mediated repressive modifications in postnatal retinal homeostasis, we generated mice carrying Ezh2 deficiency selectively in RPCs at a later time point (E11.5 and later), using Chx10 promoter15,16,17. Here we report that Chx10-Cre; Ezh2 deficient retinae developed normally to the perinatal period; however Ezh2 mutant retinae underwent progressive photoreceptor degeneration postnatally, a period during which Ezh2 is no longer expressed in wild type retinae. Taken together, these findings provide new insights into how dysregulation of Ezh2 function in development may contribute to retinal degeneration in postnatal life.
Results
Ezh2 deficiency driven by Chx10-Cre induces progressive retinal degeneration in postnatal life
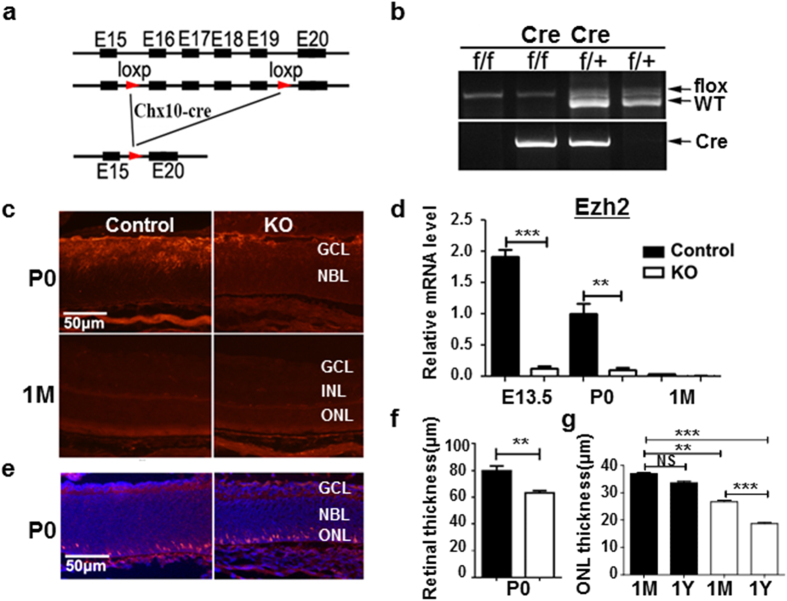
Mice with loxP sites flanking exons encoding the catalytic SET domain of Ezh2 (Ezh2flox/flox) (Fig. 1a) were crossed with mice expressing Cre-recombinase under the control of a bacterial artificial chromosome encoding Chx10 (Chx10-Cre). The Chx10-cre mouse line directs high-level Cre expression after E11.5 onward in intermediate and late RPCs, which leads to retinal-specific excision in virtually all RPC-derived cells of the retina16. All mice were genotyped by PCR of tail genomic DNA for Ezh2flox/flox alleles (Fig. 1b). In knockout (KO or Chx10-cre;Ezh2flox/flox) mice, the level of Ezh2 expression was nearly undetectable at E13 through postnatal 1 month (1 M) of age (Fig. 1c,d). Consequently, H3K27me3 deposition in retinal progenitor cells (RPCs; neuroblast layer) of KO mice was largely decreased compared to WT controls at E13.5 (Supplementary Fig. 1). In contrast, Ezh2 was detected in RPCs of the embryonic retinas of control mice (Ezh2flox/flox); at P0, this expression was limited to the ganglion cell layer (GCL; Fig. 1c) but its expression was extinguished in the retina at 1 M (Fig. 1c,d). These data confirmed that Ezh2 was successfully deleted in the retina of KO mice.
Figure 1. Ezh2 deletion induces progressive retinal degeneration.
(a) Intron/exon diagram of mice Ezh2 and Ezh2flox/flox alleles. (b) Genotyping of Ezh2 and Chx10-cre genes. (c,d) Immunohistochemistry (c) and qPCR (d) assessment of Ezh2 expression in the retinas of control and KO mice: note that Ezh2 expression was detected in the P0 control retina, but was lost in the adult retina. (e) Retinal sections labeled for nuclear marker DAPI (blue) and photoreceptor cell marker anti-Recoverin (red). (f) Retinal thickness measured in P0 retinal sections. (g) Quantification of ONL thickness in retinal sections of mice at different ages, showing progressive ONL degeneration from 1 M to 1 Y in the KO mouse retina. Bars represent the mean ± S.D. of at least six biological replicates. NS indicates no significant difference; **indicates P < 0.01; ***indicates P < 0.001.
To assess the effect of Ezh2 deletion, we compared the morphology and numbers of retinal neurons in KO mice to control mice at different stages. KO mice exhibited largely normal structured retina at birth; however, despite the normally absent expression of Ezh2 at 1 M, KO retina underwent progressive degeneration that gradually occurred from 1 M throughout adulthood. Consistent with the reported role of Ezh2 in regulating RPC proliferation and retinogenesis, the retina of KO mice (Fig. 1e,f) was slightly (∼20%) thinner than their littermate controls or those with heterozygous Ezh2 deletion (Ezh2+/− or Chx10-cre; Ezh2flox/+) at P0.
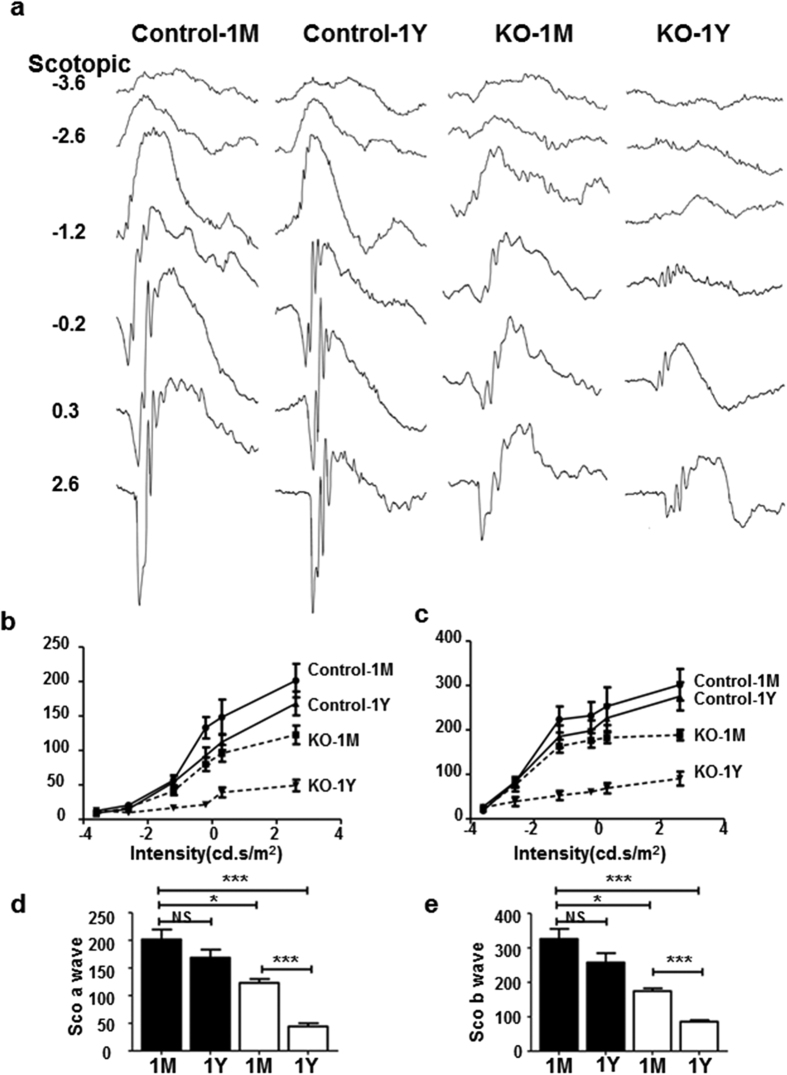
The thickness of the retina, especially the outer nuclear layer (ONL), continued to decrease from 1 M to 12 M (Fig. 1g and Supplementary Fig. 2). This was confirmed by measuring retinal thicknesses in both H&E stained retinal sections (Supplementary Fig. 2a,b) and by non-invasive imaging with high-resolution spectrum domain-optic coherence topography in live animals (Supplementary Fig. 2d,e). The ONL thickness of 1 M KO mice was ∼25% thinner than the control mice; by 12 M, and it was reduced to 50% of that in littermate controls. Ultrastructural analysis of photoreceptor outer segments (OS) with electron microscopy revealed less dense and sometimes disintegrated OS in 1 M KO mice, supporting the on-going photoreceptor degeneration in these mice; whereas, the retina of their littermate controls exhibited well-aligned OS discs (Supplementary Fig. 2f). Moreover, electroretinography (ERG) analyses showed progressive loss of photoreceptor functions as measured by scotopic a-wave and photopic b-wave, which represent rod and cone functions, respectively. From 1 M to 12 M, the amplitudes of scotopic a-wave of KO mice reduced from 65% of that of littermate controls to 20%; scotopic (Fig. 2e) and photopic (not shown) b-waves exhibited similar reduction from 1 M to 12 M (Fig. 2). Less robust, but significant, loss of bipolar cells (10% at 1 M and 25% at 12 M) was also observed in KO retinae as compared with that of littermate controls (Supplementary Fig. 3). No significant loss of RGCs was noted in the KO retina as compared to littermate control mice (not shown). Together, our data indicates that Ezh2 deficiency during development results in progressive retinal, primarily photoreceptor, degeneration throughout the postnatal period.
