Abstract
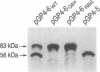
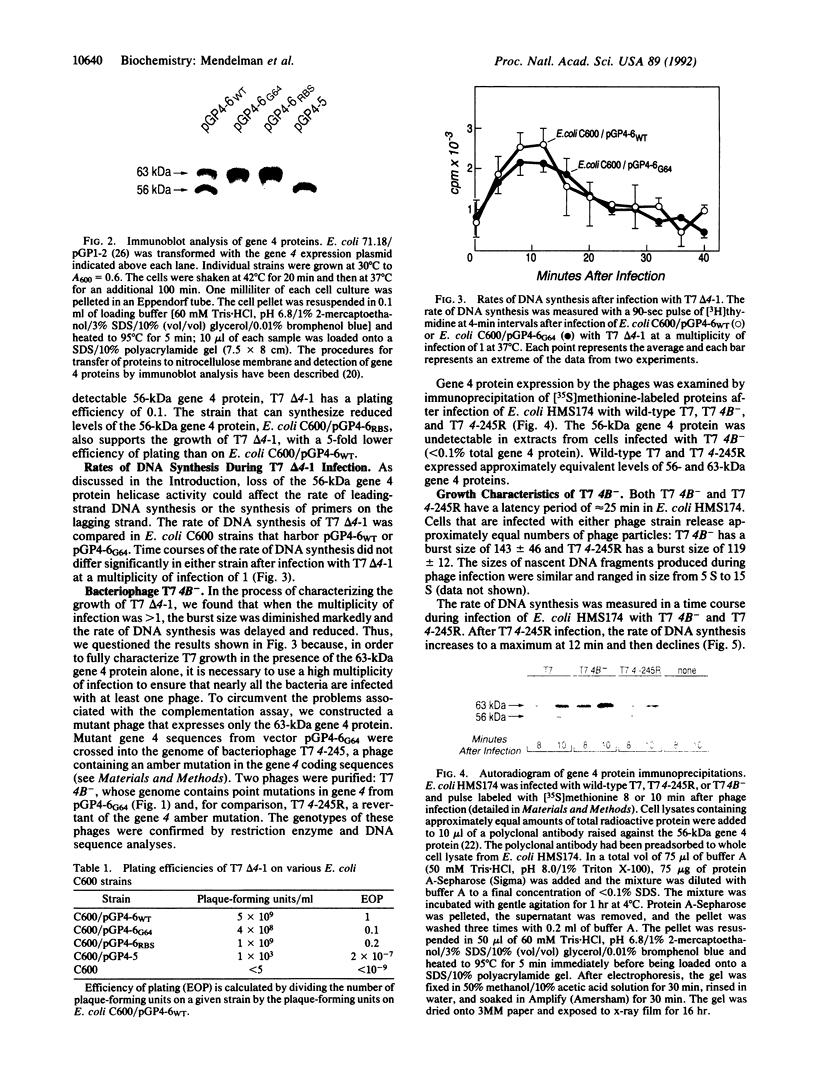
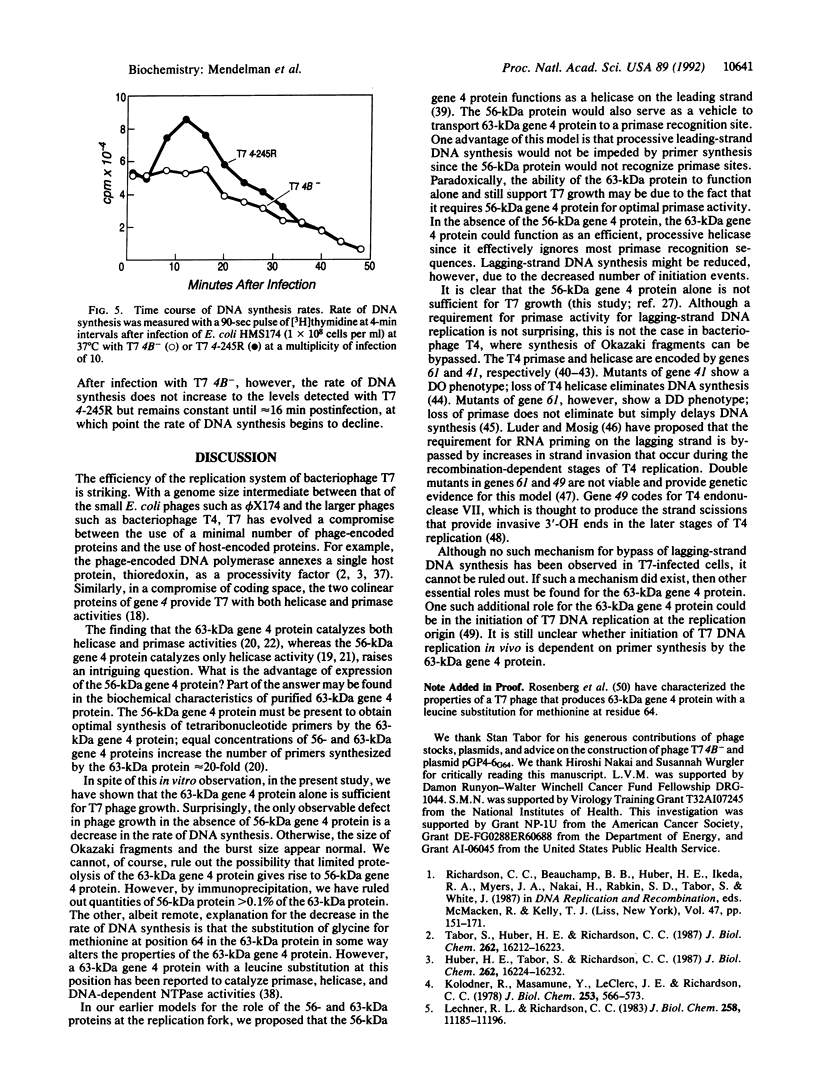
The helicase and primase activities of bacteriophage T7 are distributed between the 56- and 63-kDa gene 4 proteins. The 56-kDa gene 4 protein lacks 63 amino acids found at the N terminus of the colinear 63-kDa protein and catalyzes helicase activity. The 63-kDa gene 4 protein catalyzes both primase and helicase activities. A bacteriophage deleted for gene 4, T7 delta 4-1, has been tested for growth by complementation on Escherichia coli strains that contain plasmids expressing either one or both of the gene 4 proteins. T7 delta 4-1 cannot grow (efficiency of plating, 10(-7)) on E. coli cells that express only 56-kDa gene 4 protein. In contrast, T7 delta 4-1 has an efficiency of plating of 0.1 on an E. coli strain that expresses only 63-kDa gene 4 protein in which glycine is substituted for methionine at position 64. A bacteriophage, T7 4B-, in which methionine at residue 64 is replaced by glycine, expresses only 63-kDa gene 4 protein. The burst sizes, latency periods, and Okazaki fragment sizes of T7 4B- are similar in the presence and absence of the 56-kDa gene 4 protein; however, T7 4B- has a reduced rate of DNA synthesis when compared with a phage that synthesizes both gene 4 proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein J. A., Richardson C. C. A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc Natl Acad Sci U S A. 1988 Jan;85(2):396–400. doi: 10.1073/pnas.85.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Richardson C. C. Characterization of the helicase and primase activities of the 63-kDa component of the bacteriophage T7 gene 4 protein. J Biol Chem. 1989 Aug 5;264(22):13066–13073. [PubMed] [Google Scholar]
- Bernstein J. A., Richardson C. C. Purification of the 56-kDa component of the bacteriophage T7 primase/helicase and characterization of its nucleoside 5'-triphosphatase activity. J Biol Chem. 1988 Oct 15;263(29):14891–14899. [PubMed] [Google Scholar]
- Brown W. C., Romano L. J. Benzo[a]pyrene-DNA adducts inhibit translocation by the gene 4 protein of bacteriophage T7. J Biol Chem. 1989 Apr 25;264(12):6748–6754. [PubMed] [Google Scholar]
- Campbell J. L., Richardson C. C., Studier F. W. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci U S A. 1978 May;75(5):2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Engler M. J., Lechner R. L., Richardson C. C. Two forms of the DNA polymerase of bacteriophage T7. J Biol Chem. 1983 Sep 25;258(18):11165–11173. [PubMed] [Google Scholar]
- Fujiyama A., Kohara Y., Okazaki T. Initiation sites for discontinuous DNA synthesis of bacteriophage T7. Proc Natl Acad Sci U S A. 1981 Feb;78(2):903–907. doi: 10.1073/pnas.78.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton D. M., Nossal N. G. Bacteriophage T4 DNA primase-helicase. Characterization of oligomer synthesis by T4 61 protein alone and in conjunction with T4 41 protein. J Biol Chem. 1987 Aug 5;262(22):10873–10878. [PubMed] [Google Scholar]