Figure 2. Ezh2 deletion impairs postnatal photoreceptor cell function.
(a) Representative ERG waveforms of control and KO mice that were subjected to flashes of increasing intensities under scotopic conditions, which isolate rod photoreceptor-mediated responses. (b–e) The amplitudes of scotopic ERG a- and b-waves, which measures cone responses: Black bar, WT controls; white bar, KO. Note the abnormal and progressive reduction of ERG a- and b-wave amplitudes in KO mice from 1 M to 1 Y. Data represent the mean ± S.D. of at least six biological replicates. NS indicates no significant difference; *indicates P < 0.05; **indicates P < 0.01; ***indicates P < 0.001.
Ezh2 deficiency results in selective dysregulation of Six1 and photoreceptor-related genes
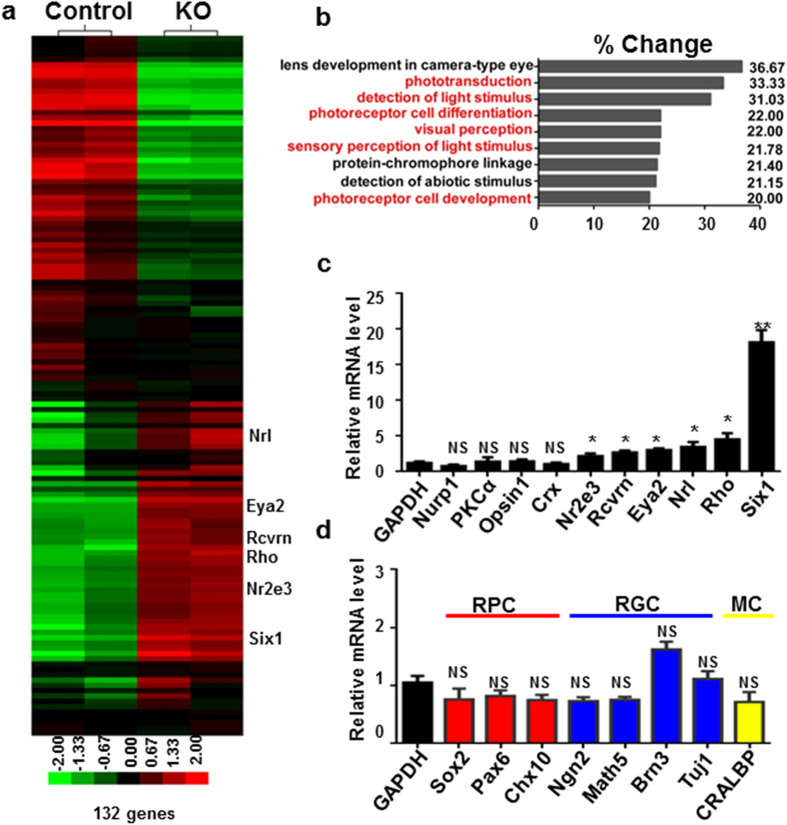
To define the genetic landscape of the KO vs control retinae, we analyzed gene transcripts in the P0 retina using Affymetrix cDNA microarray. P0 was selected because this is when Ezh2 disappears from the outer neuroblast layer or RPCs while KO retina revealed a largely normal appearance without massive degeneration. Surprisingly, instead of finding a broad change in gene expression profiles, our comparison of over 24,000 cDNA transcripts (detected from >28,000 transcripts being screened) of KO vs. control retinae of P0 mice revealed only 132 genes with over 1.5 fold change (Fig. 3a and Table 1). As Ezh2 is usually considered to deposit a repressive mark, we performed gene Ontology (GO) (http://david.abcc.ncifcrf.gov/) analysis18,19 particularly for genes that were upregulated in KO mice and likely repressive targets of Ezh2. Remarkably, the large majority of these genes were related to photoreceptor development or function (Fig. 3b,c), implicating selective and cell type-specific gene regulation or suppression in RPCs by Ezh2.
Figure 3. Ezh2 deletion promotes derepression of Six1 and photoreceptor-related genes.
(a) Heat map of clustered microarray data from P0 retina showed 132 genes changed >1.5 fold between control and KO mice. (b) GO analysis of functional categories identified a high number of genes that are directly involved in photoreceptor development, differentiation and photosensitivity. (c,d) qPCR results from P0 WT and KO retina showed the relative expression of retinal progenitor cell (RPC) and neuron-related genes in KO mice at P0. RGCs, retinal ganglion cells; MC, Müller cells. Note the significant elevation of Six1, Eya2 and rod photoreceptor-related gene expression, including Nr2e3, Recoverin (Rcvrn), Nrl and Rhodopsin (Rho), in KO mice. Bars represent the mean ± S.D. of at least six biological replicates. NS indicates no significant difference; *indicates P < 0.05; **indicates P < 0.01; ***indicates P < 0.001.
Table 1. Representative up and down-regulated genes in Chx10-cre Ezh2 flox/flox mice P0 retina from microarray hybridization.
| NM_175540 | Eda2r | ectodysplasin A2 isoform receptor | 3.95 |
| NM_009189 | Six1 | sine oculis-related homeobox 1 homolog (Drosophila) | 3.83 |
| NM_009849 | Entpd2 | ectonucleoside triphosphate diphosphohydrolase 2 | 3.19 |
| NM_008992 | Abcd4 | ATP-binding cassette, sub-family D (ALD), member 4 | 2.93 |
| NM_007723 | Cnga1 | cyclic nucleotide gated channel alpha 1 | 2.41 |
| NM_028864 | Zc3hav1 | zinc finger CCCH type, antiviral 1 | 2.32 |
| NM_013750 | Phlda3 | pleckstrin homology-like domain, family A, member 3 | 2.31 |
| NM_030707 | Fcrls | Fc receptor-like S, scavenger receptor | 2.3 |
| NM_013708 | Nr2e3 | nuclear receptor subfamily 2, group E, member 3 | 2.29 |
| NM_023048 | Asb4 | ankyrin repeat and SOCS box-containing 4 | 2.27 |
| NM_182993 | Slc17a7 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | 2.25 |
| NM_145383 | Rho | rhodopsin | 2.23 |
| NM_011978 | Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 | 2.2 |
| NM_009038 | Rcvrn | recoverin | 2.19 |
| NM_009022 | Aldh1a2 | aldehyde dehydrogenase family 1, subfamily A2 | 2.18 |
| NM_008806 | Pde6b | phosphodiesterase 6B, cGMP, rod receptor, beta polypeptide | 2.1 |
| BC086653 | Lba1 | lupus brain antigen 1 | 2 |
| NM_010720 | Lipg | lipase, endothelial | 1.89 |
| NM_010165 | Eya2 | eyes absent 2 homolog (Drosophila) | 1.88 |
| NM_011851 | Nt5e | 5′ nucleotidase, ecto | 1.88 |
| NM_001031772 | Lin28b | lin-28 homolog B (C. elegans) | 1.85 |
| NM_023456 | Npy | neuropeptide Y | 1.85 |
| NM_008630 | Mt2 | metallothionein 2 | 1.82 |
| NM_007669 | Cdkn1a | cyclin-dependent kinase inhibitor 1A (P21) | 1.81 |
| NM_133982 | Rpp25 | ribonuclease P 25 subunit (human) | 1.79 |
| NM_176844 | Chrna5 | cholinergic receptor, nicotinic, alpha polypeptide 5 | 1.78 |
| NM_008736 | Nrl | neural retina leucine zipper gene | 1.78 |
| NM_028713 | Rftn2 | raftlin family member 2 | 1.76 |
| NM_013602 | Mt1 | metallothionein 1 | 1.73 |
| NM_001033167 | Slc22a23 | solute carrier family 22, member 23 | 1.73 |
| NM_001128103 | Ano3 | anoctamin 3 | 1.72 |
| NM_031257 | Plekha2 | pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 | 1.7 |
| NM_009573 | Zic1 | zinc finger protein of the cerebellum 1 | 1.7 |
| NM_008532 | Tacstd1 | tumor-associated calcium signal transducer 1 | 1.66 |
| NM_178254 | Tcfl5 | transcription factor-like 5 (basic helix-loop-helix) | 1.66 |
| NM_013454 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 1.64 |
| NM_019397 | Egfl6 | EGF-like-domain, multiple 6 | 1.63 |
| NM_027871 | Arhgef3 | Rho guanine nucleotide exchange factor (GEF) 3 | 1.62 |
| NM_172119 | Dio3 | deiodinase, iodothyronine type III | 1.62 |
| NM_019413 | Robo1 | roundabout homolog 1 (Drosophila) | 1.58 |
| NM_153546 | Mboat1 | membrane bound O-acyltransferase domain containing 1 | −1.57 |
| NM_009657 | Aldoc | aldolase C, fructose-bisphosphate | −1.62 |
| NM_172880 | Tmprss11e | transmembrane protease, serine 11e | −1.65 |
| NM_008397 | Itga6 | integrin alpha 6 | −1.66 |
| NM_009528 | Wnt7b | wingless-related MMTV integration site 7B | −1.66 |
| NM_033268 | Actn2 | actinin alpha 2 | −1.68 |
| NM_025681 | Lix1 | limb expression 1 homolog (chicken) | −1.69 |
| NM_007515 | Slc7a3 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 | −1.7 |
| NM_015800 | Crim1 | cysteine rich transmembrane BMP regulator 1 (chordin like) | −1.71 |
| NM_001113331 | Shc1 | src homology 2 domain-containing transforming protein C1 | −1.77 |
| NM_011990 | Slc7a11 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 | −1.77 |
| NM_011419 | Jarid1d | jumonji, AT rich interactive domain 1D (Rbp2 like) | −1.79 |
| NM_173379 | Leprel1 | leprecan-like 1 | −1.81 |
| NM_009932 | Col4a2 | collagen, type IV, alpha 2 | −1.82 |
| NM_001035533 | Akap2 | A kinase (PRKA) anchor protein 2 | −1.83 |
| NM_012008 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | −1.84 |
| NM_009527 | Wnt7a | wingless-related MMTV integration site 7A | −1.84 |
| NM_027934 | Rnf180 | ring finger protein 180 | −1.88 |
| NM_013737 | Pla2g7 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | −1.89 |
| NM_007514 | Slc7a2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | −1.89 |
| NM_009484 | Uty | ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | −1.9 |
| NM_012011 | Eif2s3y | eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | −1.95 |
| NM_025760 | Ptplad2 | protein tyrosine phosphatase-like A domain containing 2 | −1.97 |
| NM_172838 | Slc16a12 | solute carrier family 16 (monocarboxylic acid transporters), member 12 | −1.97 |
| NM_009848 | Entpd1 | ectonucleoside triphosphate diphosphohydrolase 1 | −2.03 |
| NM_007933 | Eno3 | enolase 3, beta muscle | −2.05 |
| NM_026878 | Rasl11b | RAS-like, family 11, member B | −2.11 |
| NM_025769 | Efcab1 | EF hand calcium binding domain 1 | −2.18 |
| NM_146120 | Gsn | gelsolin | −2.23 |
| NM_198191 | Pip5kl1 | phosphatidylinositol-4-phosphate 5-kinase-like 1 | −2.24 |
| NM_026056 | Cap2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | −2.25 |
| NM_172868 | Palm2 | paralemmin 2 | −2.25 |
| NM_007773 | Crybb2 | crystallin, beta B2 | −2.26 |
| NM_011939 | Hsf4 | heat shock transcription factor 4 | −2.28 |
| NM_172152 | Slc24a4 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 | −2.3 |
| NM_144945 | Lgi2 | leucine-rich repeat LGI family, member 2 | −2.34 |
| ENSMUST00000108875 | Birc7 | baculoviral IAP repeat-containing 7 (livin) | −2.35 |
| NM_177780 | Dock5 | dedicator of cytokinesis 5 | −2.38 |
| NM_008180 | Gss | glutathione synthetase | −2.4 |
| NM_008010 | Fgfr3 | fibroblast growth factor receptor 3 | −2.43 |
| NM_011224 | Pygm | muscle glycogen phosphorylase | −2.43 |
| NM_025711 | Aspn | asporin | −2.49 |
| NM_013822 | Jag1 | jagged 1 | −2.68 |
| NM_146142 | Tdrd7 | tudor domain containing 7 | −2.73 |
| NM_030127 | Htra3 | HtrA serine peptidase 3 | −2.84 |
| NM_177041 | Flad1 | RFad1, flavin adenine dinucleotide synthetase, homolog (yeast) | −2.9 |
| NM_181541 | Caprin2 | caprin family member 2 | −3.1 |
| NM_001113368 | Ceacam2 | carcinoembryonic antigen-related cell adhesion molecule 2 | −3.12 |
| NM_011325 | Scnn1b | sodium channel, nonvoltage-gated 1 beta | −3.14 |
| NM_007776 | Crygd | crystallin, gamma D | −3.19 |
| NM_026439 | Ccdc80 | coiled-coil domain containing 80 | −3.34 |
| NM_138953 | Ell2 | elongation factor RNA polymerase II 2 | −3.34 |