- Hinton D. M., Silver L. L., Nossal N. G. Bacteriophage T4 DNA replication protein 41. Cloning of the gene and purification of the expressed protein. J Biol Chem. 1985 Oct 15;260(23):12851–12857. [PubMed] [Google Scholar]
- Huber H. E., Tabor S., Richardson C. C. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987 Nov 25;262(33):16224–16232. [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Masamune Y., LeClerc J. E., Richardson C. C. Gene 4 protein of bacteriophage T7. Purification physical properties, and stimulation of T7 DNA polymerase during the elongation of polynucleotide chains. J Biol Chem. 1978 Jan 25;253(2):566–573. [PubMed] [Google Scholar]
- Kolodner R., Richardson C. C. Replication of duplex DNA by bacteriophage T7 DNA polymerase and gene 4 protein is accompanied by hydrolysis of nucleoside 5'-triphosphates. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1525–1529. doi: 10.1073/pnas.74.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner R. L., Richardson C. C. A preformed, topologically stable replication fork. Characterization of leading strand DNA synthesis catalyzed by T7 DNA polymerase and T7 gene 4 protein. J Biol Chem. 1983 Sep 25;258(18):11185–11196. [PubMed] [Google Scholar]
- Luder A., Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamune Y., Frenkel G. D., Richardson C. C. A mutant of bacteriophage T7 deficient in polynucleotide ligase. J Biol Chem. 1971 Nov 25;246(22):6874–6879. [PubMed] [Google Scholar]
- Masamune Y., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. IV. DNA synthesis in E. coli infected with ligase-negative mutants of phage T4. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1328–1335. doi: 10.1073/pnas.61.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson S. W., Richardson C. C. DNA-dependent nucleoside 5'-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983 Nov 25;258(22):14009–14016. [PubMed] [Google Scholar]
- Matson S. W., Tabor S., Richardson C. C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J Biol Chem. 1983 Nov 25;258(22):14017–14024. [PubMed] [Google Scholar]
- Mendelman L. V., Richardson C. C. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J Biol Chem. 1991 Dec 5;266(34):23240–23250. [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Richardson C. C. Bacteriophage T7 deoxyribonucleic acid replication invitro. Bacteriophage T7 DNA polymerase: an an emzyme composed of phage- and host-specific subunits. J Biol Chem. 1975 Jul 25;250(14):5515–5522. [PubMed] [Google Scholar]
- Mosig G., Luder A., Ernst A., Canan N. Bypass of a primase requirement for bacteriophage T4 DNA replication in vivo by a recombination enzyme, endonuclease VII. New Biol. 1991 Dec;3(12):1195–1205. [PubMed] [Google Scholar]
- Nakai H., Richardson C. C. Leading and lagging strand synthesis at the replication fork of bacteriophage T7. Distinct properties of T7 gene 4 protein as a helicase and primase. J Biol Chem. 1988 Jul 15;263(20):9818–9830. [PubMed] [Google Scholar]