| XR_032001 | Csnk2a1-rs2 | casein kinase 2, alpha 1, related sequence 2 | −3.39 |
| NM_010442 | Hmox1 | heme oxygenase (decycling) 1 | −3.68 |
| NM_028813 | Vit | vitrin | −3.74 |
| NM_001011807 | Olfr191 | olfactory receptor 191 | −4 |
| NM_019738 | Nupr1 | nuclear protein 1 | −4.03 |
| NM_138683 | Rspo1 | R-spondin homolog (Xenopus laevis) | −4.23 |
| NM_008100 | Gcg | glucagon | −4.64 |
| NM_010917 | Nid1 | nidogen 1 | −4.7 |
| NM_013501 | Cryaa | crystallin, alpha A | −4.79 |
| NM_009604 | Chrng | cholinergic receptor, nicotinic, gamma polypeptide | −5.11 |
| NM_146405 | Olfr228 | olfactory receptor 228 | −5.4 |
| NM_145835 | Lctl | lactase-like | −5.46 |
| NM_018870 | Pgam2 | phosphoglycerate mutase 2 | −5.51 |
| NM_175013 | Pgm5 | phosphoglucomutase 5 | −5.55 |
| NM_027010 | Crygf | crystallin, gamma F | −5.58 |
| NM_144805 | Tmem40 | transmembrane protein 40 | −5.68 |
| NM_008048 | Igfbp7 | insulin-like growth factor binding protein 7 | −6.14 |
| NM_020288 | Olfr749 | olfactory receptor 749 | −6.33 |
| NM_030022 | Grifin | galectin-related inter-fiber protein | −7.11 |
| NM_023695 | Crybb1 | crystallin, beta B1 | −7.18 |
| NM_008760 | Ogn | osteoglycin | −7.83 |
| NM_009965 | Cryba1 | crystallin, beta A1 | −8.96 |
| NM_007774 | Cryga | crystallin, gamma A | −9.34 |
| NM_019957 | Dnase2b | deoxyribonuclease II beta | −9.61 |
| NM_021352 | Crybb3 | crystallin, beta B3 | −9.66 |
| NM_007777 | Cryge | crystallin, gamma E | −10.13 |
| NM_009964 | Cryab | crystallin, alpha B | −11.17 |
| NM_153076 | Crygn | crystallin, gamma N | −12.64 |
| NM_007601 | Capn3 | calpain 3 | −13.48 |
| NM_144761 | Crygb | crystallin, gamma B | −13.48 |
| NM_008123 | Gja8 | gap junction protein, alpha 8 | −14.36 |
| NM_001002896 | Bfsp2 | beaded filament structural protein 2, phakinin | −15.1 |
| NM_009751 | Bfsp1 | beaded filament structural protein 1, in lens-CP94 | −15.4 |
| NM_021541 | Cryba2 | crystallin, beta A2 | −17.03 |
| NM_016975 | Gja3 | gap junction protein, alpha 3 | −17.29 |
| NM_001082573 | Crygc | crystallin, gamma C | −17.63 |
| NM_021351 | Cryba4 | crystallin, beta A4 | −21.81 |
| NM_008600 | Mip | major intrinsic protein of eye lens fiber | −21.81 |
| NM_177693 | Lim2 | lens intrinsic membrane protein 2 | −23.85 |
| NM_009967 | Crygs | crystallin, gamma S | −25.42 |
We did not observe changes in the expression of genes related specifically to other retinal cell types, including RPCs, RGCs, bipolar cells, and Müller cells (Fig. 3d). Results of qPCR analysis in P0 retina confirmed the microarray data, and showed specific upregulation of photoreceptor related genes, such as Nrl, Nr2e3, recoverin and rhodopsin; but not genes related to RPCs (e.g. Sox2 and Pax6), RGCs (e.g., Ngn2, Math5, Brn3a, and Tuj1) and other retinal cell types (e.g. PKCα and CRALBP) (Fig. 3c,d). Along with photoreceptor degeneration in the KO retina at 1 M, the levels of expression of photoreceptor related genes were also downregulated while the expression of other non-photoreceptor-related genes remained unchanged (not shown). Despite the upregulation of Lin28b and Eya2 – markers of RPCs for early born neurons (e.g. RGCs) – no significant changes in numbers of RGCs or levels of expression of RGC related genes were noted in the postnatal retinae of KO mice. The increase of photoreceptor specific protein expression in KO mice was consistent with the previous report14 and was further verified by immunofluorescence labeling. Moderate elevation of Recoverin- and Rhodopsin-immunofluorescence intensities was detected in P3 KO retinae compared to littermate WT controls (Supplementary Fig. 4). The data suggest selective destabilization of photoreceptor gene expression associated with photoreceptor degeneration in the KO retina.
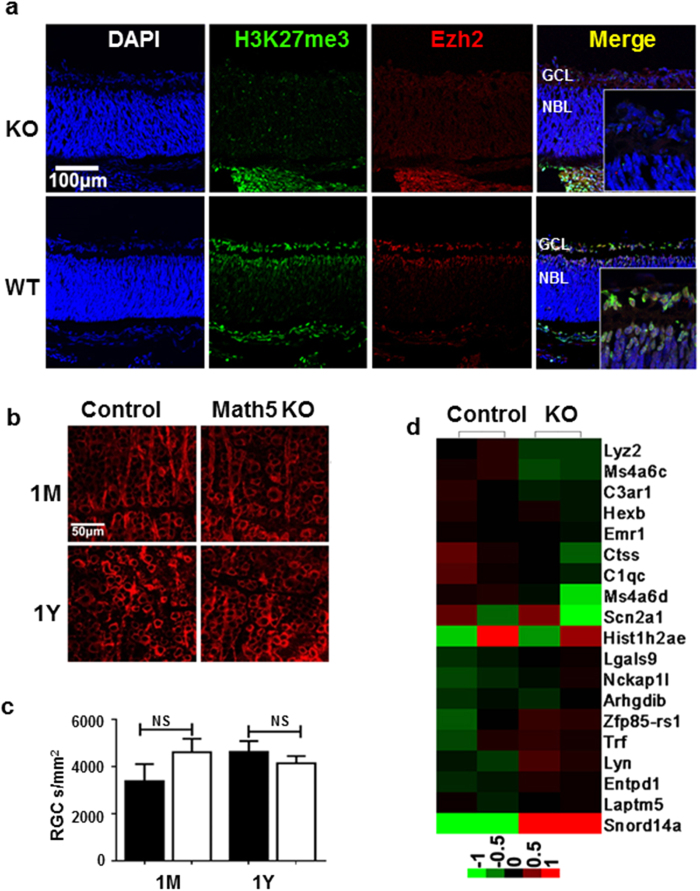
Targeted deletion of Ezh2 from RGCs does not affect cell homeostasis or RGC gene expression
The finding that RPC inactivation of Ezh2 induces primarily photoreceptor degeneration and gene expression changes suggests that Ezh2 selectively regulates photoreceptor related genes. If this hypothesis is true, Ezh2 deficiency in other retinal cell types should not directly cause retinal neurodegeneration. To test this hypothesis, we conditionally inactivated Ezh2 from all RGC progenitors, using a Math5-Cre driver (Math5-KO or Math5-Cre+; Ezh2flox/flox)20 which mediates gene excision from RPCs that are competent to generate RGCs from E11.5 onward. Ezh2 expression and H3K27me3 deposition in the outer neuroblastic layer of Math5-KO mice was maintained, but was abolished in the GCL of embryonic and P0 mice, as expected. In Math5-KO mice, we observed no RGC loss, nor morphological or functional defects throughout the adulthood (Fig. 4a–c). We next conducted an Affymetrix cDNA microarray analysis to compare gene transcripts in purified RGCs of P0 WT and Math5-KO mice. In support of the above findings that deletion of Ezh2 from intermediate or late stage RPCs selectively impact the expression of photoreceptor related genes, only a few changes were detected when compared to littermate controls (Ezh2flox/flox) (Fig. 4d). Gene Ontology analysis showed no specific association of these gene expression changes with RGCs or RPCs (Supplementary Fig. 5). Thus, disruption of Ezh2-mediated disposition of repressive marks selectively affects the gene expression and homeostasis of photoreceptor cells in the postnatal mouse retina.
Figure 4. RGC-specific Ezh2 inactivation reduces H3K27me3 deposition and gene expression of selective transcripts.
(a) Absence of H3K27me3 deposition (green) and Ezh2 expression (red) in the GCL of P0 Math5-KO retina compared to P0 control (Ezh2flox/flox) mouse retina. (b,c) Assessment of RGC (red) morphology (b) and cell counts (c) in Tuj1-immunolabeled retinal flat-mounts of adult control Black bar: WT controls; white bar: Math5-KO. (Ezh2flox/flox) and Math5-KO mice showed no significant differences in RGC morphology or cell number. (d) Heat map of microarray analysis using RNAs collected from RGCs purified in P0 Math5-KO and littermate controls. One gene was up-regulated and 18 genes were down-regulated over 1.5 fold in Math5-KO mice compared to control mice. Bars represent the mean ± S.D. of at least six biological replicates.
Forced expression of Six1 in the postnatal retina induces photoreceptor degeneration
Ezh2 mediates gene repression via catalyzing H3K27me3 on its target genes. We proposed that embryonic loss of Ezh2 resulted in aberrant sustained expression of its target genes in the postnatal period, which are normally “switched off” during the perinatal period, leading to destabilized photoreceptor gene expression and degeneration throughout the perinatal and postnatal periods. We found that the homeodomain protein Six1, which was detected in the embryonic retina, was aberrantly upregulated—from nearly 4 to 20-fold—at E13.5, P0, and 1 M, in KO vs littermate control retinae (Fig. 5a). Eya2, a co-factor of Six1, followed a similar pattern and was also upregulated at P0 and 1 M of KO retina (Fig. 5b). We next asked if Ezh2 directly deposited the repressive mark H3K27me3 onto Six1 by performing chromatin-immunoprecipitation (ChIP) assay with H3K27me3 antibody. Ezh2 deficiency resulted in H3K27me3 depletion from the Six1 and Nrl promoters, but not photoreceptor genes, that include Recoverin, Rhodopsin, and Nr2e3 (Fig. 5c–g). These results suggest that Six1, but not the majority of photoreceptor genes, is a direct target of Ezh2. We next performed the ChIP assay to determine if Six1 binds the core promoters of photoreceptor genes to regulate their expression. We noted that in KO mice, Six1 occupancy was differentially enriched at the Recoverin, Rhodopsin, Nrl and Nr2e3 promoters compared to controls (Fig. 5h–k), indicating that Six1 regulates the expression of photoreceptor-specific genes. These results suggest the following: upon loss of Ezh2 and H3K27me3, aberrantly expressed Six1, which binds Eya2 to mediate transcriptional activation, contributes to abnormally elevated expression of photoreceptor genes at P0, prior to their downregulation following postnatal photoreceptor degeneration.
Figure 5. Ezh2 deficiency causes derepression of Six1 and dysregulation of photoreceptor gene expression in the postnatal retina.
(a,b) qPCR results showed aberrantly increased expression of Six1 and Eya2 genes in the postnatal retina of KO mice. (c–g) DNA ChIP analysis showed significant reductions of Ezh2 binding to Six1 and Nrl promoters, but not to Recoverin, Rhodopsin or Nr2e3 promoters, in P0 KO retinas compared to littermate control retinas, suggesting that Ezh2 directly targets Six1 and Nrl. (h–k) KO retinae exhibited significantly increased enrichment of Six1 binding to the promoters of photoreceptor-related genes, Recoverin (Rcvrn), Rhodopsin (Rho), Nrl and Nr2e3 genes. Bars represent the mean ± S.D. of at least six biological replicates. NS indicates no significant difference; **indicates P < 0.01.
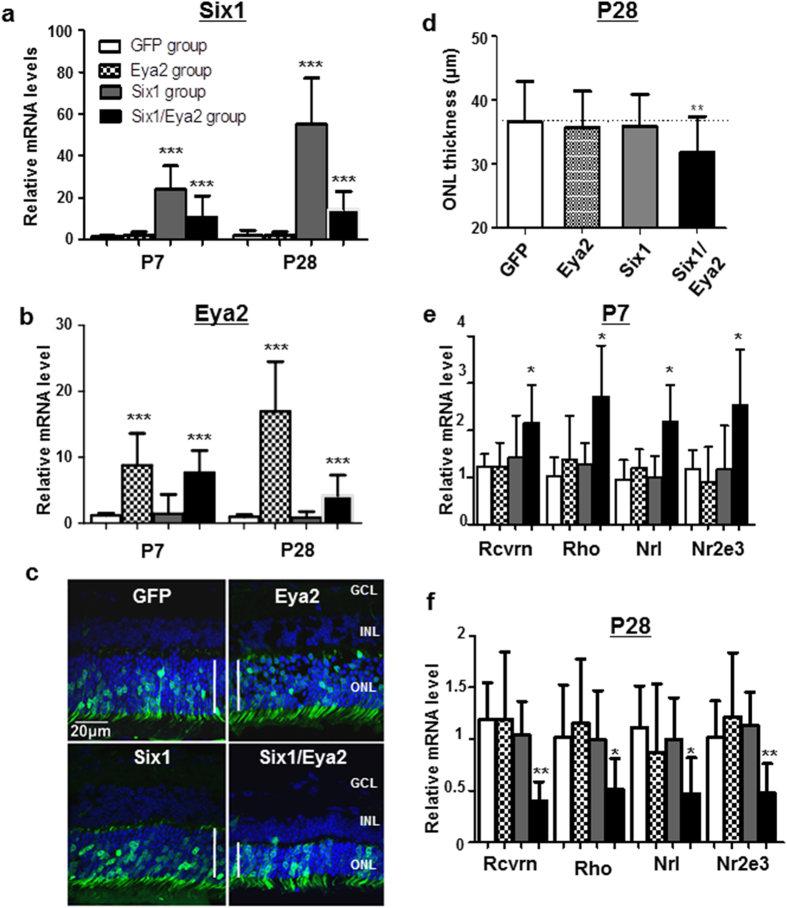
The Six family is comprised of homeodomain-containing DNA-binding proteins which play crucial roles in Drosophila and vertebrate eye formation21,22. Association between aberrant Six1 expression and photoreceptor degeneration has not been studied. We thus asked if upregulated Six1 and Eya2 in KO might phenocopy some aspects of photoreceptor degeneration seen in Ezh2 KO retinae. We electroporated plasmids encoding Six1, Eya2 or GFP (as a control) into postnatal P0 WT retinae in vivo. When analyzed at 1 M, GFP was expressed in the ONL, and qRT-PCR confirmed elevated Six1 and Eya2 expression, respectively, in electroporated WT retinae (Fig. 6a–c). Dual Six1/Eya2 electroporated retinae showed decreased ONL thickness at 1 M compared to controls (GFP), Six1 or Eya2 individually transfected groups (Fig. 6c,d). Associated with it, we noted decreased and increased levels of cell proliferation and apoptosis markers, Ki67, Caspase-3, respectively, in Six1/Eya2-electroporated group as compared to GFP, Six1, or Eya2 individually-electroporated groups (Supplementary Fig. 6). Similar to Ezh2 KO mice, recoverin, rhodopsin, Nrl and Nr2e3 were up-regulated in Six1/Eya2-co-electroporated retinae at P7 as compared to control GFP-electroporated retinae (Fig. 6e), but they were down-regulated in the Six1/Eya2 co-transfected group retina at P28 due to photoreceptor degeneration (Fig. 6f). The levels of recoverin, rhodopsin, Nrl and Nr2e3 in Six1 or Eya2 individually transfected groups remained unchanged in both time points measured when were compared to control GFP-electroporated retinae. The data further support that Six1, which requires the presence of Eya2, directly mediates the expression of photoreceptor-related genes. Together, our data suggests that adult photoreceptor degeneration secondary to Ezh2 inactivation during retinal development is mediated by aberrant expression of derepressed Six1 and Eya2, which induces degeneration through decreased cell proliferation and increased apoptosis.
Figure 6. Forced expression of Six1 and Eya2 in postnatal retina induces photoreceptor degeneration.
(a,b) qPCR analyses of Six1 (a) and Eya2 (b) mRNA levels in wild-type mouse retinae taken at P7 and P28 following P0 in vivo electroporation of Six1, Eya2, or Six1 and Eya2 simultaneously (Six1/Eya2) to the mouse retinae (n = 12/group). (c) Detection of GFP expression in 1 M mouse retinal sections after P0 in vivo electroporation of GFP, Six1, Eya2, or Six1 and Eya2 simultaneously. Note the robust detection of GFP expression in the ONL and the thinner ONL in Six1/Eya2 electroporated retina compared to the other electroporated retina groups. Vertical lines mark the range of the ONL. (d) Quantification of ONL thicknesses in retinal sections of 1 M old mice that received in vivo electroporation of GFP, Six1, Eya2, or Six1 and Eya2 simultaneously at P0. (e,f) Results of qPCR showed increased mRNA levels of photoreceptor genes only in Six1 and Eya2 co-electroporated retinas at P7 (e) but reduced mRNA levels of photoreceptor genes at P28, when compared to the GFP-electroporated group. Bars represent the mean ± S.D. of at least six biological replicates. *Indicates P < 0.05, ** indicates P < 0.01.