- Rabkin S. D., Richardson C. C. In vivo analysis of the initiation of bacteriophage T7 DNA replication. Virology. 1990 Feb;174(2):585–592. doi: 10.1016/0042-6822(90)90111-4. [DOI] [PubMed] [Google Scholar]
- Romano L. J., Richardson C. C. Characterization of the ribonucleic acid primers and the deoxyribonucleic acid product synthesized by the DNA polymerase and gene 4 protein of bacteriophage T7. J Biol Chem. 1979 Oct 25;254(20):10483–10489. [PubMed] [Google Scholar]
- Romano L. J., Richardson C. C. Requirements for synthesis of ribonucleic acid primers during lagging strand synthesis by the DNA polymerase and gene 4 protein of bacteriophage T7. J Biol Chem. 1979 Oct 25;254(20):10476–10482. [PubMed] [Google Scholar]
- Rosenberg A. H., Patel S. S., Johnson K. A., Studier F. W. Cloning and expression of gene 4 of bacteriophage T7 and creation and analysis of T7 mutants lacking the 4A primase/helicase or the 4B helicase. J Biol Chem. 1992 Jul 25;267(21):15005–15012. [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Genetic analysis of gene 1.2 of bacteriophage T7: isolation of a mutant of Escherichia coli unable to support the growth of T7 gene 1.2 mutants. J Virol. 1981 Jan;37(1):343–351. doi: 10.1128/jvi.37.1.343-351.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lanka E., Hillenbrand G. Role of bacteriophage T7 DNA primase in the initiation of DNA strand synthesis. Nucleic Acids Res. 1977 Dec;4(12):4151–4163. doi: 10.1093/nar/4.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lanka E., Morelli G., Seiffert D., Yuki A. Bacteriophage-T7-induced DNA-priming protein. A novel enzyme involved in DNA replication. Eur J Biochem. 1977 Feb;72(3):543–558. doi: 10.1111/j.1432-1033.1977.tb11278.x. [DOI] [PubMed] [Google Scholar]
- Silver L. L., Nossal N. G. Purification of bacteriophage T4 gene 61 protein. A protein essential for synthesis of RNA primers in the T4 in vitro DNA replication system. J Biol Chem. 1982 Oct 10;257(19):11696–11705. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H. Genetic and physical mapping of the late region of bacteriophage T7 DNA by use of cloned fragments of T7 DNA. J Mol Biol. 1981 Dec 15;153(3):503–525. doi: 10.1016/0022-2836(81)90405-8. [DOI] [PubMed] [Google Scholar]
- Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987 Nov 25;262(33):16212–16223. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci U S A. 1981 Jan;78(1):205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M., Silver L. L., Nossal N. G. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J Biol Chem. 1982 Oct 25;257(20):12426–12434. [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Yegian C. D., Mueller M., Selzer G., Russo V., Stahl F. W. Properties of the DNA-delay mutants of bacteriophage T4. Virology. 1971 Dec;46(3):900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]