Discussion
We have identified an epigenetic mechanism by which Ezh2 expression during embryonic and early postnatal retinal development regulates photoreceptor homeostasis in the adult, a stage when Ezh2 is not normally expressed. In the absence of Ezh2, Six1 is aberrantly upregulated by Ezh2-mediated loss of H3K27me3 at its promoter. This causes misexpression of photoreceptor related genes that is, at least in part, contributing to progressive photoreceptor degeneration in this model. Our findings relate transiently expressed, developmental regulators to adult retinal neuron survival and function, and for the first time reveal an epigenetic link by which in utero events lead to juvenile and adult-onset vision loss.
Here we uncovered that, in addition to its role in stem cell self-renewal and lineage specification in development23, Ezh2-mediated gene repression orchestrates postnatal retinal neuron survival and function. During retinal development, a delicate balance between proliferation and differentiation of progenitors is essential for ensuring proper cell fate development and establishment of cell survival and function. Our data suggests that Ezh2 governs this process through deposition of repressive modification H3K27me3 on early transcription factors, such as Six1, to allow stabilized expression of photoreceptor-related genes in the differentiated cell decedents24. The Six family of homeodomain-contain DNA-binding proteins is a group of transcription factors, which participate in eye formation during early development25. Six1 acts together with its coactivators Eya1 and Eya2 to drive cell proliferation, tissue growth and cell fate specification in early and mid-eye development in Drosophila26. Yet, its involvement in mammalian retinal development has not been documented. Our results suggest that Six1 is transiently expressed in the embryonic mouse retina and binds to the promoter regions of photoreceptor specific genes, including rhodopsin and recoverin, to mediate their levels of expression before birth. Overexpression of Six1 in the postnatal stage results in destabilized photoreceptor gene expression in the mature retina and causes postnatal onset of retinal degeneration. These findings indicate that epigenetic dysregulation in retinal development is a predisposing factor for congenital photoreceptor dystrophy and postnatal photoreceptor degeneration. Future study investigating photoreceptor rescue in KO mice by deletion of Six1 or Eya2 (through crossing with Six1 or Eya2 deficient mice) may provide direct testimony if derepression of Six1 is essential for photoreceptor degeneration following Ezh2 loss-of-function.
Our findings suggest a previously unappreciated role of Ezh2 in retinal homeostasis. Report by Aldiri et al.27 showed that in Xenopus, knockdown of Ezh2 constrains the generation of retinal neurons and promotes a Müller glial cell fate, partially due to upregulation of Cdk inhibitor p15. In agreement with this finding, Iida et al.13 reported accelerated differentiation of Müller glia and rod photoreceptors, and impaired cell proliferation in Dkk3-Cre driven Ezh2 deficient retinae. Using Pax6-αCre, Zhang et al.14 observed similarly reduced proliferation of RPCs and diminished numbers of retinal ganglion cells and amacrine cells, which is accompanied by increased photoreceptor and Müller cell differentiation, following Ezh2 deletion. Their RT-PCR and RNA sequencing results from the Ezh2-deficient retina revealed drastically upregulated Six1, in addition to other genes regulating early RPC development. Nevertheless, the developmental defects in their retinae prevented further elucidation of a role for Ezh2 in retinal homeostasis. The difference between the phenotypes observed in the present study and those reported by others is presumably due to the fact that Pax6-α Cre28 and Dkk3-Cre12 induce Cre recombinase in early proliferating RPCs before E10.5; whereas, Chx10-Cre drives Cre expression in RPCs at a differentiation stage slightly later than that by Pax6-α Cre and Dkk3-Cre16. Thus, Ezh2 deletion driven by Chx10-Cre results in gene expression changes that occur likely at the nexus point of cell proliferation and differentiation, persisting into adulthood, and lead to postnatal retinal degeneration.
It is intriguing that although highly expressed in RPCs and RGCs during perinatal period, Ezh2 loss in all retinal progenitors (by use of the Chx10-Cre strain) selectively affects gene expression and homeostasis of photoreceptors, but not RGCs or other retinal cell types. This result is further confirmed by using Math5-Cre, which drives gene deletion from RPCs that are competent to generate RGCs20. The data reveal a unique and cell type specific regulation of gene expression by Ezh2, rather than genome-wide repression. While its underlying mechanisms remain uncertain, in part, this is unlikely to be caused solely by the spatiotemporal dynamics and cooperative functionalities of other HMTs and demethylases. Among the two HMTs that regulate H3K27 trimethylation, Ezh1 and Ezh2, and the demethylase Jmjd3, Ezh2 and Jmjd3 are widely expressed in retinal progenitors during development; Ezh1 is expressed only weakly in the postnatal retina29. The transient expression of Ezh2 during development, which is completely repressed postnatally in the retina, was also confirmed during human retinal development11. A cell type specific effect was also observed with Jmjd3 deficiency, which resulted in impaired differentiation of subsets of bipolar cells, despite strong Jmjd3 expression in the RGCs9. Previous studies have reported a model of bidirectional and mutually reinforcing crosstalk between Ezh2 and another repressive HMT, G9a, on histone hypermethylation in other tissues30,31. In support of a possible crosstalk between these two epigenetic silencing regulators in RGCs, G9a (which catalyzes H3K9me2, another repressive histone mark) is highly expressed in RGCs of the mouse retina during a similar period when Ezh2 is detected10. It would be interesting to investigate the possible interactions of these repressive epigenetic modifications in RGCs such as by analyzing conditional knockouts of both G9a and Ezh2 in the future.
In summary, our observations point to a central role for Ezh2-mediated histone methylation in the epigenetic repression of Six1 and photoreceptor homeostasis in the adult. Six1 and the targetable HMT epigenetic enzyme regulating Six1, Ezh2, thus may represent novel therapeutic targets in congenital photoreceptor degeneration.
Methods
Generation and genotyping of KO mice
All animal procedures were performed in accordance with the statement of the Association for Research in Vision and Ophthalmology, and the protocols were approved by Institutional Animal Care Committee (IACUC) of the Schepens Eye Research Institute. Mice were housed in a temperature- controlled room with a 12 h light/dark cycle. Fresh water and rodent diet were available at all times.
Mice carrying deletion of Ezh2 specifically in all retinal progenitors was produced using the Chx10-Cre mice line (from the Jackson Laboratory) crossed with conditional allele of Ezh2flox/flox mice (from the Mutant Mouse Regional Resource Centers (MMRRC) (https://www.mmrrc.org/catalog/sds.php?mmrrc_id=15499). Chx10-cre; Ezh2flox/flox (KO) mice were eventually generated and allowed to investigate its function in retina development.
Mice were genotyped by PCR analysis. Mice were anesthetized with 2–4% isoflurane (Vedco Inc., MO, USA) and less than 0.5 cm of tail tissues were cut by razor blade. The tail tissue was incubated in direct PCR buffer solution (Viagen Biotech, CA, USA) containing proteinase K (Invitrogen Life Technologies, Carlsbad, CA, USA), and 56 °C overnight according to the manufacturer’s instruction. PCR genotyping was performed using the following primers: Chx10-cre forward primer 5′-GGGCACCTGGGACCAACTTCACGA-3′ and reverse primer 5′-CGGCGGCGGTCACGAACTCC-3′ (750 bp PCR product)16; Ezh2 forward primer 5′-CTGCTCTGAATGGCAACTCC-3′ and reverse primer 5′-TTATTCATAGAGCCACCTGG-3′ (Control 430 bp and KO 470 bp). The PCR was performed using Apex Taq DNA Polymerase (Genesse Scientific, San Diego, CA). The primers were designed to detect the expression of Ezh2 gene at P0 and 1 M. The primers information as follows: forward primer 5′-TCAGGATGAAGCAGACAGAAG-3′ and reverse primer 5′-TTTGTTGCCCTTTCGGGTTG-3′. The PCR reaction was carried out according to the manufacturer’s instruction.
Hematoxylin and Eosin (HE) Staining and Immunohistochemistry (IHC)
Fixation, sectioning, HE staining were carried out as described32. For immunohistochemistry, mouse embryos and eyecups were fixed in 4% paraformaldehyde in PBS for either 30 min to 3 h. The samples were cryoprotected, embedded, frozen, and sectioned (8 μm thick). Slides were incubated with blocking solution (4% normal donkey serum and 0.5% Triton X-100 in PBS) for 1 h, and then with the primary antibodies overnight at 4 °C degree. Slides were washed with PBS three times for 10 min each time and incubated with the secondary antibodies for 1 h at room temperature. The specimens were observed under a laser confocal microscope (Leica TCS SP5).
Primary antibodies used in this study were Beta-III Tubulin (1:1000; AB9354, Millpore), Recoverin Antibody (1:1000; AB5585, Millpore), PNA Antibody (1:200; Sigma), Ezh2 (1:1000; 3147s, cell signaling) and H3K27me3 (1:500, 9733S, Cell Signaling). Second antibodies were Cy3-labeled donkey anti-mouse antibody (1:1500; Vector Laboratories), Cy3-labeled donkey anti-rabbit antibody (1:1500; Vector Laboratories) and Alexa Fluor 488 Conjugated antibody (1:400; cell signaling). For all quantifications, KO mice and controls were used for all analyses. Quantification of the thickness of nuclear staining for each antibody was carried out on fluorescence microscope images (Leica, Mannheim, Germany) with ImageJ software (National Institutes of Health, Bethesda, MD). Student’s t tests were used to measure differences between KO mice and controls.
Transmission Electron Microscopy (TEM)
Mice were euthanized with CO2 and the eyes were enucleated and the posterior segments fixed in 1% glutaraldehyde/1% paraformaldehyde in PBS and postfixed in veronal acetate buffered osmium tetroxide (2%), dehydrated in ethanol and water, and embedded in Epon. Ultrathin sections were cut from blocks and mounted on copper grids. The specimens were examined by using a transmission electron microscope (Tecnai Spirit, FEI).
Optical Coherence Tomography (OCT)
Retinal morphology and thickness of the ONL of live mice were assessed non-invasively with spectrum domain optical coherence tomography (SD-OCT). Mice were anesthetized with an intraperitoneal injection of ketamine (120 mg/kg)/xylazine (12 mg/kg) mixture and pupils were dilated using tropicamide (1%; Falcon Pharmaceuticals, Fort Worth, TX). Lubricant gel drops (Novartis Pharmaceuticals Corp, East Hanover, NJ) were applied to maintain the moisture of cornea. Images were acquired using SD-OCT (Bioptigen Inc, Research Triangle Park, NC). Radial volume scan (centered on optic disc, diameter 1.3 mm) was used, and ONL thickness within the 200–400 μm area from the central of optic disk were measured and analyzed for the ONL thickness as previously described33.
Electroretinogram (ERG)
Retinal functions of control and KO mice were assessed by ERG. Mice were dark-adapted 5 h, and then anesthetized with intraperitoneal injection. Pupils were dilated with topical 1% tropicamide and 0.5% phenylephrine HCl, and the mice were placed on a heating pad for the duration of the ERG recordings. The mice were stimulated with stroboscopic stimuli with six levels of stimuli 0.00022, 0.00215, 0.06455, 0.64545, 2.15 and 500 cd-s/m2 to elicit the scotopic ERGs. After the scotopic measurement, mice were exposed to a rod-saturating background light (40 cd/m2) for 10 min, then the mice with stimuli 10 cd-s/m2 for the photopic ERGs. The data were recorded and processed by the ERG system (Espion Electroretinography System; Diagnosys LLC, Lowell, MA). The amplitudes of a-wave and b-wave of ERG response were determined.
Microarray Expression Analysis
Total RNA was extracted from both cortices from three KO mice and controls at P0 stage. Total retinal RNA (5 μg) was isolated using RNeasy Plus Mini Kit (QIAGEN, Valencia, CA, USA) and converted to cDNA using the One-Cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s instructions. Gene Expression of KO mice and controls were detected using the Affymetrix Mouse Gene 1.0 ST Array (Dana-Farber Cancer Institute) and the microarray data were analyzed using the dChip software (www.dchip.org). Gene Ontology (GO) analysis of genes up-regulated and down-regulated in the KO mice retina was performed using DAVID tools (http://david.abcc.ncifcrf.gov).
Quantitative Real-Time PCR (qPCR)
Total retina RNA was extracted using RNeasy Plus Mini Kit (QIAGEN, Valencia, CA, USA) and cDNA synthase using SuperScript™ III First-Strand Synthesis System (Invitrogen Life Technologies, Carlsbad, CA, USA). QPCR was performed using SYBR FAST qPCR Kits (Kapa Biosystems, Woburn, MA, USA) and LightCycler® 480 (Roche Biosystems; Indianapolis, IN) according to the manufacturer’s instructions. The primer sequences are listed in Supplementary Table 1. For relative comparison of each gene, we analyzed the Ct value of real-time PCR data with the ∆∆Ct method normalizing by an endogenous control (glyceraldehyde 3-phosphate dehydrogenase, GAPDH).
Chromatin immunoprecipitation (ChIP)
The P0 retina tissues of KO mice and controls were cut into small pieces (between 1–3 mm3) and the tissues were cross-linked with 1% paraformaldehyde (#252549, Sigma) in PBS for 1 hour at 37 °C. Then chromatin isolation and ChIP assay were performed according to the manufacturer’s protocol using a commercially available kit (EZ-Zyme Chromatin prep kit, EZ-ChIP; Millipore). The immunoprecipitated chromatin DNA was incubated with rabbit anti-H3K27me3 antibody (#9756s, Cell Signaling) and goat anti-six1 antibody (Santa Cruz) and was analyzed by qPCR using gene-specific primers (Supplementary Table 1).
RGC qualification
The mouse retinas were dissected to remove the cornea, lens, iris, sclera, and choroid. The vitreous was carefully removed and the flat mounted retina was placed on the super-frost sections with ganglion cell layer facing up. RGCs were double immunolabeled with β-III tubulin (Tuj1) and DAPI. Our previous studies showed that Tuj1 specifically labels RGCs in retinal flat-mounts34. The specimens were visualized under the confocal microscope at 800× magnification. The flat mounts were divided into quadrants: superior, temporal, nasal and inferior and total 16 square regions of each eye were photographed and all β-III tubulin positive cells in the GCL were quantified. The average RGC densities of the whole retina were calculated and the percentage of RGC loss was determined by comparing RGC densities with that obtained from the contralateral control eyes.
In vivo Electroporation
Three of the expression vectors were purchased from GeneCopoeia (Germantown, MD, USA), including pEZ-M90 (GFP report), pEZ-M90-Six1 and pEZ-M90-Eya2. The experiment was divided into four groups: pEZ-M90 (GFP group), pEZ-M90-Six1 (Six1 group), pEZ-M90-Eya2 (Ezh2 group), and pEZ-M90-Six1 mixed with pEZ-M90-Eya2 (Six1/Ezh2 group). The plasmids were amplified using QIAGEN Plasmid Maxi Kit (Valencia, CA, USA). Newborn ICR mouse pups were anesthetized by chilling on ice, and a small incision was made in the eyelid and sclera near the lens with a 32-gauge needle. 0.5–1 μl DNA solutions (1 μg/μl) in PBS containing 0.1% blue dye as a tracer were injected into the subretinal space through the incision by using a Hamilton syringe (Reno, Nevada, USA) with a 32 gauge blunt-ended needle under a dissecting microscope. After DNA injection, 10 mm diameter tweezer-type electrodes (Nepagene, Chiba, Japan) briefly soaked in PBS were placed to softly hold the heads of the pups. Five square pulses of 80 volts and 50 ms duration with 950 ms intervals were applied by using a NEPA21 Electroporator (Nepagene, Chiba, Japan).
The eyeballs were harvested at 7 and 28 days, respectively, after electroporation. The eyeballs were fixed with 4% paraformaldehyde, cryoprotected with 30% sucrose, embedded in OCT compound (Sakura Finetek, Tokyo, Japan) and cryosectioned into 8 μm section. The GFP-positive retina sections were selected under a Leica DM3000 fluorescent microscope. Only retinal sections containing 1/3 or more GFP expressing cells were selected for scoring. Mouse eyes that developed cataract, small eyes, or apparent inflammation were excluded. For each group, at least 12 GFP-positive retinae were scored. Quantification of the thickness of ONL for each group was carried out on fluorescence microscope images with NIH ImageJ software. At 7 and 28 days after electroporation, electroporated retinae were dissected under a fluorescent microscope to select the GFP-positive retinae. Total retina RNA was extracted and cDNA was synthesized. QPCR was performed and the results were analyzed with the ∆Ct method.
Statistical Analysis
The statistical significance of IHC, OCT, ERG and qPCR results were analyzed by using using GraphPad Prism 5 statistical software (GraphPad Software, La Jolla, CA). All the numerical variables in this article were presented as Mean ± S.E.M. Two-tailed student’s t-test was used on numerical variables of independent samples; one-way ANOVA analysis was applied for comparisons of data among groups.
Additional Information
How to cite this article: Yan, N. et al. Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1. Sci. Rep. 6, 33887; doi: 10.1038/srep33887 (2016).
Supplementary Material
Acknowledgments
This research was supported by funding from the Department of Veterans Affairs (1I01RX000110), NIH/NEI (R41 EY025913 and R01EY025259) and Curing Kids Fund to D.F.C.; the Core Grant for Vision Research from NIH/NEI to the Schepens Eye Research Institute (P30EY03790); the National Natural Science Foundation of China (81371024) and Science and Technology Department of Sichuan Province (2014SZ0030) to N.H.Y.; and National Institutes of Health (K12EY022299) and Research to Prevent Blindness to R.C.R.
Footnotes
Author Contributions N.Y. performed the large parts of the experiments described in Figures 1–6 and SI Figures 1–6, analyzed the data, and wrote the manuscript. L.C. performed the study described in Figure 5, as well as part of the ERG and OCT. N.Y., K.C., R.C.R. and D.F.C. devised the experimental plans and wrote the manuscript. L.X. and H.Y. prepared Figure 6 and SI Figure 3. M.T.A.M., C.G. and R.Z. performed part of the studies in Figures 4 and 5 and SI Figures 3 and 5. All authors discussed the results and commented to the final paper.
References
- Michaelides M., Hardcastle A. J., Hunt D. M. & Moore A. T. Progressive cone and cone-rod dystrophies: phenotypes and underlying molecular genetic basis. Surv Ophthalmol 51, 232–258, 10.1016/j.survophthal.2006.02.007 (2006). [DOI] [PubMed] [Google Scholar]
- Moore A. T. Childhood macular dystrophies. Curr Opin Ophthalmol 20, 363–368, 10.1097/ICU.0b013e32832f8002 (2009). [DOI] [PubMed] [Google Scholar]
- Nakamura M., Lin J. & Miyake Y. Young monozygotic twin sisters with fundus albipunctatus and cone dystrophy. Arch Ophthalmol 122, 1203–1207, 10.1001/archopht.122.8.1203 (2004). [DOI] [PubMed] [Google Scholar]
- Berghmans L. V. et al. Discordance for retinitis pigmentosa in two monozygotic twin pairs. Retina 31, 1164–1169, 10.1097/IAE.0b013e3181fbcf2b (2011). [DOI] [PubMed] [Google Scholar]
- Walia S. et al. Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene. Arch Ophthalmol 126, 379–384, 10.1001/archophthalmol.2007.72 (2008). [DOI] [PubMed] [Google Scholar]
- Gluckman P. D., Hanson M. A., Cooper C. & Thornburg K. L. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359, 61–73, 10.1056/NEJMra0708473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R. & Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14, 155–164, 10.1016/j.gde.2004.02.001 (2004). [DOI] [PubMed] [Google Scholar]
- Pereira J. D. et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America 107, 15957–15962, 10.1073/pnas.1002530107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A. et al. Histone demethylase Jmjd3 is required for the development of subsets of retinal bipolar cells. Proc Natl Acad Sci USA 111, 3751–3756, 10.1073/pnas.1311480111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. C. et al. Dynamic patterns of histone lysine methylation in the developing retina. Invest Ophthalmol Vis Sci 51, 6784–6792, 10.1167/iovs.09-4730 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. et al. Characterization and pharmacologic targeting of EZH2, a fetal retinal protein and epigenetic regulator, in human retinoblastoma. Lab Invest 95, 1278–1290, 10.1038/labinvest.2015.104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. et al. Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis 45, 502–507, 10.1002/dvg.20318 (2007). [DOI] [PubMed] [Google Scholar]
- Iida A. et al. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Developmental neurobiology 75, 947–960, 10.1002/dneu.22261 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation. Dev Biol 403, 128–138, 10.1016/j.ydbio.2015.05.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N. S. et al. Absence of chx10 causes neural progenitors to persist in the adult retina. Invest Ophthalmol Vis Sci 47, 386–396, 10.1167/iovs.05-0428 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S. & Cepko C. L. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol 271, 388–402, 10.1016/j.ydbio.2004.03.039 (2004). [DOI] [PubMed] [Google Scholar]
- Liang L. & Sandell J. H. Focus on molecules: homeobox protein Chx10. Experimental eye research 86, 541–542, 10.1016/j.exer.2007.03.004 (2008). [DOI] [PubMed] [Google Scholar]
- Huang D. W. et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research 35, W169–W175, 10.1093/nar/gkm415 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57, 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- Yang Z., Ding K., Pan L., Deng M. & Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol 264, 240–254 (2003). [DOI] [PubMed] [Google Scholar]
- Oliver G. & Gruss P. Current views on eye development. Trends Neurosci 20, 415–421, S0166-2236(97)01082-5 (1997). [DOI] [PubMed] [Google Scholar]
- Kumar J. P. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci 66, 565–583, 10.1007/s00018-008-8335-4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly M. L., MacQuarrie K. L., Fong A. P., Yao Z. & Tapscott S. J. Polycomb-mediated repression during terminal differentiation: what don’t you want to be when you grow up? Genes Dev 25, 997–1003, 10.1101/gad.2054311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R. & Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349, 10.1038/nature09784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. M., Weasner B. M., Weasner B. P. & Kumar J. P. Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139, 991–1000; 10.1242/dev.077255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heanue T. A. et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev 13, 3231–3243 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I., Moore K. B., Hutcheson D. A., Zhang J. & Vetter M. L. Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/beta-catenin signaling. Development 140, 2867–2878, 10.1242/dev.088096 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T. et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55 (2001). [DOI] [PubMed] [Google Scholar]
- Watanabe S. & Murakami A. Regulation of Retinal Development via the Epigenetic Modification of Histone H3. Adv Exp Med Biol 854, 635–641, 10.1007/978-3-319-17121-0_84 (2016). [DOI] [PubMed] [Google Scholar]
- Coward W. R., Feghali-Bostwick C. A., Jenkins G., Knox A. J. & Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J 28, 3183–3196, 10.1096/fj.13-241760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C. et al. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 53, 277–289, 10.1016/j.molcel.2013.12.005 (2014). [DOI] [PubMed] [Google Scholar]
- Fischer A. H., Jacobson K. A., Rose J. & Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008, pdb prot4986, 10.1101/pdb.prot4986 (2008). [DOI] [PubMed] [Google Scholar]
- Yang Q. et al. Microbead-induced ocular hypertensive mouse model for screening and testing of aqueous production suppressants for glaucoma. Invest Ophthalmol Vis Sci 53, 3733–3741, 10.1167/iovs.12-9814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Optic neuropathy due to microbead-induced elevated intraocular pressure in the mouse. Invest Ophthalmol Vis Sci 52, 36–44, 10.1167/iovs.09-5115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.